Abstract
The rate of evolution in terms of the number of mutant substitutions in a finite population is investigated assuming a quantitative character subject to stabilizing selection, which is known to be the most prevalent type of natural selection. It is shown that, if a large number of segregating loci (or sites) are involved, the average selection coefficient per mutant under stabilizing selection may be exceedingly small. These mutants are very slightly deleterious but nearly neutral, so that mutant substitutions are mainly controlled by random drift, although the rate of evolution may be lower as compared with the situation in which all the mutations are strictly neutral. This is treated quantitatively by using the diffusion equation method in population genetics. A model of random drift under stabilizing selection is then applied to the problem of "nonrandom" or unequal usage of synonymous codons, and it is shown that such nonrandomness can readily be understood within the framework of the neutral mutation--random drift hypothesis (the neutral theory, for short) of molecular evolution.
Full text
PDF
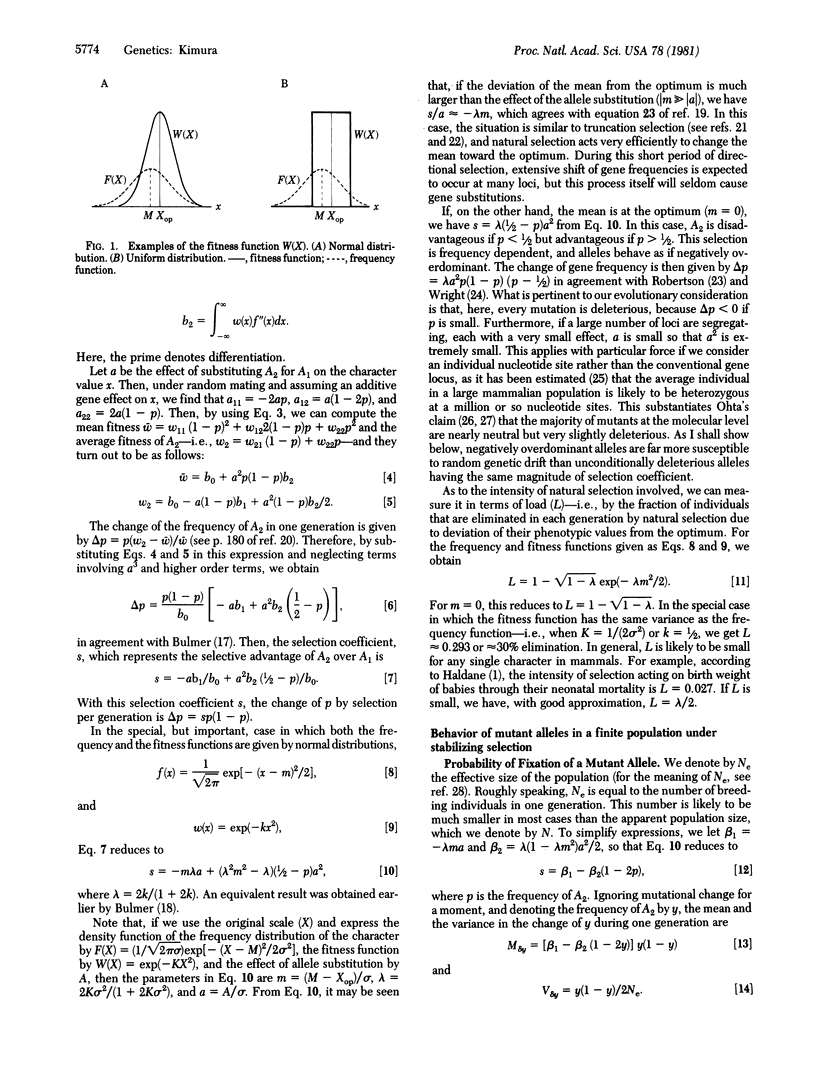



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bulmer M. G. The genetic variability of polygenic characters under optimizing selection, mutation and drift. Genet Res. 1972 Feb;19(1):17–25. doi: 10.1017/s0016672300014221. [DOI] [PubMed] [Google Scholar]
- Bulmer M. G. The stability of equilibria under selection. Heredity (Edinb) 1971 Oct;27(2):157–162. doi: 10.1038/hdy.1971.81. [DOI] [PubMed] [Google Scholar]
- Crow J. F., Kimura M. Efficiency of truncation selection. Proc Natl Acad Sci U S A. 1979 Jan;76(1):396–399. doi: 10.1073/pnas.76.1.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Mercier R., Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980 Jan 11;8(1):r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Codons and nearest-neighbor nucleotide pairs in mammalian messenger RNA. J Mol Evol. 1978 Jun 20;11(2):121–127. doi: 10.1007/BF01733888. [DOI] [PubMed] [Google Scholar]
- KARN M. N., PENROSE L. S. Birth weight and gestation time in relation to maternal age, parity and infant survival. Ann Eugen. 1951 Sep;16(2):147–164. [PubMed] [Google Scholar]
- KIMURA M. On the probability of fixation of mutant genes in a population. Genetics. 1962 Jun;47:713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Crow J. F. Effect of overall phenotypic selection on genetic change at individual loci. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6168–6171. doi: 10.1073/pnas.75.12.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Estimation of evolutionary distances between homologous nucleotide sequences. Proc Natl Acad Sci U S A. 1981 Jan;78(1):454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968 Feb 17;217(5129):624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. Gene pool of higher organisms as a product of evolution. Cold Spring Harb Symp Quant Biol. 1974;38:515–524. doi: 10.1101/sqb.1974.038.01.056. [DOI] [PubMed] [Google Scholar]
- Kimura M. The rate of molecular evolution considered from the standpoint of population genetics. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1181–1188. doi: 10.1073/pnas.63.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latter B. D. Influence of selection pressures on enzyme polymorphisms in Drosophila. Nature. 1975 Oct 16;257(5527):590–592. doi: 10.1038/257590a0. [DOI] [PubMed] [Google Scholar]
- Milkman R. Selection differentials and selection coefficients. Genetics. 1978 Feb;88(2):391–403. doi: 10.1093/genetics/88.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973 Nov 9;246(5428):96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Ota T. Mutational pressure as the main cause of molecular evolution and polymorphism. Nature. 1974 Nov 29;252(5482):351–354. doi: 10.1038/252351a0. [DOI] [PubMed] [Google Scholar]
- Post L. E., Strycharz G. D., Nomura M., Lewis H., Dennis P. P. Nucleotide sequence of the ribosomal protein gene cluster adjacent to the gene for RNA polymerase subunit beta in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1697–1701. doi: 10.1073/pnas.76.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The Distribution of Gene Frequencies Under Irreversible Mutation. Proc Natl Acad Sci U S A. 1938 Jul;24(7):253–259. doi: 10.1073/pnas.24.7.253. [DOI] [PMC free article] [PubMed] [Google Scholar]


