Abstract
Mycobacterium ulcerans is the causative agent of Buruli ulcer, one of the most common mycobacterial diseases of humans. Recent studies have implicated aquatic insects in the transmission of this pathogen, but the contributions of other elements of the environment remain largely unknown. We report here that crude extracts from two green algae added to the BACTEC 7H12B culture medium halved the doubling time of M. ulcerans and promoted biofilm formation. Using the 7H12B medium, modified by the addition of the algal extract, and immunomagnetic separation, we also demonstrate that M. ulcerans is associated with aquatic plants in an area of the Ivory Coast where Buruli ulcer is endemic. Genotype analysis showed that plant-associated M. ulcerans had the same profile as isolates recovered in the same region from both aquatic insects and clinical specimens. These observations implicate aquatic plants as a reservoir of M. ulcerans and add a new potential link in the chain of transmission of M. ulcerans to humans.
Mycobacterium ulcerans is an emerging environmental pathogen (2). It is the etiologic agent of Buruli ulcer, a necrotic skin disease highly prevalent in many countries throughout west Africa and one of the most common mycobacterial diseases in humans after tuberculosis and leprosy (2). M. ulcerans is the only mycobacterium known to produce a toxin, a polyketide-derived macrolide called mycolactone (10, 11). The infection begins with a painless nodule or papule that spreads over the surrounding tissue. Ischemic and necrotized tissue disappears and is replaced by centralized ulceration of the limb. At present, the only effective treatment consists of excision of the lesions, and this is often followed by extensive skin grafting (6).
Until very recently M. ulcerans had never been isolated in culture from the environment. Indirect evidence from epidemiological studies suggests that M. ulcerans is an environmental mycobacterium present in swampy areas. Humans are thought to be infected through minor wounds or skin abrasions via contact with mycobacterium-containing water (33). In a previous study we demonstrated that aquatic insects are a possible mode of transmission of M. ulcerans to humans. We showed that, in an experimental mouse model, aquatic insects (Naucoridae) were able to transmit an infection by biting, thereby inoculating bacilli that had accumulated in the salivary glands of the insects (16). We were then able to isolate in pure culture M. ulcerans from the salivary glands of Naucoridae captured in a region of endemicity in the Ivory Coast. However it seems unlikely that these insects are the only environmental source of M. ulcerans. In another recent study, data gathered using an M. ulcerans-specific PCR to survey environments of endemicity in southeastern Australia identified aquatic plants as a possible reservoir of this pathogen (30).
In this report, we test the hypothesis that M. ulcerans is associated with aquatic plants by studying the effects of crude aquatic plant extracts on the growth of M. ulcerans in vitro and then by attempting to culture the organism from material obtained from an area of endemicity.
MATERIALS AND METHODS
Bacterial strains.
M. ulcerans 1G897 was originally isolated from a skin biopsy sample from a human patient from French Guiana (7). Strains 7ICEF99 and 75ICO99 were isolated from patients in the Ivory Coast, and strain Nau. CI. 002 was isolated from an aquatic insect (16) also collected in the Ivory Coast. M. ulcerans strains O1EIHGA99 and 1615 (Trudeau Collection Strain, Lake Saranac, N.Y.) were originally isolated from human skin biopsy samples from Ghana and Malaysia, respectively. The reference strain of M. ulcerans, NCTC 10417, was obtained from the American Type Culture Collection bacteriology collection (ATCC 19423). Mycobacterium kansasii (11B0014), Mycobacterium fortuitum (10B0345), and Mycobacterium chelonae (6B0139) were isolated from tap water, while Mycobacterium tuberculosis (7B0143) and Mycobacterium marinum (8B0432) were isolated from French patients. A1l strains were passaged in BACTEC 7H12B medium (Becton Dickinson) at 30°C to a concentration of approximately 105 bacilli/ml. Aliquots of 0.2 ml were immediately inoculated with a syringe fitted with a 25-gauge needle into BACTEC 12B vials containing 4 ml of 7H12B medium.
Bacterial counting.
The total number of bacteria was determined by DAPI (4′,6′-diamidino-2-phenylindole) staining (28). The bacterial suspension was first dispersed by using a syringe fitted with a 25-gauge needle and then shaken with glass beads (300 rpm for 10 min). One milliliter of suspension was deposited in glass tubes containing 8 ml of water, 1 ml of DAPI (Sigma-Aldrich, St. Louis, Mo.), and 0.1% Triton X-100 (Prolabo, Nogent-sur-Marne, France). The tubes were mixed for 1 min and left to stand for 10 min at 30°C. The samples were filtered through a black polycarbonate membrane (0.22-μm pore size; Millipore Corporation, Bedford, Mass.). The filters were rinsed with 100 ml of distilled water, dried, and hydrated with a drop of buffered glycerin (Sanofi Diagnostics Pasteur, Marnes la Coquette, France). The bacteria were counted under UV light with an epifluorescence microscope (Olympus; BX 60) and an oil immersion objective. The bacteria were counted in 40 fields, and the results were expressed as cells per milliliter. This method was used to determine the doubling time during the exponential phase of bacterial growth in vials.
Measurement of M. ulcerans growth.
Different amounts of algal preparation were added to the BACTEC vials by a syringe fitted with a 23-gauge needle; the final total volume was adjusted to 6 ml with double-distilled water. The BACTEC vials contained Middlebrook 7H12B medium with 14C-labeled palmitic acid as a carbon source. Substrate consumption generates 14CO2 in the airspace of the sealed vial. The BACTEC TB-460 instrument detects the amount of radioactivity and records it as a growth index (GI) on a scale from 0 to 999. The vials were incubated at 30°C, and every 5 days the GI was registered. Each experiment was performed five times.
Collection of two freshwater green algae.
A Rhizoclonium sp., a member of the tropical Chlorophyceae, was cultured at 28°C and exposed to a photoperiod of 12 h of light and 12 h of dark in an aquarium with standard medium (1) with some modifications as follows: 34 μM KNO3, 87 μM NaCl, 38 μM K2HPO, 50 μM KH2PO4, 1.7 μM CaCl2, 0.1 μM HBO3, 0.2 μM MnSO4, 28 μM CuSO4, 20 μM ZnSO4, and 18 μM CoCl2.
Hydrodictyon reticulatum was collected in swamps of western France, transported to the laboratory in sterile water containers at 4°C, and treated within 4 h of reception. This species was chosen because the genus is ubiquitous throughout temperate and tropical freshwater systems.
Preparation of crude and filtered alga extracts.
Five hundred grams of freshly collected wet algae was washed twice in distilled water for 5 min and then placed in 500 ml of distilled water and autoclaved at 121°C for 20 min. These preparations were filtered through sterile gauzes of 1-mm porosity before being transferred to BACTEC vials. Two hundred milliliters of algal extract was filtered through a washed 0.22-μm-pore-size membrane (Millipore) to separate particulate matter. The particulate matter was then recovered from the membrane and resuspended in sterile distilled water.
Organic carbon.
The total organic carbon (TOC) and dissolved organic carbon (DOC) of each preparation were measured as previously described (15) with a carbon analyzer (O.I. Corporation; model 700 TOC analyzer) calibrated with 5 mg of potassium phthalate (Prolabo)/ml. The biodegradable fraction of DOC (BDOC) was determined as previously described (15).
Protein assay.
The quantity of proteins in the dissolved fraction was established with the BC assay kit (Interchim, Montluçon, France) by a bicinchoninic acid method. Results were expressed in milligrams per liter with a bovine serum albumin standard curve.
Carbohydrate assay (algal filtrate).
The carbohydrate was measured by a colorimetric method (9). One milliliter of sample (triplicate) was added to 1 ml of 5% phenol (Sigma-Aldrich)-5 ml of concentrated sulfuric acid. The mixture was heated at 95°C for 10 min. After 30 min of cooling in darkness, the optical density at 492 nm was obtained with a spectrophotometer. Results were determined with a standard curve and expressed in milligrams of glucose per liter.
Preparation for scanning electron microscopy.
The samples were prepared as previously described (13) with minor modifications. The samples were fixed for 2 h at 20°C in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4; Sigma-Aldrich). After centrifugation (3,500 × g) for 30 min, the pellet was resuspended in the same buffer and left for 12 h at 20°C. The samples were postfixed for 2 h at 20°C with 2% osmium tetroxide (Sigma-Aldrich) and rinsed in double-distilled water. The specimens were dehydrated in a graded ethanol series (50, 70, and 95%) for 10 min each, followed by three changes in 100% ethanol (10 min each). The specimens were metallized with a fine layer of carbon (5 nm thick) by ion sputter-coating (MED 020; Baltec, Balzers, Lichtenstein) and examined by scanning electron microscopy with a JEOL 6301F field emission microscope. Examinations were done with a tension at 5 kV.
Collection of aquatic plants from areas of endemicity and areas of nonendemicity and detection of M. ulcerans.
One hundred grams of a species of Scrophulariaceae, which is the predominant family of aquatic plants, was collected at five points on the right bank of the Lobo River downstream from the bridge of the Daloa-Zoukougbeu road, Ivory Coast, in March 2001 just before the rainy season. Other specimens were collected in areas of nonendemicity in the Adzopé region. The specimens were transported in sterile double-distilled water. They were minced with disposable scalpels in a petri dish and ground with a Potter-Elvehjem homogenizer (size 22; Kimble/Kontes, Vineland, N.J.) in distilled water with 1% Triton X-100. The suspension was shaken with 5-mm glass beads for 30 min at 300 rpm and then processed by immunomagnetic separation (IMS) for the detection of M. ulcerans by culture or by PCR of the insertion sequence IS2404 (16, 25).
For all attempts to isolate M. ulcerans in culture, a replicate BACTEC vial containing 20% (vol/vol) Rhizoclonium sp. crude extract, was prepared as described above. M. ulcerans PCR genotype analysis was performed as previously described (31).
Mycobacterial virulence.
Ten BALB/c mice (Iffa Credo) were subcutaneously inoculated in the tail with 0.1 ml of culture that was PCR positive. Culture and PCR were performed on the tissues when an inflammatory lesion appeared (16).
Identification of other mycobacteria.
The mycobacteria were identified in BACTEC 7H12B medium by PCR and restriction enzyme analysis of the hsp65 gene as previously described (8).
Statistical analysis.
The nonparametric Mann-Whitney U test was used to compare the kinetics of growth and the doubling time values. A P value of <0.05 was considered significant.
RESULTS
Growth of M. ulcerans in the presence of algae.
The characteristics of the two freshwater algae used are summarized in Table 1. The fraction of DOC relative to the total amount was 72.3% for the Rhizoclonium sp. and 80% for H. reticulatum. The material was almost entirely biodegradable (>96%). The content of carbohydrate relative to that of proteins was slightly higher for H. reticulatum.
TABLE 1.
Characterization of organic matter of two green algaea
| Alga | Concn of:
|
||||
|---|---|---|---|---|---|
| TOC (mg of C/liter) | DOC (mg of C/liter) | BDOC (mg of C/liter) | Protein (mg/liter) | Carbohydrate (mg/liter) | |
| Rhizoclonium sp. | 360.5 ± 15 | 260.6 ± 12 | 250 ± 11 | 20.4 ± 2 | 121 ± 6 |
| H. reticulatum | 470.7 ± 16 | 380.3 ± 14 | 372.5 ± 10 | 30.6 ± 1.2 | 203 ± 8 |
TOC was determined after filtration through 1-mm gauze. The other analytes (DOC, BDOC, proteins, and carbohydrates) were tested after filtration through membranes of 0.22-μm porosity. The values are means ± standard deviations of three measurements of the same sample.
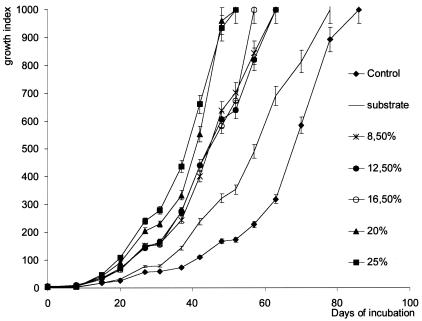
The growth kinetics of M. ulcerans (strain 1G897) were monitored with the BACTEC system and by direct-count microscopy after the addition of increasing amounts of a crude extract from the Rhizoclonium sp. to the culture medium. The doubling time during the exponential phase of growth was established at 3.3 ± 0.56 days with 25% (vol/vol) extract and at 4.8 ± 0.3 days for the no-extract control. Extract concentrations greater than 20% decreased the time required to obtain the value 999 for the GI (GI 999) by 39.5% (P = 0.007) (Fig. 1).
FIG. 1.
Kinetics of growth of M. ulcerans (strain 1G897) in 7H12B medium in the presence of various quantities of algal extract from the Rhizoclonium sp. (3.5 to 25%, vol/vol). Each point is the mean ± standard error for five vials per time point.
A comparison of the midpoints of the growth curves shows that the growth rate of M. ulcerans increased as the amount of algae added to the medium increased. Between the addition of 8.5 and 25% algal extract, the midpoint shifted from 44 days to 38 days, and under these conditions the time required to obtain the same amount of bacilli as determined by microscopy shortened from 67 days to 38 to 44 days (Fig. 1). At GI 500, the number of bacilli was evaluated at 1 × 104 to 5 × 104 bacilli per ml.
The addition of more than 25% algal extract did not further alter the kinetics of growth. When M. ulcerans was cultured in media supplemented by a crude extract obtained from another plant (H. reticulatum), the growth-stimulating effect was not significantly different (P = 0.126) from that produced by the addition of the same quantity of crude extract from the Rhizoclonium sp. (data not shown).
Similar patterns of M. ulcerans growth were observed under the same conditions with the M. ulcerans strains from Australia (105.425) and the Ivory Coast (7ICEF99) (P = 0.125) (data not shown).
Fractionation of the algal extract.
The use of 7H12B medium supplemented with filtered Rhizoclonium sp. extracts produced slower growth rates than the unfiltered extracts (time to reach GI 500: 45 versus 34 days; P = 0.006), but the growth rates remained faster than those for the negative controls without any algal extract (time to reach GI 500: 60 days; P = 0.124). Supplementing the medium with the particulate fraction produced an effect equivalent to that seen with the dissolved fraction (time to reach GI 500: 40 versus 45 days; P = 0.128) (Fig. 2).
FIG. 2.
Kinetics of M. ulcerans (strain 1G897) growth, comparing filtered with nonfiltered preparations of the Rhizoclonium sp. Twenty-five percent (vol/vol) extract obtained from the Rhizoclonium sp. was added to 7H12B medium after filtration through membranes of 0.22-μm porosity. Each point is the mean ± standard error of five vials per time point.
Growth of other mycobacteria in the presence of algae.
A study was then undertaken to compare the effects of the addition of 25% (vol/vol) Rhizoclonium sp. extract to the BACTEC 12B medium on the growth rates of three other mycobacterial species. The time required to obtain the maximum GI did not change between experiments conducted in the presence and absence of algal extract for M. tuberculosis (time to reach GI 999: 10 ± 0 days), M. kansasii (time to reach GI 999: 28 ± 2 days), and M. marinum (time to reach GI 999: 5 ± 0 days). The results represent the means and standard errors of five replicates. Growth curves were also plotted for these experiments, but no significant differences were observed for any time point (data not shown).
Biofilm formation.
Scanning electron microscopy was used to observe the formation of biofilms of M. ulcerans on particulates within the culture medium containing the crude extract from the Rhizoclonium sp. By 12 h cells were fixed in filament formations. After 10 days, the cells had multiplied and formed small cell clusters. After 40 days, a biofilm composed of large cell clusters that measured more than 100 μm in diameter had formed. Higher-magnification micrographs indicated that material resembling an extracellular matrix was present (Fig. 3). The same experiment was then performed for four other environmental mycobacteria, and, for each species, biofilm formation was observed (Fig. 4).
FIG. 3.
Formation of M. ulcerans biofilm on the Rhizoclonium sp. M. ulcerans was cultured in 7H12B vials supplemented by 25% (vol/vol) Rhizoclonium sp. extract. (A) Filaments of algae alone. (B) After 12 h, some individual bacilli (arrows) were attached on alga filaments. (C) After 15 days, the bacteria had multiplied and formed small clusters (arrow). (D) By day 40, large clusters of bacilli were forming (arrow) and evolving into microcolonies. Arrows in the insets, extracellular matrix. Scale bars: 30 (A), 2.5 (B, C, and insets), and 10 (D) μm.
FIG. 4.
Formation of biofilms by four environmental mycobacteria on the Rhizoclonium sp. The mycobacteria were cultured in 7H12B vials supplemented by 25% (vol/vol) Rhizoclonium sp. extract for 25 days. (A) M. fortuitum biofilm; (B) M. chelonei biofilm; (C) M. marinum biofilm; (D) M. kansasii biofilm. Scale bars: 4 (A to C) and 10 (D) μm.
PCR, culture, and virulence of mycobacteria isolated from algae in the Daloa region.
It is known that the Daloa region of the Ivory Coast is an area where Buruli ulcer is highly endemic (17). Many rural villages in this area are situated among swamps, close to the principal waterway, the river Lobo. This prompted us to focus on collecting aquatic plants in the area of endemicity between Daloa and Zoukougbeu. In the Lobo River the predominant family of aquatics plant was Scrophulariaceae. Other aquatic plants collected from areas where Buruli ulcer was not endemic were not identified. IS2404 PCR and culture were performed on all specimens. Two samples from the river Lobo were PCR positive, whereas all samples collected in the areas of nonendemicity were negative. One positive BACTEC culture was detected 2 months after inoculation. The IS2404 PCR performed on this culture was positive, suggesting the presence of M. ulcerans, but this culture was contaminated with M. szulgai. This M. szulgai- and possibly M. ulcerans-positive culture was inoculated subcutaneously into the tails of five mice, and 4 months later the same signs of infection (inflammatory lesions with edema) were observed for three mice. PCRs performed on infected tissues were positive for M. ulcerans. Unfortunately all attempts to separate M. ulcerans from M. szulgai in pure culture were unsuccessful. This was despite several rounds of IMS and passaging through mice. The M. szulgai isolate was, however, readily isolated in pure culture. The IS2404 PCR was performed on this pure culture, and no amplified product was detected; when 105 M. szulgai bacilli were inoculated subcutaneously into the tails of five mice, no sign of infection was observed 6 months later.
Genotype analysis.
M. ulcerans-specific PCR genotype analysis was then performed on DNA extracted from the M. szulgai-contaminated M. ulcerans culture and from DNA extracted from the M. szulgai pure culture. This system uses outward-facing primers to amplify between two different, high-copy-number M. ulcerans insertion sequences. Only DNA from the contaminated M. ulcerans culture produced a profile, and this profile matched exactly the profile obtained from another recent M. ulcerans environmental isolate (Fig. 5). This profile was also identical to that obtained from two local M. ulcerans clinical isolates and a clinical isolate from Ghana. Thus the environmental strains have the same “epidemic” genotype as other west African clinical isolates. As has been shown previously with this method, M. ulcerans isolates from different geographic origins such as Malaysia, Australia, and French Guiana produced different profiles, demonstrating the ability of this technique to discriminate between strains. No bands were detected with DNA from M. szulgai (Fig. 5).
FIG. 5.
Silver-stained polyacrylamide gel showing the results of IS2426 PCR genotype analysis for M. ulcerans strains isolated from environmental and clinical sources. Lane identification is as follows. Lane 1, M. szulgai plant isolate; lane 2; M. ulcerans or M. szulgai plant isolate; lane 3, M. ulcerans insect isolate (Nau. CI. 002); lane 4, M. ulcerans clinical isolate (7ICEF99; Ivory Coast); lane 5, M. ulcerans clinical isolate (75ICO99; Ivory Coast); lane 6, M. ulcerans clinical isolate (01EIHGA99; Ghana); lane 7, M. ulcerans clinical isolate (1615; Malaysia); lane 8, M. ulcerans clinical isolate (NCTC 10417; Australian); lane 9, M. ulcerans clinical isolate (1G897; French Guiana); lane M, molecular size marker (Eurogentec).
DISCUSSION
In 1998 the World Health Organization (WHO) launched in Yamoussokro, Ivory Coast, the Buruli Ulcer Initiative, with the specific aim of trying to combat the alarming increase of Buruli ulcer throughout central and west Africa. Two of the key research priorities identified by the WHO were to understand the environmental ecology and mode(s) of transmission of M. ulcerans. However, the environmental ecology of M. ulcerans has remained obscure for more than 50 years because of the failure to isolate the bacteria in culture from the environment. This situation changed dramatically with the development of a highly specific and sensitive PCR assay (12, 27, 31, 32) which has enabled the accumulation of significant evidence to implicate water, plants, and insects in the ecology of M. ulcerans (25, 26, 30; F. Portaels, P.-A. Fonteyne, and W. M. Meyers, Letter, Lancet 353:986, 1999).
Guided by the PCR evidence of M. ulcerans in aquatic insects, we experimentally infected species of Naucoridae and observed that M. ulcerans localized exclusively within the salivary glands of these insects (16). In this niche M. ulcerans could both survive and multiply. Other mycobacterial species such as M. marinum, M. fortuitum, and M. kansasii could not occupy this niche (16). This then led us to examine the salivary glands of aquatic insects collected from a Buruli ulcer in the Daloa region, an zone of endemicity in the Ivory Coast. By dissecting out the salivary glands and using IMS to enrich for M. ulcerans in the glands, we were able to isolate the bacteria in culture from the environment for the first time. The difficulties associated with primary culture of M. ulcerans from environmental samples are multifactorial. Most signficantly, M. ulcerans is a slow-growing mycobacterium and samples are readily overgrown by other faster-growing bacteria. In addition, the procedures for sample decontamination are too aggressive (22). Mycobacterial isolation methods have been developed for the detection of mycobacteria from clinical samples, where bacterial populations are noncomplex and their concentrations are high. The reverse situation is found in environmental specimens; thus it is not surprising that these methods fail to detect M. ulcerans. We set out to ameliorate this situation by using IMS as previously developed to selectively enrich for M. ulcerans and by attempting to enhance the growth of M. ulcerans in culture by the addition of aquatic plant extracts to the media.
Aquatic plants, such as algae, are able to secrete many organic compounds, such as amino acids and polysaccharides, which are in turn used by bacteria as substrates for growth (14, 19, 20, 23). For example, low-molecular-mass products (<700 Da) are an important source of carbon for heterotrophic bacteria (19). We used two cosmopolitan freshwater green algae, Rhizoclonium and Hydrodictyon, which are found unattached, lying on mud or rocks, in both temperate and tropical environments. They are able to synthesize and secrete many BDOCs (18), and these may provide nutrients for mycobacteria. We reasoned that, when added to culture media, extracts from these algae may accelerate and facilitate the primary culture of M. ulcerans.
In this study we have shown that crude organic extracts from both a Rhizoclonium sp. and H. reticulatum were able to stimulate the growth of M. ulcerans. The addition of 20% crude extract caused the doubling time to decrease by half (Fig. 1), a significant result for a bacterium with a doubling time of 80 days. This effect was not observed with other slow-growing mycobacteria, suggesting that M. ulcerans uses specific components of the extract to augment its growth.
Simple fractionation of the algal extract by filtration showed that it was the combination of particulate and dissolved material that produced the shortest doubling times (Fig. 2). The addition of the individual fractions also reduced doubling times, but the effect was only one-half as great as that of the complete extract. One explanation may be that M. ulcerans has a deficiency in a particular metabolic pathway that is compensated for by a component of the dissolved fraction. The presence of particulate matter probably enhances growth nonspecifically by acting as a solid support for the formation of biofilms. It has been shown previously that the doubling time for biofilm-attached bacteria is less than the doubling time for planktonic bacteria (3, 4, 13). Electron micrography showed that M. ulcerans produces prolific biofilms (Fig. 3). However, it is important that biofilm formation is not a feature unique to M. ulcerans. We tested four other environmental mycobacteria under the same conditions, and they all produced biofilms on the algal particulate matter (Fig. 4). In addition, it has been previously shown that the growth rate of biofilm-attached M. fortuitum is approximately twice that of nonattached cells (13). Thus the algal extract probably has both specific and nonspecific growth-enhancing influences on M. ulcerans.
The extreme hydrophobicity of their cell wall makes the mycobacteria adhesion specialists within a biofilm (29), but bacteria growing in biofilms often produce an extracellular matrix that is also involved in adhesion or protection against predation (5, 21, 24, 34). Production of an extracellular matrix was also observed in M. ulcerans, but only when it was growing in a biofilm, in association with aquatic plant material. Perhaps this matrix also plays a role in adhesion and protection of M. ulcerans in an aquatic environment.
The combination of IMS and the improved media allowed us to detect and cultivate M. ulcerans in plant samples collected from a region in the Ivory Coast where Buruli ulcer is endemic. However, we were unable to isolate M. ulcerans in pure culture. All samples were contaminated by M. szulgai, a more rapidly growing mycobacterium. Nevertheless PCR genotype analysis provided unequivocal evidence that M. ulcerans was indeed present and that the strain present in the aquatic plant samples had the same profile as strains isolated from aquatic insects and from Buruli ulcer patients in the region. This is the first report that directly implicates both aquatic plants and insects in the chain of transmission of M. ulcerans to humans. It is not clear by what means both plants and carnivorous aquatic insects harbor M. ulcerans. It is unlikely that the insects are contaminated by direct contact with aquatic plants. It is possible, however, that they are contaminated after preying on other organisms such as snails, fishes, and insect larvae, as these organisms have a strictly vegetarian diet and may be colonized by M. ulcerans via contact with aquatic plants.
More research is now needed to test these hypotheses, to try and identify other environmental reservoirs of M. ulcerans, and to determine how this pathogen is transmitted to humans.
Acknowledgments
We thank B. Denizot, S. Sourice, and M. Leitao from Bi-eau for technical assistance and F. Portaels and J. Grosset for critically reading the manuscript.
We are grateful for the doctoral scholarship from the Association Française Raoul Follereau (L.M.). This work was supported by grants from the Association Française Raoul Follereau.
REFERENCES
- 1.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 2.Asiedu, K., R. Sherpbier, and M. C. Raviglione. 2000. Buruli ulcer Mycobacterium ulcerans infection. W. H. O. Global Buruli Ulcer Initiative. Report 2000. World Health Organization, Geneva, Switzerland.
- 3.Charackis, W. G. 1990. Kinetics of microbial transformation, p. 233-264. In W. G. Charackis and K. C. Marshall (ed.), Biofilms. Wiley, New York, N.Y.
- 4.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 5.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darie, H., S. Djakeaux, and A. Cautoclaud. 1994. Therapeutic approach in Mycobacterium ulcerans infections. Bull. Soc. Pathol. Exot. 87:19-21. [PubMed] [Google Scholar]
- 7.De Gentile, P. L., C. Mahaza, F. Rolland, B. Carbonnelle, J. L. Verret, and D. Chabasse. 1992. Cutaneous ulcer from Mycobacterium ulcerans. Apropos of 1 case in French Guyana. Bull. Soc. Pathol. Exot. 85:212-214. [PubMed] [Google Scholar]
- 8.Devallois, A., K. S. Goh, and N. Rastogi. 1997. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J. Clin. Microbiol. 35:2969-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois, M., K. A. Gilles, J. K. Hamiltin, P. A. Rebers, and F. Smith. 1956. Colorometric method for determination of sugars and related substances. Ann. Chem. 28:350-356. [Google Scholar]
- 10.George, K. M., D. Chatterjee, G. Gunawardana, D. Welty, J. Hayman, R. Lee, and P. L. Small. 1999. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283:854-857. [DOI] [PubMed] [Google Scholar]
- 11.George, K. M., L. Pascopella, D. M. Welty, and P. L. Small. 2000. A Mycobacterium ulcerans toxin, mycolactone, causes apoptosis in guinea pig ulcers and tissue culture cells. Infect. Immun. 68:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guimares-Peres, A., F. Portaels, P. D. Rijk, K. Fissete, S. R. Pattyn, P.-P. V. Vooren, and P.-A. Fonteyne. 1999. Comparison of two PCRs for detection of Mycobacterium ulcerans. J. Clin. Microbiol. 37:206-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall-Stoodley, L., and H. Lappin-Scott. 1998. Biofilm formation by the rapidly growing mycobacterial species Mycobacterium fortuitum. FEMS Microbiol. Lett. 168:77-84. [DOI] [PubMed] [Google Scholar]
- 14.Jensen, L. M. 1991. Phytoplankton release of extracellular organic carbon, molecular weight composition and bacterial assimilation. J. Plankton Res. 11:39-48. [Google Scholar]
- 15.Joret, J. C., Y. Levi, and M. Gilbert. 1989. The measurement of bioeliminable dissolved organic carbon (BDOC): a tool in water treatment. Water Supply 7:1-5. [Google Scholar]
- 16.Marsollier, L., R. Robert, J. Aubry, J. P. Saint André, H. Kouakou, P. Legras, A. L. Manceau, C. Mahaza, and B. Carbonnelle. 2002. Aquatic insects as a vector for Mycobacterium ulcerans. Appl. Environ. Microbiol. 68:4623-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marston, B. J., M. O. Diallo, C. R. Horsburgh, Jr., I. Diomande, M. Z. Saki, J. M. Kanga, G. Patrice, H. B. Lipman, S. M. Ostroff, and R. C. Good. 1995. Emergence of Buruli ulcer disease in the Daloa region of Côte d'Ivoire. Am. J. Trop. Med. Hyg. 52:219-224. [DOI] [PubMed] [Google Scholar]
- 18.Mathay, A. 1993. Etude biologique et écotoxicologique de la chlorophycée Hydrodictyon reticulatum. Ph.D. thesis. INPL, Université de Nancy I, Nancy, France.
- 19.Maurin, N., C. Amblard, and G. Bourdier. 1997. Phytoplanktonic excretion and bacterial reassimilation in an oligotrophic lake: molecular weight fractionation. J. Plankton Res. 18:1045-1068. [Google Scholar]
- 20.Moore, B. G. 1964. Extracellular polysaccharides of algae: effects on life-support system. Science 145:586-587. [DOI] [PubMed] [Google Scholar]
- 21.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 22.Palomino, J. C., and F. Portaels. 1998. Effects of decontamination methods and culture conditions on viability of Mycobacterium ulcerans in the BACTEC system. J. Clin. Microbiol. 36:402-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulet, S. A., and V. Martin-Jezequel. 1983. Relation between dissolved free amino acids, chemical composition and growth of marine diatom Chaetoceros debile. Mar. Biol. 77:93-100. [Google Scholar]
- 24.Raad, I. I., S. Vartivarian, A. Khan, and G. P. Bodey. 1991. Catheter-related infections caused by the Mycobacterium fortuitum complex: 15 cases and review. Rev. Infect. Dis. 13:1120-1125. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, B., and R. Hirst. 1997. Immunomagnetic separation and PCR for detection of Mycobacterium ulcerans. J. Clin. Microbiol. 35:2709-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross, B. C., P. D. Johnson, F. Oppedisano, L. Marino, A. Sievers, T. Stinear, J. A. Hayman, M. G. Veitch, and R. M. Robins-Browne. 1997. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl. Environ. Microbiol. 63:4135-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross, B. C., L. Marino, F. Oppedisano, R. Edwards, R. M. Robins-Browne, and P. D. R. Jonhson. 1997. Development of a PCR assay for rapid diagnosis of Mycobacterium ulcerans infection. J. Clin. Microbiol. 35:1696-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saby, S., I. Sibille, L. Mathieu, J. L. Paquin, and J. C. Block. 1997. Influence of water chlorination on the counting of bacteria with DAPI (4′,6-diamidino-2-phenylindole). Appl. Environ. Microbiol. 63:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze Röbbecke, R. 1993. Mycobacteria in the environment. Immun. Infekt. 21:126-131. [PubMed] [Google Scholar]
- 30.Stinear, T., J. K. Davies, G. A. Jenkin, J. A. Hayman, F. Oppedisano, and P. D. R. Johnson. 2000. Identification of Mycobacterium ulcerans in the environment from regions in southeast Australia in which it is endemic with sequence capture-PCR. Appl. Environ. Microbiol. 66:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stinear, T., J. K. Davies, G. A. Jenkin, F. Portaels, B. C. Ross, F. Oppedisano, M. Purcell, J. A. Hayman, and P. D. Johnson. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stinear, T., B. C. Ross, J. K. Davies, L. Marino, R. M. Robins-Browne, F. Oppedisano, A. Sievers, and P. D. Johnson. 1999. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 37:1018-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uganda Buruli Group. 1971. Epidemiology of Mycobacterium ulcerans infection at Kinyara, Uganda. Trans. Soc. Trop. Med. Hyg. 65:763-775. [DOI] [PubMed] [Google Scholar]
- 34.Vess, R. W., R. L. Anderson, J. H. Carr, W. W. Bond, and M. S. Favero. 1993. The colonization of solid PVC surfaces and the acquisition of resistance to germicides by water microorganisms. J. Appl. Bacteriol. 74:215-221. [DOI] [PubMed] [Google Scholar]