Abstract
The marine dinoflagellate genus Alexandrium includes a number of species which produce neurotoxins responsible for paralytic shellfish poisoning (PSP), which in humans may cause muscular paralysis, neurological symptoms, and, in extreme cases, death. A. minutum is the most widespread toxic PSP species in the western Mediterranean basin. The monitoring of coastal waters for the presence of harmful algae also normally involves microscopic examinations of phytoplankton populations. These procedures are time consuming and require a great deal of taxonomic experience, thus limiting the number of specimens that can be analyzed. Because of the genetic diversity of different genera and species, molecular tools may also help to detect the presence of target microorganisms in marine field samples. In this study, we developed a real-time PCR-based assay for rapid detection of all toxic species of the Alexandrium genus in both fixative-preserved environmental samples and cultures. Moreover, we developed a real-time quantitative PCR assay for the quantification of A. minutum cells in seawater samples. Alexandrium genus-specific primers were designed on the 5.8S rDNA region. Primer specificity was confirmed by using BLAST and by amplification of a representative sample of the DNA of other dinoflagellates and diatoms. Using a standard curve constructed with a plasmid containing the ITS1-5.8S-ITS2 A. minutum sequence and cultured A. minutum cells, we determined the absolute number of 5.8S rDNA copies per cell. Consequently, after quantification of 5.8S rDNA copies in samples containing A. minutum cells, we were also able to estimate the number of cells. Several fixed A. minutum bloom sea samples from Arenys Harbor (Catalan Coast, Spain) were analyzed using this method, and quantification results were compared with standard microscopy counting methods. The two methods gave comparable results, confirming that real-time PCR could be a valid, fast alternative procedure for the detection and quantification of target phytoplankton species during coastal water monitoring.
The occurrence of harmful algal bloom (HAB) events in marine waters along temperate coasts throughout the world is an important and expanding threat to human health, fishery resources, and tourism industries (2, 23, 53). Several phytoplankton species have the capacity to produce toxins that can be accumulated through the food web. The consumption of shellfish and other grazers that have accumulated these algal toxins causes a variety of poisoning syndromes in humans. The global increase in HAB phenomena has involved the western Mediterranean basin in enclosed sea sites such as harbors, beaches, and near-shore waters along the coasts (40, 58). In these HAB episodes, several dinoflagellates (which include the genus Alexandrium) proliferated, with a number of species producing potent neurotoxins responsible for paralytic shellfish poisoning, which in humans may cause neurological disorders and, in extreme cases, death (3, 54). Three species of Alexandrium (A. minutum, A. catenella, and A. taylori) have often caused bloom events in the western Mediterranean over the last decade (M. Masó, E. Garcés, M. G. Giacobbe, A. Penna, M. Vila, G. Basterretxea, A. Orfila, J. Camp, and J. Tintore, Abstr. Xth Int. Conf. Harmful Algae, abstr. 189, 2002). A. minutum is the most widespread toxic species in the Mediterranean basin. This species formed toxic blooms along the northern part of the French Brittany coast (32), along the Catalan coast (Spain) (M. Vila, E. Garcés, M. Masò, and J. Camp, Abstr. Xth Int. Conf. Harmful Algae, abstr. 291, 2002), in the South Tyrrhenian Sea, and along the northwestern coast of the Adriatic Sea (Italy), where it contaminated mussels with paralytic shellfish poisoning toxin (27). Widespread blooms of A. catenella and A. taylori were described during recent years along the Spanish, French, and Italian coasts (15, 18, 34, 59). A. taylori is not a toxin producer, but it can proliferate, with high biomass densities.
Monitoring coastal waters for the presence of HAB species is essential in assessing the potential for bloom formation. Normally, this type of monitoring involves accurate morphology identification and enumeration of target phytoplankton species by using light or electron microscopy examinations in addition to toxicity tests of shellfish. Unfortunately, these identification methods can be difficult for long-term monitoring, because the microscopic analyses are time consuming and require a great deal of taxonomic experience to identify the species (47).
Because of the genetic diversity of different genera and species (30, 33, 38, 41), molecular tools may be very useful for the detection of a microorganism, both in seawater and in sediment (5, 22, 43, 49, 51). Because of their rapidity, PCR-based methods are used more and more. The PCR amplification technique of targeting ribosomal DNA (rDNA) regions has been successfully employed for the detection of various toxic dinoflagellates in seawater samples (8, 11, 19, 44, 48). The rDNA genes have been the regions of choice for the development of molecular assays because these sequences are repeated in tandem in high copy numbers and are highly conserved (24, 36, 37, 50, 55). Different regions of the rDNA cluster have been selected as targets for PCR amplification. These regions include the small subunit, the large subunit, the 5.8S region, two internal transcribed spacers (ITS1 and ITS2), and the nontranscribed spacer (7). The choice of a particular region as the target was based mostly on the level of variability of each region within a particular species of interest and the requirements of assay specificity and sensitivity (1, 9, 10, 16, 29, 35).
Real-time PCR offers all the advantages of conventional PCR, such as high sensitivity and specificity, and also allows for the quantification of PCR product formation during the exponential phase of the reaction. The detection of PCR products is monitored by measuring the increase in fluorescence caused by the binding of SYBR Green dye (25) to double-stranded (dsDNA). Even though real-time PCR was originally developed for clinical applications, it has recently been applied to microbial ecology. For example, it has been used to detect small-subunit rRNA in natural microbial communities (56) and for rapid detection of Pfiesteria piscicida in culture and environmental water samples (8).
In this study, we developed a real-time PCR-based assay using the 5.8S region as the target for the detection of Alexandrium species and quantification of A. minutum cells in both fixative-preserved environmental samples and cultures. Based on several rDNA sequences obtained from different Alexandrium spp. strains in GenBank, genus-specific primers were designed and used in real-time PCR assays, first on Alexandrium species cultures and then on field samples from one bloom spatial series collected in winter inside the Arenys harbor (Catalan coast).
MATERIALS AND METHODS
Algal cultures, sample collection, and species identification.
The A. minutum strain used in this study (CNR-AMI-A4) was kindly provided by M. G. Giacobbe (CNR, Istituto per l'Ambiente Marino Costiero, Messina, Italy). A. minutum cultures were maintained in f/2 medium (21) at 17 ± 1°C. Light was provided by cool-white fluorescent bulbs (photon flux of 100 μE m−2 s−1) on a standard 14-h-light, 10-h-dark cycle. Between 7 and 17 days after inoculation, when cultures were in exponential growth phase, algal cell density was accurately determined by enumeration of Lugol's iodine-stained cells (57) by using a hemacytometer (Neubauer; Hausser Scientific, Horsham, Pa.). To ensure reliability, the counts were repeated several times until the total number of cells counted was greater than 100. The average of these values was considered the concentration of cells in culture. Samples of culture equivalent to 1 × 105 or 5 × 104 cells were harvested by centrifugation at 4,000 × g for 15 min. Cell pellets were carefully washed with 800 μl of sterile artificial seawater and stored at −80°C.
Field samples were collected from Arenys Harbor (Spain) during a huge A. minutum bloom in February 2002 (E. Garcés, I. Bravo, M. Vila, R. I. Figueroa, M. Masó, and N. Sampedro, submitted for publication) and another one in April 2003. A volume of 150 ml was fixed with Lugol's iodine solution; a 50-ml volume of preserved samples was settled in Utermöhl chambers for 24 h and counted by using an inverted microscope. After being counted, cells were concentrated by centrifugation, resuspended in 2 to 5 ml of seawater, and stored in the dark at 4°C. Cells were also counted before real-time PCR. For the identification of A. minutum, the samples were dyed with calcofluor white (14) and observed under an epifluorescence microscope. Identification was based on cell shape and size but especially on the thecal plates tabulation (6).
DNA preparation from cultures and Lugol-fixed samples.
Frozen pellets of 1 × 105 or 5 × 104 cells, respectively, were resuspended in 800 μl or 400 μl of lysis buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.5% Nonidet P-40, 0.5% Tween 20, 0.1 mg of proteinase K/ml), briefly sonicated with Ultrasonic Homogenizer LABSONIC L with needle probe tip 40T (B. Braun Biotech International) for 10 seconds at 50 to 100 W, and incubated at 60°C for 3 h. The samples were vortexed every 30 min. The samples were then incubated at 100°C for 5 min to inactivate proteinase K and centrifuged at 12,000 × g for 1 min at room temperature. The supernatants containing DNA were immediately used in real-time PCR assays. Alternatively, the supernatants were aliquoted, stored at −80°C, and used for PCR within 2 to 3 weeks.
Lugol-fixed samples were centrifuged at 4,000 × g for 15 min at room temperature, and pellets were washed with 10 ml of sterile artificial seawater (0.4 M NaCl, 10 mM KCl, 20 mM MgSO4, 20 mM MgCl2, 10 mM CaCl2, 2 mM NaHCO3, 0.4 mM boric acid). DNA was prepared by resuspending the pellets in 400 μl of lysis buffer and following the procedure described above.
Alternatively, DNA was extracted by using a Dynabeads DNA DIRECT Universal kit (Dynal) or DNeasy Plant Mini kit (QIAGEN) according to the manufacturer's instructions.
DNA quantification.
Purified DNA and DNA contained in crude extract were quantified by using a PicoGreen dsDNA quantification kit (Molecular Probes) and a spectrofluorophotometer RF-5301PC (Shimadzu). The standard curve was made with the bacteriophage λ DNA provided in the kit. PicoGreen is an ultrasensitive fluorescent nucleic acid stain for the quantification of dsDNA in solution with minimal interference by RNA, single-stranded DNA, nucleotides, salts, proteins, and detergents. PicoGreen provides a linear correlation between dsDNA concentration and fluorescence, with a detection range from 25 pg to 1 μg of dsDNA/ml (52). For the quantification of DNA in cell lysates, to avoid denaturation of DNA we did not boil the samples after incubation at 60°C. Alternatively, purified DNA was quantified on agarose gel by using serially diluted DNA marker 2 (MBI Fermentas) and a Gel Doc apparatus (Bio-Rad).
rDNA cloning and sequencing.
The 5.8S rDNA and flanking internal transcribed spacers (ITS1 and ITS2) were amplified from purified A. minutum total DNA and cloned into the pMOSBlue vector (Amersham) as previously described (44). Several clones were sequenced by using the BigDye Terminator cycle sequencing kit (Applied Biosystems) in an ABI PRISM 310 instrument. Plasmid DNA was purified by using a Qiaprep Spin Miniprep kit (QIAGEN).
Primer design.
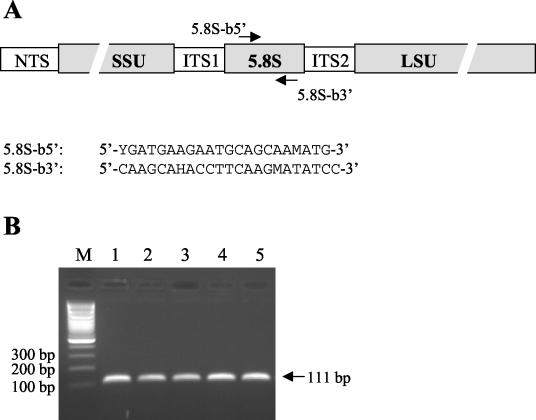
The primers were designed by using Primer Express software (Applied Biosystems) and an alignment of all available ITS1-5.8S-ITS2 ribosomal DNA sequences for the genus Alexandrium. Sequences were either downloaded from GenBank or obtained from our laboratory. The alignment was constructed by using DIALIGN 2.0 (42) and CLUSTALW. The alignment included sequences of several strains of A. catenella, A. minutum, A. taylori, and A. tamarense (Table 1). The 5.8S region is very conserved among these species. Primers 5.8S-b5′ (5′-YGATGAAGAATGCAGCAAMATG-3′) and 5.8S-b3′ (5′-CAAGCAHACCTTCAAGMATATCC-3′) were designed to target consensus sequences specific for the genus Alexandrium. In order to confirm the sequence specificity, we used BLAST (Basic Local Alignment Search Tool) to search in the EMBL and GenBank databases for published sequences identical to the primers 5.8S-b5′ and 5.8S-b3′. At present, the databases contain about 480 5.8S sequences belonging to the Dinophyceae class and more than 800 belonging to the group Alveolata. The primers were purchased from Sigma-Genosys Ltd. The primers and PCR conditions were tested in a qualitative PCR assay to ensure the specificity of the reaction.
TABLE 1.
Isolates and strains of Alexandrium species, collection sites, and EMBL ITS1-5.8S-ITS2 rDNA sequence accession numbers
| Species | Strain | Collection site | EMBL accession no. |
|---|---|---|---|
| A. catenella | CSICa-4 | Catalan Sea, Mediterranean, Spain | AJ298900 |
| A. catenella | M17 | Harima Nada, Japan | AB006990 |
| A. taylori | CSICa-AV8 | Catalan Sea, Mediterranean, Spain | AJ251654 |
| A. taylori | CNRb-AT4 | Ionian Sea, Mediterranean, Italy | AJ251653 |
| A. taylori | CBAc-ATAYB2 | Tyrrhenian Sea, Mediterranean, Italy | AJ300451 |
| A. taylori | CYSTS | Tyrrhenian Sea, Mediterranean, Italy | AJ291785 |
| A. tamarense | CCMPd 116 | European Atlantic, Spain | AJ005047 |
| A. tamarense | AT-A | Chinhae Bay, Korea | AF374224 |
| A. tamarense | AT-2 | Chinhae Bay, Korea | AF374227 |
| A. tamarense | AT-6 | Chinhae Bay, Korea | AF374228 |
| A. tamarense | AT-B | Chinhae Bay, Korea | AF374225 |
| A. minutum | CNRb-AMIA4 | Ionian Sea, Mediterranean, Italy | AJ318460 |
| A. minutum | LAC-27 | Adriatic Sea, Mediterranean, Italy | AJ005050 |
Institut de Ciències del Mar, Barcelona, Spain.
Consiglio Nazionale delle Ricerche, Messina, Italy.
Centro Biologia Ambientale, Urbino University, Italy.
Provasoli-Guillard National Center for Culture of Marine Phytoplankton, Maine.
Theoretical basis of real-time PCR.
In real-time PCR, reactions are analyzed during the initial exponential phase rather than at the end point. By using SYBR Green (an intercalating fluorescent dye) in the PCR mixture, it is possible to detect double-stranded PCR product formation after each cycle. For each reaction tube, the fluorescence signal of the reporter dye (SYBR) is divided by the fluorescence signal of the passive reference dye (ROX) to obtain a ratio defined as the normalized reporter signal (Rn). The higher the starting quantity of the target molecule, the earlier a significant increase in fluorescence is observed. The threshold cycle parameter (Ct) is defined as the fractional cycle number at which the fluorescence crosses a fixed threshold above the baseline. The amount of target sequence in an unknown sample is calculated by plotting the Ct value on the standard curve.
Real-time PCR.
Real-time PCR assays were performed on an ABI PRISM 7000 SDS (Applied Biosystems) in a final volume of 25 μl containing 1× SYBR Green PCR Master Mix (Applied Biosystems) and primers at a final concentration of 300 nM. Two microliters of each appropriately diluted sample (standard curve points and algal lysates) was added to 23 μl of the PCR master mixture. The following quantification cycling protocol was used: 95°C for 10 min, followed by 35 or 40 cycles at 95°C for 15 s and 60°C for 1 min. At the end of each run, a dissociation protocol was performed to ensure that only the specific PCR product was present. Experiments were performed with duplicates for each lysate sample and with triplicates for each standard curve point. Each PCR run included the standard curve, established by serially diluted plasmid containing the target sequence, and a no-template control.
Standard curve construction.
The standard curve was constructed with 10-fold serial dilutions of plasmid containing the ITS1-5.8S-ITS2 sequence of the target. The curve range was from 106 to 10 copies. Dilutions were freshly prepared for each experiment from one aliquot of plasmid stock solution stored at −20°C.
RESULTS
A. minutum identification in field bloom samples.
The algal cells in field samples were rounded to ovoid and small sized (average dimensions, 21 to 24 μm wide and 22.5 to 24 μm long; n = 110). Plate 1′ was narrow and contacted the apical pore plate directly. It showed a small ventral pore on the anterior right margin. Plate 6′′ was narrow, and the posterior sulcal plate was slightly wider than it was long. All these features are typical for the species A. minutum (6).
Specificity and sensitivity of real-time PCR assay.
Selected primers for the conserved 5.8S rDNA region of the Alexandrium genus gave a 111-bp PCR product. Because real-time PCR was carried out in the presence of SYBR Green, a fluorescent dsDNA intercalating dye, it was important to avoid unspecific PCR product or primer dimer formation. The specificity of the primers and the absence of unspecific products or primer dimers were tested by analyzing the reactions in 2% agarose gel stained with ethidium bromide. The molecular weight of the amplified product was as expected, and no other bands were visible when DNA from A. minutum, A. taylori, or A. catenella was used (Fig. 1). Moreover, all real-time PCRs were followed by a dissociation protocol consisting of measuring fluorescence in a temperature gradient from 60 to 95°C. This protocol allowed establishment of the melting temperature of the PCR product, at the same time excluding the presence of other (shorter or longer) unspecific amplicons. The melting temperature of the amplicon was in agreement with the theoretical melting temperature calculated by using Primer Express software. The primers' specificity was also confirmed in silico by using BLAST: the results of the BLAST search showed that the sequences of selected primers matched exactly only with the 5.8S sequences belonging to the Alexandrium genus. Furthermore, the primers' specificity was tested by real-time amplification of representative DNA samples from other dinoflagellates and diatoms. Table 2 shows a list of DNAs screened and their phylogenetic affinities. Only the PCRs containing DNA from the genus Alexandrium were positive, while amplification products were undetectable in reactions containing the DNA of other dinoflagellates and diatoms. Real-time PCR sensitivity was tested by using a plasmid containing the ITS1-5.8S-ITS2 rDNA A. minutum sequence (see below). This assay was sensitive enough to detect 10 copies of 5.8S rDNA sequences in less than 35 amplification cycles.
FIG. 1.
(A) Organization of Alexandrium species rDNA and location of primers for the Alexandrium genus-specific PCR. (B) Amplification of target DNA in real-time PCR followed by analysis on standard agarose gel. Lanes: 1, template DNA from A. catenella CSIC-4; 2, template DNA from A. taylori CBA-1; 3, template DNA from A. minutum CNR-AMIA4; 4, template plasmid containing A. catenella ITS1-5.8S-ITS2 rDNA; 5, template plasmid containing A. taylori ITS1-5.8S-ITS2 rDNA; M, 100-bp ladder molecular size marker. A clean no-template control was included in the real-time assay (not shown on the gel). NTS, nontranscribed spacer; SSU, small subunit; LSU, large subunit.
TABLE 2.
Specificity of real-time PCR
| Species | Strain | Phylogenetic association | Amplificatione |
|---|---|---|---|
| Pseudonitzschia pseudodelicatissima | IEOa-PS 50V | Diatom (Bacillariaceae) | − |
| Gyrodinium corsicum | GC1 CSICb-1 | Dinoflagellate (Gymnodiniaceae) | − |
| Karlodinium micrum | KM1 CSICb-1 | Dinoflagellate (Gymnodiniaceae) | − |
| Coolia monotis | IEOa-CM7C | Dinoflagellate (Ostreopsidaceae) | − |
| Ostreopsis sp. | IEOa-OSBR3 | Dinoflagellate (Ostreopsidaceae) | − |
| Dinophysis sacculus (ITS1-5.8S-ITS2 cloned sequence) | No strain | Dinoflagellate (Dinophysidae) | − |
| Alexandrium minutum | CNRc-AMIA4 | Dinoflagellate (Gonyaulacaceae) | + |
| Alexandrium catenella | CSICb-4 | Dinoflagellate (Gonyaulacaceae) | + |
| Alexandrium taylori | CBAd-1 | Dinoflagellate (Gonyaulacaceae) | + |
Instituto Español de Oceanografia, Vigo, Spain.
Institut de Ciències del Mar, Barcelona, Spain.
Consiglio Nazionale delle Ricerche, Messina, Italy.
Centro Biologia Ambientale, Urbino University, Italy.
−, amplification products not detected; +, amplification products detected.
Standard curve and dynamic range of real-time PCR.
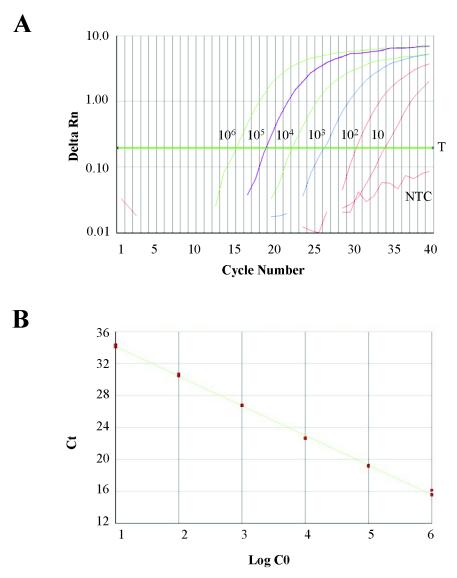
The standard curve was constructed with 10-fold serial dilutions of plasmid containing the ITS1-5.8S-ITS2 sequence of A. minutum. The dynamic range was wide: from 106 to 10 copies (up to 5 orders of magnitude); also, a strong linear relationship between the Ct and the log of the starting copy number was always demonstrated (R2 ≥ 0.996). The efficiency of the reaction (E) calculated by the formula E = 10(1/m) − 1, where m is the slope of the standard curve, ranged from 80 to 90% in the different assays (Fig. 2).
FIG. 2.
Example of standard curve with plasmid containing ITS1-5.8S-ITS2 rDNA sequence of A. minutum. (A) Amplification plot for standard curve with plasmid copy number from 106 to 10. The cycle number is plotted versus the Delta Rn. The Delta Rn represents the Rn minus the baseline signal established in the early PCR cycles. Three replicates were performed for each reference DNA sample, but data for only one are shown here. (B) Calibration curve plotting log starting copy number (C0) versus Ct. Slope, −3.71; correlation coefficient (R2), 0.9986. NTC, no-template control; T, threshold.
Total DNA quantification in cells of A. minutum.
In general, DNA content per algal cell is very heterogeneous, varying from 0.1 pg for Navicula pelliculosa to 200 pg for Gonyaulax poliedra (26). Nothing is known about the DNA content in Alexandrium species cells. We tried to establish the DNA content in A. minutum to support and validate data obtained with real-time PCR (see below). At first, we purified DNA from cultured algal cells by using commercial kits (QIAGEN or Dynal). DNA yields were variable, ranging from 89 to 144 ng per 100,000 cells. Due to the variability and poor DNA yield that result using commercial kits, we tried to quantify DNA directly in cell lysates. Because of its sensitivity and specificity, we used PicoGreen dye for quantification of DNA in crude cell lysates. Different scalar dilutions of cell lysate (from 1:50 to 1:5,000) were done to ensure the absence of inhibitors during quantification. We prepared the lysates as described in Materials and Methods, but we did not boil the samples after incubation at 60°C, to avoid denaturation of DNA and loss of sensitivity of PicoGreen, which binds exclusively to dsDNA. In fact, it was observed that the quantity of DNA in the boiled samples was underestimated by about three times compared to that for the nonboiled samples (data not shown). The PicoGreen excitation-emission wavelength does not interfere with chlorophyll a, but excitation at 480 nm could interfere with beta carotene and xanthophyll absorbance. In order to assess the possible interference from pigments or other cell components, we compared the fluorimetric dosages of cell lysate diluted 1:20 with or without the PicoGreen component. The fluorescence intensity of the lysate without PicoGreen was about 0.2% compared to that for lysate with PicoGreen, suggesting that lysate auto-fluorescence was completely negligible.
The values obtained from the fluorimetric quantifications were expressed in pg/μl. Dividing these values for cell concentration in the lysate (125 cells/μl), we calculated the amount of DNA per cell. In this way we established that the DNA content per cell was 11.4 ± 2.6 pg for A. minutum. This average value was obtained from five experiments, in duplicate or triplicate, including two experiments in which we used the lysis buffer and protocol from a DNeasy Plant kit (QIAGEN) or a lysis buffer containing 1% SDS and 25 mM EDTA.
Application of real-time PCR for quantification of 5.8S rDNA in A. minutum.
To avoid variability and low yields in DNA purifications, we used crude cell lysates (prepared as described in Materials and Methods) as DNA templates in PCRs. These lysates contained cell components which could interfere with PCRs. To find an amount of template which can be amplified without any inhibitory effect, cell lysates containing DNA were diluted fivefold until quantification results were proportional to sample dilutions. We found that A. minutum lysate dilutions equal to or higher than 1:50 (corresponding to five cells per reaction) were always amplified without inhibition. This approach was possible because of the abundance of 5.8S rDNA sequences in dinoflagellate cells and because of the sensitivity of this PCR. The amount of 5.8S rDNA copies in the PCR mixtures was calculated for each diluted lysate sample by plotting the Ct values on a standard curve obtained using a known copy number of the plasmid containing the cloned target sequence (see Materials and Methods). The lysate dilutions used for the PCR amplifications were 1:50, 1:250, and 1:1,250, corresponding to 5, 1, and 0.2 cells per reaction, respectively. The number of 5.8S rDNA copies obtained for each sample after real-time PCR was normalized to that for sample dilutions corresponding to 1 cell per reaction. In this way, we found that the absolute number of 5.8S rDNA copies per cell of A. minutum was 1,084 ± 120.3. This value resulted from the average of 16 different determinations, ranging from 860 to 1,284, obtained from three different sample collections. Moreover, the absolute number of 5.8S rDNA copies normalized to 1 pg of purified DNA was also calculated, and the result was 104 ± 32.1. Total DNA was purified with Dynabeads from A. minutum cells either in the exponential growth phase (8 days after inoculation) or in the stationary phase (1 month after inoculation). The ratio between the number of 5.8S rDNA copies per cell and the number of 5.8S rDNA copies per picogram of purified DNA gave us the theoretical amount of total DNA per cell in A. minutum. The DNA content calculated in this manner was 10.42 pg/cell, in agreement with the DNA quantification with PicoGreen on cell lysates. Thus, we confirmed the real-time PCR data, and at the same time, we validated the DNA content per cell measured with PicoGreen.
Quantification of A. minutum in field bloom samples fixed with Lugol.
Based on the number of 5.8S gene copies per cell, the concentrations of A. minutum in environmental field samples were determined by using the real-time PCR assay. A. minutum field bloom samples were collected and fixed with Lugol as described in Materials and Methods. The samples were stored at 4°C for more than 1 year prior to being analyzed. Lysates of Lugol-fixed bloom samples were diluted 10-fold (1:10 to 1:1,000) to eliminate inhibitory effects of fixative or cell components and used in real-time PCR assays. Dilutions of 1:10 partially inhibited the PCR and were not considered in the analysis of results. Quantification results were compared with those for standard counting methods (Table 3). The results showed that the two methods were comparable in all tested bloom samples. In fact, one-way analysis of variance showed that, with a P value of 0.01, the means of the standard and real-time counting methods for each sample were not significantly different. The cell concentration in sample 3/0b is much lower than that for other bloom samples because some of the vial content was lost during transportation.
TABLE 3.
Quantification of A. minutum cells in bloom field samples from Arenys harbor (Spain): comparison between standard and real-time PCR counting methods
| Sample IDb | Collection date |
A. minutum cells/litera
|
|
|---|---|---|---|
| Standard method | Real-time method | ||
| 2 | April 2003 | 3,964,000 ± 313,500 | 3,276,450 ± 527,290 |
| 6 | February 2002 | 2,500,000 ± 256,500 | 2,497,000 ± 74,500 |
| 9 | February 2002 | 1,845,000 ± 156,000 | 2,475,000 ± 336,150 |
| 11 | February 2002 | 2,125,000 ± 247,500 | 1,317,600 ± 153,250 |
| 12 | February 2002 | 2,171,000 ± 239,000 | 2,255,000 ± 180,650 |
| 13 | February 2002 | 2,950,000 ± 279,000 | 2,566,000 ± 353,140 |
| 1/1b | February 2002 | 1,832,000 ± 158,500 | 1,198,300 ± 99,000 |
| 3/0b | February 2002 | 286,000 ± 31,100 | 188,800 ± 20,750 |
| 4/2a | February 2002 | 1,894,000 ± 161,200 | 1,389,000 ± 137,950 |
Values are ± standard deviations.
ID, identification.
DISCUSSION
A. minutum is a small and widely distributed toxin-producing species responsible for HAB events in the western Mediterranean basin (13, 17, 59). The real-time PCR-based assay described in this study has proven to be specific and sensitive for the detection of Alexandrium spp. cells in cultured and environmental samples. Bowers et al. (8) previously developed a real-time PCR assay for rapid detection of Pfiesteria piscicida 18S rDNA, but no quantitative approaches were attempted due to the lack of information concerning the number of 18S gene copies per cell. In our study, which was focused on the 5.8S rDNA region of the genus Alexandrium (dinoflagellates), we attempted to overcome this limit by determining the number of rDNA copies per A. minutum cell.
DNA extraction from marine algae is hampered by the large quantity of polysaccharides and polyphenolics produced in many species. There are several DNA extraction methods that work well with marine algae, but DNA yield and reproducibility are sometimes poor. Because of this, for quantification purposes we decided to perform PCRs using crude cell lysates from A. minutum cultures and field bloom samples. These lysates were diluted until the potential inhibitory effects of contaminants were eliminated. This approach was possible because in most eukaryotes the rDNA gene family is repeated in tandem in a high copy number. For example, 100 to 200 copies are present in yeast, and 50 to 10,000 copies are present in mammals (49). Within the group Alveolata, the rDNA copy number ranges from 2 to >100. Two copies have been reported in Theileria parva (28) and Cryptosporidium parvum (31), 3 copies have been reported in Babesia bovis (12), 110 copies have been reported in Toxoplasma gondii (20), and 100 to 200 copies have been reported in Pfiesteria piscicida (49). Nothing is known about rDNA copies in the Alexandrium genus. In this work, for the first time we provided information regarding genome size and rDNA sequence amount in A. minutum cells. This information allowed us to determine the number of A. minutum cells in environmental seawater samples without doing laborious microscopic counting.
In oceanographic field work, fixation of samples is an important step in preserving archived material until laboratory processing. Fixation methodology should have minimal impact on downstream analysis. Lugol's solution is a widely used fixative that, like glutaraldehyde and formaldehyde, can introduce artifacts like reduction in the abundance of particular taxa and changes in phytoplankton cell size. In the dinoflagellates, for example, fixation could induce species-specific cell swelling or shrinking (39). In this work, we demonstrated that the real-time PCR approach can be applied as a quantitative assay to environmental A. minutum samples fixed with Lugol's solution even after 14 months of storage without significant loss of cell number and 5.8S rDNA sequence information. In samples 11, 1/1b, and 3/0b, the more evident variability between standard counting methods and real-time methods could be due to some cell loss during sample concentration from 50 ml to 2 to 5 ml and/or during long-term storage in Lugol's solution. However, after statistical analysis, the resulting values from the two counting methods were not significantly different. Generally, there are two main factors that could affect the variability between standard and real-time PCR-based counts. The first is the cellular DNA content during a bloom event. A. minutum cells are generally haploid (n), only the planozygotes/resting cysts being diploid (2n). It has been stated that one of the causes of the bloom decline might be sexual processes and subsequent encystment (4, 45, 46). The high percentage of diploid cells in a bloom could lead to an overestimation of cell number when the real-time PCR-based count is used. However, during the intensive monitoring of the A. minutum bloom in Arenys Harbor in February 2002, it was observed that the encystment rate was very low (<1%) (E. Garcés et al., submitted). The second factor concerns the presence of species morphologically similar to the target in the phytoplankton community. In the Arenys area, A. minutum usually blooms together with other dinoflagellates such as Prorocentrum micans, P. triestinum, and Scrippsiella spp., among others. In some cases, the Scrippsiella species have the same size as A. minutum. In those cases, the position in which Scrippsiella is observed under light microscopy is critical for correct identification. The possible misidentification of some specimens could increase the counting error when these species are highly abundant. However, in Arenys Harbor during the bloom in February 2002, Scrippsiella spp. represented less than 0.3% of the dinoflagellate concentration (M. Vila, unpublished data).
In this work, we demonstrated the feasibility of detection and quantification of A. minutum in high-density bloom samples. It is noteworthy that this real-time PCR assay was able to amplify ≤ 0.2 cell equivalent of algal lysate and up to 10 copies of 5.8S rDNA. Therefore, the sensitivity level of this molecular method will allow detection of Alexandrium species not only at bloom concentrations but also in field samples containing a low number of cells, which will be extremely useful for long-term monitoring programs, during which high-density blooms could be exceptional events and low-density blooms could occur commonly.
The real-time PCR assay was optimized with primers specific for the genus Alexandrium. For this reason, we were able to quantify only A. minutum bloom samples, where other Alexandrium species were not significantly represented. In fact, up to 96% of the microphytoplankton abundance in the bloom samples collected for this study was A. minutum. Optimization of real-time PCR with A. minutum-specific primers will allow detection and quantification of these algae in environmental samples containing mixed Alexandrium species populations. Moreover, we established that A. minutum rDNA quantification data cannot be extended to other Alexandrium species. In fact, preliminary data indicate that the quantity of 5.8S rDNA in A. catenella and A. taylori is different from that in A. minutum, generally appearing proportional to the cell's DNA content. After we complete and validate these data, we may have the tools for fast monitoring of toxic and harmful Alexandrium algae in the Mediterranean sea.
Acknowledgments
We thank M. G. Giacobbe, S. Fraga, and M. Montresor for supplying culture strains and Mercedes Masó for the contribution in field sampling.
This work was partially supported by the EU STRATEGY project, contract no. EVK3-CT-2001-00046, and by EU CHEMAG project PPC00-3045 (2000).
REFERENCES
- 1.Adachi, M., Y. Sako, and Y. Ishida. 1996. Analyses of Alexandrium (Dinophyceae) species using sequences of the 5.8S ribosomal DNA and internal transcribed spacer regions. J. Phycol. 32:424-432. [Google Scholar]
- 2.Anderson, D. M. 1989. Toxic algal blooms and red tides: a global perspective, p. 11-16. In T. Okaichi, D. M. Anderson, and T. Nemoto (ed.), Red tides: biology, environmental science and toxicology. Elsevier, New York, N.Y.
- 3.Anderson, D. M. 1997. Turning back the harmful red tide. Nature 388:513. [Google Scholar]
- 4.Anderson, D. M. 1998. Physiology and bloom dynamics of toxic Alexandrium species, with emphasis on life cycle transitions, p. 29-48. In Anderson et al (ed.), Physiological ecology of harmful algal blooms. NATO ASI series. Springer-Verlag, Berlin, Germany.
- 5.Anderson, D. M., D. M. Kulis, B. A. Keafer, and E. Berdalet. 1999. Detection of the toxic dinoflagellate Alexandrium fundyense (Dinophyceae) with oligonucleotide and antibody probes: variability in labeling intensity with physiological conditions. J. Phycol. 35:870-883. [Google Scholar]
- 6.Balech, E. 1995. The genus Alexandrium Halim (Dinoflagellata). Sherkin Island Marine Station, Cork, Ireland.
- 7.Bena, G., M. F. Jubier, I. Olivieri, and B. Lejeune. 1998. Ribosomal external and internal transcribed spacers: combined use in the phylogenetic analysis of Medicago (Leguminosae). J. Mol. Evol. 46:299-306. [DOI] [PubMed] [Google Scholar]
- 8.Bowers, H. A., T. Tengs, H. B. Glasgow, Jr., J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connell, L. 2002. Rapid identification of marine algae (Raphidophyceae) using three-primer PCR amplification of nuclear internal transcribed spacer (ITS) regions from fresh and archived material. Phycologia 41:15-21. [Google Scholar]
- 10.Costas, E., R. Zardoya, J. Bautista, A. Gurrido, C. Rojo, and V. López-Rodas. 1995. Morphospecies vs. genospecies in toxic marine dinoflagellates: an analysis of Gimnodinium catenatum/Gyrodinium impudicum and Alexandrium minutum/A. lusitanicum using antibodies, lectins, and gene sequences. J. Phycol. 31:801-807. [Google Scholar]
- 11.Coyne, K. J., D. A. Hutchins, C. E. Hare, and S. C. Cary. 2001. Assessing temporal and spatial variability in Pfiesteria piscicida distributions using molecular probing techniques. Aquat. Microb. Ecol. 24:275-285. [Google Scholar]
- 12.Dalrymple, B. P. 1990. Cloning and characterization of the rRNA genes and flanking regions from Babesia bovis: use of the genes as strain discriminating probes. Mol. Biochem. Parasitol. 43:117-124. [DOI] [PubMed] [Google Scholar]
- 13.Forteza, V., G. Quetglas, M. Delgado, I. Reyero, S. Fraga, J. M. Franco, and E. Cacho. 1998. Toxic Alexandrium minutum bloom in Palma de Mallorca harbour (Balearic Islands, western Mediterranean), p. 58-59. In Reguera, et al. (ed.), Harmful algae. Xunta de Galicia and IOC of UNESCO, Santiago de Compostela, Spain.
- 14.Fritz, L., and R. E. Triemer. 1985. A rapid simple technique utilizing calcofluor white M2R for the visualization of dinoflagellate thecal plates. J. Phycol. 21:662-664. [Google Scholar]
- 15.Garcés, E., M. Masò, and J. Camp. 1999. A recurrent and localized dinoflagellate bloom in a Mediterranean beach. J. Plankton Res. 21:2373-2391. [Google Scholar]
- 16.Giacobbe, M. G., A. Penna, A. Ceredi, A. Milandri, R. Poletti, and X. M. Yang. 2000. Toxicity and ribosomal DNA of the dinoflagellate Dinophysis sacculus (Dinophyta). Phycologia 39:177-182. [Google Scholar]
- 17.Giacobbe, M. G., and G. Maimone. 1994. First report of Alexandrium minutum Halim in a Mediterranean lagoon. Cryptogam. Algol. 15:47-52. [Google Scholar]
- 18.Giacobbe, M. G., and X. M. Yang. 1999. The life history of Alexandrium taylori (Dinophyceae). J. Phycol. 35:331-338. [Google Scholar]
- 19.Godhe, A., S. K. Otta, A.-S. Rehnstam-Holm, I. Karunasagar, and I. Karunasagar. 2001. Polymerase chain reaction in detection of Gymnodium mikimotoi and Alexandrium minutum in field samples from southwest India. Mar. Biotechnol. 3:152-162. [DOI] [PubMed] [Google Scholar]
- 20.Guay, J. M., A. Huot, S. Gagnon, A. Tremblay, and R. C. Levesque. 1992. Physical and genetic mapping of cloned ribosomal DNA from Toxoplasma gondii: primary and secondary structure of the 5S gene. Gene 114:165-711. [DOI] [PubMed] [Google Scholar]
- 21.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 26-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 22.Guillou, L., E. Nezan, V. Cueff, E. Erard-Le Denn, M. A. Cambon-Bonavita, P. Gentien, and G. Barbier. 2002. Genetic diversity and molecular detection of three toxic dinoflagellate genera (Alexandrium, Dinophysis, and Karenia) from French coasts. Protistologica 153:223-238. [DOI] [PubMed] [Google Scholar]
- 23.Hallegraeff, G. M., and C. J. Bolch. 1992. Transport of diatom and dinoflagellate resting spores via ship's ballast water: implication for plankton biogeography and aquaculture. J. Plankton Res. 14:1067-1084. [Google Scholar]
- 24.Hershkovitz, M. A., and L. A. Lewis. 1996. Deep-level diagnostic value of the rDNA-ITS region. Mol. Biol. Evol. 13:1276-1295. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi, R., G. Dollinger, P. S. Walsh, and R. Griffith. 1992. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology 10:413-417. [DOI] [PubMed] [Google Scholar]
- 26.Holm-Hansen, O. 1969. Algae: amounts of DNA and organic carbon in single cells. Science 163:87-88. [DOI] [PubMed] [Google Scholar]
- 27.Honsell, G., R. Poletti, M. Pompei, L. Sidari, A. Milandri, C. Casadei, and R. Viviani. 1996. Alexandrium minutum Halim and PSP contamination in the northern Adriatic Sea (Mediterranean Sea), p. 77-83. In T. Yasumoto, T. Oshima, and Y. T. Fukuyo (ed.), Harmful and toxic algal blooms. UNESCO, Paris, France.
- 28.Kibe, M. K., O. K. ole-MoiYoi, V. Nene, B. Khan, B. A. Allsopp, N. E. Collins, S. P. Morzaria, E. I. Gobright, and R. P. Bishop. 1994. Evidence for two single copy units in Theileria parva ribosomal RNA genes. Mol. Biochem. Parasitol. 66:249-259. [DOI] [PubMed] [Google Scholar]
- 29.La Du, J., D. Erdner, S. Dyhrman, and D. M. Anderson. 2002. Molecular approaches to understanding population dynamics of the toxic dinoflagellate Alexandrium fundyense. Biol. Bull. 203:244-245. [DOI] [PubMed] [Google Scholar]
- 30.LaJeunesse, T. C. 2001. Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinum using the ITS region: in search of a “species” level marker. J. Phycol. 37:866-880. [Google Scholar]
- 31.Le Blancq, S. M., N. V. Khramtsov, F. Zamani, S. J. Upton, and T. W. Wu. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463-478. [DOI] [PubMed] [Google Scholar]
- 32.Le Doux, M., J. M. Frémy, E. Nézan, and E. Erard. 1990. Recent occurrences of paralytic shellfish poisoning (PSP) toxins from the northwestern coasts of France. J. Shellfish Res. 8:486. [Google Scholar]
- 33.Leaners, G., C. Scholin, Y. Bhaud, D. Saint-Hilaire, and M. Herzog. 1991. A molecular phylogeny of dinoflagellate protists (Pyrrhophyta) inferred from the sequence of 24S rRNA divergent domains D1 and D8. J. Mol. Evol. 32:53-63. [DOI] [PubMed] [Google Scholar]
- 34.Lilly, E. L., D. M. Kulis, P. Gentien, and D. M. Anderson. 2002. Paralytic shellfish poisoning toxins in France linked to a human-introduced strain of Alexandrium catenella from western Pacific: evidence from DNA and toxin analysis. J. Plankton Res. 24:443-452. [Google Scholar]
- 35.Marin, I., A. Aguilera, B. Reguera, and J. P. Abad. 2001. Preparation of DNA suitable for PCR amplification from fresh or fixed single dinoflagellate cells. BioTechniques 30:88-93. [DOI] [PubMed] [Google Scholar]
- 36.Medina, M., A. G. Collins, J. D. Silberman, and M. L. Sogin. 2001. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc. Natl. Acad. Sci. USA 98:9707-9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 38.Medlin, L., M. Lange, U. Wellbrock, G. Donner, M. Elbrächter, C. Hummert, and B. Luckas. 1998. Sequence comparisons link toxic European isolates of Alexandrium tamarense from the Orkney Islands to toxic North American stocks. Eur. J. Protistol. 34:329-335. [Google Scholar]
- 39.Menden-Deuer, S., E. J. Lessard, and J. Satterberg. 2001. Effect of preservation on dinoflagellate and diatom cell volume and consequences for carbon biomass predictions. Mar. Ecol. Prog. Ser. 222:41-50. [Google Scholar]
- 40.Montresor, M., D. Marino, A. Zingone, and G. Dafnis. 1990. Three Alexandrium species from coastal Tyrrhenian waters, p. 82-87. In E. Graneli, B. Sundström, L. Edler, and D. A. Anderson (ed.), Toxic marine phytoplankton. Elsevier, New York, N.Y.
- 41.Moon-van der Stay, S., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 42.Morgenstern, B., T. Werner, and A. W. M. Dress. 1996. Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc. Natl. Acad. Sci. USA 93:12098-12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen, J. L. 1990. Nucleic acids in algal systematics. J. Phycol. 26:209-214. [Google Scholar]
- 44.Penna, A., and M. Magnani. 1999. Identification of Alexandrium (Dinophyceae) species using PCR and rDNA-targeted probes. J. Phycol. 35:615-621. [Google Scholar]
- 45.Probert, I. 1999. Sexual reproduction and ecophysiology of the marine dinoflagellate Alexandrium minutum Halim. Ph.D. thesis. University of Westminister, London, England.
- 46.Probert, I., J. Lewis, and E. Erard-le Denn. 2002. Morphological details of the life history of Alexandrium minutum (Dinophyceae). Cryptogamie 23:343-355. [Google Scholar]
- 47.Rehnstam-Holm, A. S., A. Godhe, and A. M. Anderson. 2002. Molecular studies of Dinophysis (Dinophyceae) species from Sweden and North America. Phycologia 41:348-357. [Google Scholar]
- 48.Rollo, F., S. Sassaroli, L. Boni, and L. Marota. 1995. Molecular typing of the red-tide dinoflagellate Gonyaulax polyedra in phytoplankton suspensions. Aquat. Microb. Ecol. 9:55-61. [Google Scholar]
- 49.Saito, K., T. Drgon, J. A. Robledo, D. N. Krupatkina, and G. R. Vasta. 2002. Characterization of the rRNA locus of Pfiesteria piscicida and development of standard and quantitative PCR-based detection assays targeted to the nontranscribed spacer. Appl. Environ. Microbiol. 68:5394-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholin, C. A., G. M. Hallegraeff, and D. M. Anderson. 1995. Molecular evolution of the Alexandrium tamarense “species complex” (Dinophyceae): dispersal in the North American and West Pacific regions. Phycologia 34:472-485. [Google Scholar]
- 51.Scholin, C. A., R. Marin, P. E. Miller, G. J. Doucette, C. L. Powell, P. Haydock, J. Howard, and J. Ray. 1999. DNA probes and a receptor-binding assay for detection of Pseudonitzschia (Bacillariophyceae) species and domoic acid activity in cultured and natural samples. J. Phycol. 35:1356-1367. [Google Scholar]
- 52.Singer, V. L., I. J. Jones, S. T. Yue, and R. P. Haugland. 1997. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantification. Anal. Biochem. 249:228-238. [DOI] [PubMed] [Google Scholar]
- 53.Smayda, T. J. 1990. Harmful algal blooms: their ecophysiology and general relevance to phytoplankton blooms in the sea. Limnol. Oceanogr. 42:1137-1153. [Google Scholar]
- 54.Steidinger, K. A., and K. Tangen. 1997. Dinoflagellates, p. 387-584. In C. R. Tomas (ed.), Identifying marine phytoplankton. Academic Press, Inc., Orlando, Florida.
- 55.Stryer, L. 1995. Genes for ribosomal RNAs are tandemly repeated several hundred times, p. 992-993. In L. Stryer (ed.), Biochemistry, 4th ed. W. H. Freeman and Company, New York, N.Y.
- 56.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Throndsen, J. 1978. Preservation and storage, p. 69-74. In A. Sournia (ed.), Phytoplankton manual. UNESCO, Paris, France.
- 58.Vila, M., E. Garcés, M. Masó, and J. Camp. 2001. Is the distribution of the toxic dinoflagellate Alexandrium catenella expanding along the NW Mediterranean coast? Mar. Ecol. Prog. Ser. 222:73-83. [Google Scholar]
- 59.Vila, M., J. Camp, E. Garcés, M. Masó, and M. Delgado. 2001. High resolution spatio-temporal detection of HABs in confined waters of the NW Mediterranean. J. Plankton Res. 23:497-514. [Google Scholar]