Abstract
Antibiotic interactions are believed to be significant to microbial fitness in soil, yet little is known of the frequency, intensity, and diversity of antibiotic inhibition and resistance among indigenous microbes. To begin to address these issues, we studied the abilities of streptomycete isolates from prairie soil to inhibit growth and display resistance to antibiotics produced by a test collection of 10 streptomycete isolates. Wide variations in antibiotic inhibition and resistance for prairie isolates among three locations and four soil depths within a 1-m2 plot were revealed. Fewer than 10% of 153 prairie isolates inhibited all 10 test isolates, while more than 40% of the isolates did not inhibit any of the test isolates. No field isolate was resistant to all of the test isolates, nor was any isolate susceptible to all of the test isolates. No correlation between inhibition and resistance phenotypes was found, suggesting that inhibition and resistance are under independent selection. The significant spatial variation in the frequency and intensity of antibiotic inhibition implies that the fitness benefits of antibiotic production are not the same among locations in soil. In contrast, the consistency of resistance over space indicates that its significance to fitness across locations is stable or the costs of maintaining resistance in the absence of selection are small or nonexistent. The spatial clustering of antibiotic inhibitory activity suggests a variable matrix of selection pressures and microbial responses across the soil landscape.
Although antibiotic activity may significantly affect interactions among soil microbes, information on the ecology of antibiotic-producing microbial populations in soil is limited. Indeed, the factors that predict the presence of strong antibiotic inhibitory or resistance activities within the soil microbial community are poorly understood. Streptomycetes (order Actinomycetales, family Streptomycetaceae, referred to here as the streptomycetes; 72) are gram-positive, filamentous bacteria that are ubiquitous in soil and produce the majority (>70%) of known antibiotics (63). These microbes are excellent saprophytes and prolific producers of extracellular enzymes. The taxonomy of this group has undergone substantial revision in recent years on the basis of genomic DNA and rRNA sequence analyses, although taxospecies definitions remain unresolved (2).
Among the streptomycetes, both the quantity and types of antibiotics produced vary widely among individuals of the same species (25, 67). Many streptomycetes produce more than one antibiotic. For example, in Streptomyces coelicolor, genes involved in the biosynthesis of several antibiotics are found on the chromosome (reviewed in reference 24). There is limited evidence of horizontal transfer of antibiotic gene clusters among streptomycetes in soil (17, 18). In some cases, antibiotic biosynthesis genes have been found on giant linear plasmids that could be transferred not only among streptomycete strains but also between species (46). Similarly, many streptomycetes possess resistance to multiple antibiotics (22, 48). Antibiotic resistance may spread readily among streptomycetes via horizontal transfer of plasmids (reviewed in references 47 and 58).
Antibiotic-producing streptomycetes can inhibit a broad range of soilborne microbes, including gram-positive and gram-negative bacteria, fungi, and nematodes (10, 13, 15, 19, 28, 32, 37, 39, 51, 73). As a result, the potential for inoculated antibiotic-producing streptomycetes to control soilborne plant pathogens on diverse crop species has been widely investigated (28, 30, 37, 38, 50, 51, 52). However, despite the apparent significance of antibiotic activity to the soil microbial community and the potential effects of antibiotic-producing bacteria on plant health in both agricultural and nonagricultural soils, there have been few studies of the dynamics of antibiotic activity within the soil microbial community. Although there is a growing body of work on the spatial distribution of genetic diversity in soil (4, 21, 68), we lack an understanding of the spatial variation in antibiotic activities, including antibiotic inhibition and resistance, among microbial populations in nondisturbed soils. Further, the absence of specific information on the extent to which antibiotic inhibition and resistance capabilities are correlated among individuals from the same location in soil constrains our ability to identify factors, such as nutrient utilization capabilities and genetic relatedness, that may influence antibiotic phenotypes in soil.
The objectives of this research were to (i) quantify the frequency and mean intensity of antibiotic inhibition and resistance for streptomycetes among locations and depths in a prairie soil, (ii) characterize the spatial distribution of antibiotic inhibition and resistance phenotypes of streptomycetes from a prairie soil, and (iii) evaluate the relationship between antibiotic inhibition and resistance capabilities among individual streptomycetes from a prairie soil.
MATERIALS AND METHODS
Soil sampling.
Research was conducted at the Cedar Creek Natural History Area (CCNHA), a National Science Foundation long-term ecological research site (www.lter.umn.edu). The plots studied in this work are part of a long-term investigation of the effects of nitrogen on plant productivity, diversity, and succession (E001 at the CCNHA website). These plots (field C) were abandoned from row crop production in 1934. The present study focuses on fine-scale spatial variation in phenotypic diversity within one plot (plot 8A; 4 by 4 m). Within this plot, a grid (1 by 1 m) located 1.2 m south and 1.2 m east of the northwest corner was delineated. Within this grid, three sampling locations were randomly selected with (x-y) coordinates. To facilitate fine-scale sampling of the soil microbial community, small soil corers were created at the University of Minnesota scientific apparatus shop. Sections of aluminum pipe, 30 by 1 cm with a 1-mm-thick wall (K & S Manufacturing, Chicago, Ill.), were milled into soil corers by removing one-half of the outer wall of a 10-cm segment 6 mm from the tip. Corer tips were tapered to facilitate insertion into the ground. In December 1999 at each sampling location, three soil corers were bundled together and pounded gently into the soil with a hammer. Soil was left in the corers, which were wrapped in plastic, placed on ice, and transported back to the laboratory for immediate processing.
Soil processing.
Cores were unwrapped and placed against an 8-cm guide. Each core was divided into 2-cm subsections corresponding to soil depths of 0 to 2, 2 to 4, 4 to 6, and 6 to 8 cm (depths 1 to 4, respectively). Therefore, within the single plot there were a total of 36 samples (four depths times three cores times three locations).
Soil from each sample was placed in a plastic weighboat under two layers of sterile cheesecloth to dry overnight on the bench. Samples were transferred into 50-ml centrifuge tubes containing 10 ml of buffered phosphate solution (0.5 M K2HPO4, 0.4 M KH2PO4, pH 7.0). Tubes were shaken for 1 h on a reciprocal shaker (4°C, 250 rpm). Resulting soil suspensions were dilution plated onto oatmeal agar amended with antibiotics (73). Plates were incubated at 28°C for 7 days. Densities of total culturable bacteria and streptomycetes were estimated for every sample. Colonies exhibiting characteristic streptomycete colony morphology (6) were selected randomly and pure cultured for further study. For each of the 36 samples, we sought to obtain five pure culture isolates. In some cases, we were not successful because of either a low streptomycete density or an inability to obtain pure cultures. From the 36 soil samples, a total of 153 streptomycete isolates were purified for further study. Purified spore suspensions of each isolate were maintained in 20% glycerol in a −80°C freezer.
Antibiotic assays.
All field isolates were tested in all possible pairwise combinations for the ability to inhibit and for resistance to members of a collection of 10 streptomycete test isolates, which included an isolate pathogenic to potato (Streptomyces scabies), as well as nonpathogenic isolates (A. L. Davelos, K. Xiao, J. M. Flor, and L. L. Kinkel, submitted for publication). A modified form of the antibiotic assay of Vidaver et al. (66) was used to determine the inhibition of and resistance to each of the test isolates by the indigenous isolates. Specifically, spore suspensions (approximately 108 spores/ml) of individual isolates were dotted (10 μl per spot) onto 15-ml starch casein agar plates (100 by 15 mm), five dots per plate. Plates were incubated at 28°C for 3 days. Dotted isolates were killed by inverting the uncovered petri plates over 4 ml of chloroform in a watch glass for 1 h. Watch glasses were removed, and plates were aerated in a fume hood for 30 min to permit evaporation of chloroform. Plates were subsequently overlaid with 15 ml of 1% water agar and inoculated with 100 μl of a test isolate (approximately 108 spores/ml) spread uniformly over the surface of the agar. Plates were incubated at 28°C for 3 days. The size of any zone of growth inhibition of the overlaid isolate surrounding any dotted isolate was measured in millimeters from the edge of the dotted colony to the edge of the cleared zone. Each indigenous isolate was both dotted (to measure inhibition) and overlaid (to measure resistance). Specifically, 1,530 (153 field isolate dots × 10 test isolate overlays) potentially inhibitory interactions caused by the indigenous isolates against the test isolates and 1,530 (10 test isolate dots × 153 field isolate overlays) potentially resistant interactions for the indigenous isolates were evaluated. Each interaction was replicated on three separate plates. Thus, a total of 9,180 antibiotic interactions were examined here.
Analyses. (i) Characterization of antibiotic activity. (a) Classification of individual isolates.
The frequency and intensity of the antibiotic interactions between the indigenous isolates and the collection of 10 test isolates were examined. The mean zone size for each interaction between a field isolate and a test isolate was determined (three replicates for each of 10 inhibition and 10 resistance interactions). All analyses were performed on means. Frequency summarizes the number of test isolates that an indigenous isolate inhibited or was resistant to (presence or absence of an inhibition zone). Intensity is used to describe the quantitative inhibition or resistance to inhibition of indigenous isolates when paired with test isolates as determined by the size of the inhibition zone. When quantifying the intensity of inhibition for the field isolates, a small zone size indicates little inhibition of the test isolate. In contrast, when quantifying resistance of the field isolates, a small zone size indicates a high resistance to inhibition by the test isolate.
(b) Antibiotic phenotypic diversity.
For the frequency data (presence or absence of inhibition of or resistance to each of the 10 test isolates), simple matching criteria (62) were used to generate similarity matrices (NT-SYS) (49). Cluster analyses were performed by the unweighted pair-group method using arithmetic averages (60), and dendrograms were constructed (NT-SYS) (49). Phenotypic groups were determined on the basis of 100% similarity of antibiotic phenotypes. Three dendrograms were created, one each for inhibition and resistance data separately and one based on the combined inhibition and resistance data matrix.
The phenotypic groupings resulting from dendrograms of antibiotic frequency were used to calculate richness (S = the number of phenotypic groups) and the Shannon diversity index [H = −Σ(xi/x0)ln(xi/x0), where xi is the number of isolates in a phenotypic group and x0 is the total number of isolates] (40). Diversity indices and standard deviations were calculated with EstimateS (12).
(ii) Spatial variation.
To assess spatial variation in the antibiotic activity of indigenous CCNHA streptomycete isolates, the frequency and intensity of inhibition and resistance were examined among the three field locations and the four depths (0 to 2, 2 to 4, 4 to 6, and 6 to 8 cm). The frequency of antibiotic interactions that showed either inhibition (n = 4,590 [153 field isolates times 10 test isolates times three replicates]) or resistance (n = 4,590) and the frequency of test isolates that a field isolate inhibited or was resistant to were compared among locations and among depths with a binomial model (PROC GENMOD) (1, 55, 57). Significant differences were determined with the likelihood ratio statistic. Differences among mean inhibition and resistance zone sizes were evaluated by analysis of variance (PROC GLM) (54). Significant differences among means were determined with least significant differences. Distributions of inhibition and resistance phenotypes of the field isolates, based on qualitative inhibition or resistance matrices, among locations and depths were examined with a multinomial model (PROC GENMOD) (1, 55, 57).
Because the above-described analyses revealed substantial variation among the three field locations (see Results), antibiotic frequency and intensity were evaluated among cores and depths nested within cores at each location separately.
(iii) Correspondence between inhibition and resistance.
To examine the association between inhibition and resistance among prairie isolates, the correlation between mean zone sizes for each field isolate when inhibiting the test isolates and when resisting the test isolates was determined overall, among locations and depths, and within locations (PROC CORR) (53). A positive correlation indicates that prairie isolates that produce a large mean inhibition zone size (i.e., inhibit the test isolates) also have a large mean resistance zone size (i.e., are not resistant to the test isolates). Further, correlations between the number of test isolates that a field isolate resisted and the mean inhibition zone size and between the number of test isolates that a field isolate inhibited and the mean resistance zone size among all isolates were examined.
Distance matrices based on inhibition and resistance data were compared to determine whether the matrices were correlated with Mantel's test (NT-SYS) (49) with the significance of the correlation determined as described by Lapointe and Legendre (34). Analyses evaluated the correspondence between prairie isolate inhibition and resistance groupings. A positive correlation indicates that field isolates in a single inhibition phenotypic group are likely to cluster in the same resistance group.
(iv) Correlations between density and antibiotic activity.
Differences in density of culturable bacteria and streptomycetes among locations and depths were examined by analysis of variance (PROC GLM) (54). Further, correlations among densities and mean zone sizes and number of test isolates inhibited and resisted were evaluated (PROC CORR) (53).
RESULTS
Characterization of antibiotic activity. (i) Classification of individual isolates
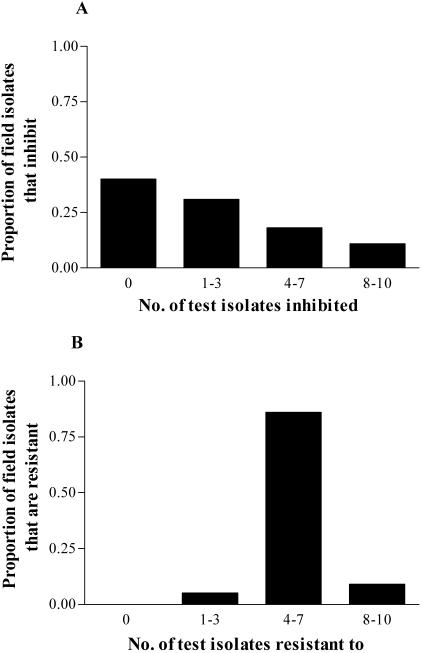
There was wide variation in the ability of prairie streptomycetes to inhibit or resist the 10 test isolates. Of the 4,590 interactions examining the ability of the field isolates to inhibit the test isolates, 769 (17%) were inhibitory. In 3,056 (67%) of the 4,590 interactions examining the resistance of the field isolates to the 10 test isolates, the interaction showed that the field isolate was resistant to the test isolate. Inhibitory and resistance profiles of individual prairie isolates differed substantially (Fig. 1). The majority of field isolates inhibited a small number of test isolates (fewer than four). Forty percent of the field isolates (61 out of 153) did not inhibit any of the test isolates, while 14 field isolates (9.2%) inhibited all of the test isolates (Fig. 1A). In contrast, most of the field isolates showed resistance to at least half of the test isolates (Fig. 1B). No field isolate was resistant to all 10 test isolates. All prairie isolates were resistant to at least one of the test isolates; one isolate was resistant to two test isolates, and seven isolates were resistant to three test isolates.
FIG. 1.
Inhibition (A) and resistance (B) of 153 streptomycete isolates, indigenous to CCNHA prairie soil, to 10 test streptomycete isolates. The proportion of field isolates that inhibited or resisted 0, 1 to 3, 4 to 7, or 8 to 10 test isolates are presented.
(ii) Antibiotic phenotypic diversity.
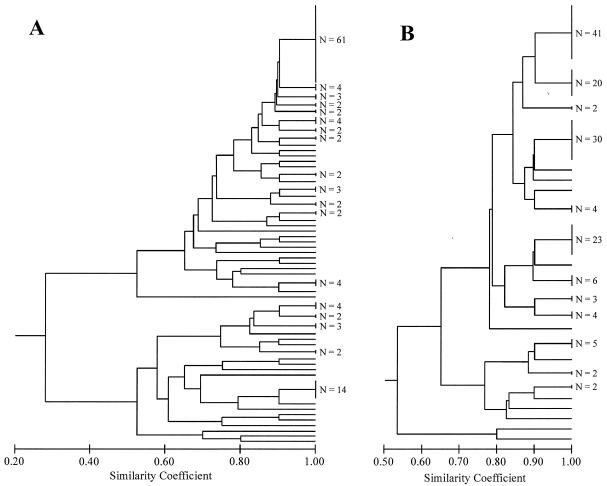
Field isolates with 100% similarity in qualitative (presence or absence) inhibition or resistance profiles were assigned to a phenotypic group (Fig. 2). Thirty-five isolates had unique inhibition phenotypes, and the remaining isolates were placed into 18 inhibition phenotypes, for a total of 53 phenotypic groups (Fig. 2A). Sixty-one isolates had the most common inhibition phenotype; these prairie isolates did not inhibit any of the test isolates. Eleven isolates had unique resistance phenotypes, while the remaining isolates were grouped into 12 resistance phenotypes, for a total of 23 phenotypic groups (Fig. 2B). Forty-one isolates had the most common resistance phenotype (resistance to the same five test isolates). There was greater diversity in inhibition than resistance phenotypes in the prairie streptomycete community (2.86 ± 0.01 versus 2.26 ± 0.01, respectively). When inhibition and resistance data were combined in a single dendrogram, there were 79 isolates with unique phenotypes, with the remaining isolates clustering into 14 phenotypic groups, for a total of 93 phenotypic groups. Twenty-two isolates formed the largest phenotypic group; none of these field isolates inhibited any of the test isolates, and all were resistant to the same five test isolates. Phenotypic diversity based on the combined data was higher than when measured with inhibition or resistance data alone (4.02 ± 0.01).
FIG. 2.
Dendrograms of inhibition (A) and resistance (B) phenotypes generated from the frequency (presence or absence of an inhibition zone) of antibiotic interactions between CCNHA streptomycete isolates and 10 test streptomycete isolates.
Spatial variation.
To evaluate spatial variation in antibiotic activity among prairie isolates, differences in proportions of inhibitory or resistant interactions, frequency of test isolates a prairie isolate inhibited or was resistant to, mean zone sizes, and the distribution of phenotypes among locations and depths were examined.
The proportion of inhibitory interactions varied significantly among locations (χ2 = 22.37, df = 2, P < 0.0001) and depths (χ2 = 22.33, df = 3, P < 0.0001). The proportion of interactions in which the field isolates inhibited the test isolates was significantly greater in location 1 than in the other two locations. The proportion of inhibitory interactions was significantly lower at depth 2 (2 to 4 cm) than at other depths. In contrast, the proportion of resistance interactions (the ability of field isolates to resist the 10 test isolates) was remarkably stable among locations (χ2 = 6.93, df = 2, P < 0.04) and depths (χ2 = 6.40, df = 3, P < 0.10). These results were largely consistent with those found when the frequency of test isolates that a prairie isolate inhibited or was resistant to was examined among locations and depths (data not shown).
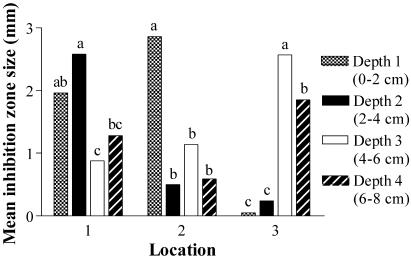
Mean inhibition zone sizes of field isolates against the set of 10 test isolates varied significantly among locations (F2,1524 = 6.89, P < 0.002). Streptomycete isolates from location 1 had significantly larger mean inhibition zones than isolates from the other two locations, indicating that these prairie isolates were relatively better at inhibiting the test isolates (Table 1). No significant differences in mean inhibition zone size were found among depths (F3,1524 = 2.33, P < 0.08; Table 1). To examine spatial variation in mean inhibition zone size within locations, differences in cores and depths within cores were evaluated. No significant differences among cores within locations were found (results not presented); thus, further analyses focused on depths. Significant differences among depths within each location were found (Fig. 3). However, the pattern of differences in inhibitory activity depended on the location. For example, prairie isolates from depths 3 and 4 had significantly greater inhibition zone sizes (higher activity) at location 3 (F3,486 = 32.16, P < 0.0001) while at location 1 isolates from depths 1 and 2 had the greatest zone sizes (F3,486 = 4.47, P < 0.005). At location 2, field isolates from depth 1 also produced a significantly greater mean inhibition zone size (F3,546 = 15.87, P < 0.0001) than isolates from other depths.
TABLE 1.
Variation in inhibition and resistance, Shannon diversity, and density of culturable bacteria and streptomycetes among locations and depthsa
| Location or depth | N | Mean inhibition zone size (mm) | Mean resistance zone size (mm) | Diversity (H) | Density of culturable bacteria (CFU/g) | Density of streptomycetes (CFU/g) |
|---|---|---|---|---|---|---|
| Location 1 | 49 | 1.73a | 3.87ab | 3.41 ± 0.02 | 1.7 × 106a | 1.5 × 105b |
| Location 2 | 55 | 1.30b | 3.22b | 3.38 ± 0.02 | 1.4 × 106a | 2.6 × 105a |
| Location 3 | 49 | 0.94b | 4.32a | 2.94 ± 0.03 | 1.2 × 106a | 1.1 × 105b |
| Depth 1 | 44 | 1.60a | 3.14b | 2.66 ± 0.03 | 1.7 × 106a | 1.9 × 105a |
| Depth 2 | 41 | 1.02a | 4.04ab | 3.17 ± 0.03 | 1.3 × 106a | 2.1 × 105a |
| Depth 3 | 34 | 1.44a | 4.22a | 3.44 ± 0.02 | 1.5 × 106a | 1.5 × 105a |
| Depth 4 | 34 | 1.21a | 3.87ab | 2.97 ± 0.03 | 1.2 × 106a | 1.4 × 105a |
Depths 1 to 4 correspond to soil depths of 0 to 2, 2 to 4, 4 to 6, and 6 to 8 cm, respectively. N is the sample size of CCNHA streptomycete isolates. Raw mean zone sizes, measured in millimeters, are reported. For resistance, a smaller mean zone size indicates greater resistance. Values followed by the same letter(s) are not significantly different at P < 0.05 (least significant difference). Standard deviations for Shannon diversity (H) are presented.
FIG. 3.
Mean inhibition zone sizes of field isolates against the 10 test isolates for each depth at each location. Significant differences among means within a location are indicated by different letters above the bars (least significant difference, P < 0.05).
Prairie streptomycete resistance to inhibition by the test isolates varied significantly as a function of both location (F2,1524 = 4.55, P < 0.02) and depth (F3,1524 = 2.65, P < 0.05). Isolates from location 2 were more resistant to the test isolates than were isolates from the other two locations. When examining variation in mean resistance zone size within locations, in general neither cores nor depths varied significantly. The greatest resistance to antibiotic inhibition (smallest mean resistance zone size) was shown by isolates from the shallowest depth (0 to 2 cm) (Table 1). This same trend for prairie isolates from depth 1 (0 to 2 cm) having the greatest resistance was found within each location.
Different inhibition phenotypes (Fig. 2A) were found at each depth (χ2 = 12.04, df = 3, P < 0.008) but not at each location (χ2 = 5.87, df = 2, P < 0.06). More than two-thirds of the field isolates that did not inhibit any of the 10 test isolates were at depths of 0 to 4 cm, while more than half of field isolates with unique inhibition phenotypes were found at depths of 4 to 8 cm. The distribution of resistance phenotypes (Fig. 2B) did not vary among locations (χ2 = 0.39, df = 2, P < 0.83) or depths (χ2 = 7.48, df = 3, P < 0.06).
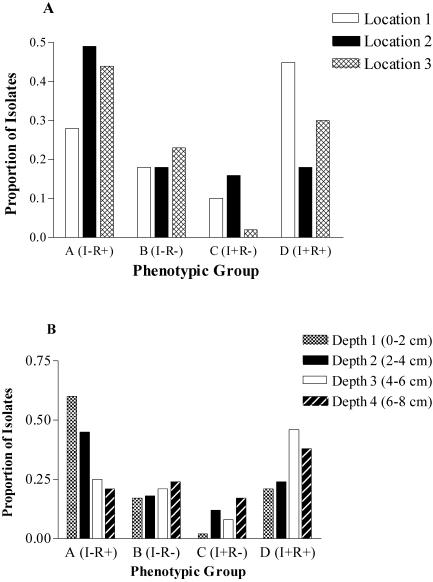
On the basis of the combined inhibition and resistance qualitative data (presence or absence of inhibition of or resistance to the 10 test isolates), each prairie isolate was assigned to one of four groups. Group A isolates (I− R+) inhibited fewer than four test isolates and were resistant to more than six test isolates. Group B isolates (I− R−) inhibited fewer than four test isolates and were resistant to fewer than six test isolates. Group C isolates (I+ R−) inhibited more than four test isolates and were resistant to fewer than six test isolates. Group D isolates (I+ R+) inhibited more than four test isolates and were resistant to more than six test isolates. In the dendrogram generated from the combined inhibition and resistance profiles, indigenous CCNHA streptomycete isolates generally clustered in the above-defined groups, with 63 to 88% similarity within a cluster. Twenty-five field isolates did not fall into any of these broad clusters.
The distribution of phenotypic groups A to D defined above varied among locations (χ2 = 6.85, df = 2, P < 0.04) and with depth (χ2 = 13.04, df = 3, P < 0.005). Location 1 had a low frequency of group A (I− R+) prairie isolates. Location 3 had a low frequency of group C (I+ R−) isolates, while location 2 had a low frequency of group D (I+ R+) isolates. Group B (I− R−) isolates were fairly evenly distributed among locations (Fig. 4A). Group B (I− R−) field isolates were represented at similar frequencies among the depths. Group C (I+ R−) isolates were common at 6 to 8 cm but occurred rarely at 0 to 2 cm. Depths 1 and 2 (0 to 4 cm) were dominated by group A (I− R+) isolates, while group D (I+ R+) isolates were found at depths 3 and 4 (4 to 8 cm) (Fig. 4B).
FIG. 4.
(A) Frequencies of CCNHA streptomycete isolates among locations from phenotypic groups A to D. (B) Frequencies of CCNHA streptomycete isolates among depths from phenotypic groups A to D.
Correspondence between inhibition and resistance.
There was a negative but marginally significant relationship between mean inhibition and resistance zone sizes for individual streptomycete field isolates (r = −0.13, n = 153, P < 0.10); isolates that inhibit strongly tend to be more resistant to inhibition by the test isolates. Within location 2, where field isolates had significantly greater resistance, mean inhibition and resistance zone sizes were significantly negatively correlated (r = −0.40, n = 55, P < 0.003) while mean inhibition and resistance zone sizes were poorly correlated at the other two locations (results not shown). Mean inhibition and resistance zone sizes of prairie isolates were poorly correlated at depths 2 and 3 (2 to 6 cm) (data not presented). At depth 1 (0 to 2 cm), mean inhibition and resistance zone sizes were significantly negatively correlated (r = −0.36, n = 44, P < 0.02). At depth 4 (6 to 8 cm), the trend was in the opposite direction (r = 0.40, n = 34, P < 0.02), indicating that field isolates that produced large inhibition zones tended to be less resistant to the test isolates.
There was a marginally significant negative relationship between the number of test isolates that an isolate was resistant to and its mean inhibition zone size (r = −0.15, n = 153, P < 0.07). Therefore, the more test isolates that a field isolate was resistant to, the smaller the intensity of its inhibition of the test isolates. However, the number of test isolates that an isolate inhibited and its mean resistance zone size were poorly correlated (r = 0.05, n = 153, P < 0.51).
Qualitative inhibition and resistance profiles of CCNHA prairie isolates against the test isolates were significantly positively correlated (Mantel's test: r = 0.13, P < 0.001). This association appears to be driven by the largest inhibition phenotypic group (61 isolates), field isolates that did not inhibit any of the 10 test isolates. Forty-five isolates in this group clustered in one of three resistance phenotypes. However, the remaining 16 isolates in this group were distributed among 10 resistance phenotypes. Indeed, multiple inhibition phenotypes were found frequently within a single resistance phenotype (results not shown). Although isolates found within a resistance phenotype often clustered with at least one other isolate in the same inhibition phenotype, a number of isolates showed unique inhibition phenotypes.
In general, there was no consistent spatial relationship between inhibition and resistance. However, there was an overall trend for isolates with greater inhibition to also have greater resistance.
Correlations between density and antibiotic activity.
Total culturable bacterial densities were an order of magnitude greater than streptomycete densities at all locations and depths (Table 1). There was no significant variation in bacterial density among locations (F2,33 = 1.07, P < 0.36) or depths (F3,33 = 1.11, P < 0.36). However, there was a clear correspondence between antibiotic activity and bacterial density. At location 1, where the bacterial density was greatest, the highest diversity and greatest mean inhibition zone size were also found, and location 3, which had the lowest bacterial density, had the lowest diversity and smallest mean inhibition zone size (Table 1).
Streptomycete density did not vary with depth (F3,33 = 0.51, P < 0.69). However, there were significant differences in streptomycete density among locations (F2,33 = 5.88, P < 0.01), with location 2 having higher streptomycete densities than location 1 or 3 (Table 1). Streptomycete and total bacteria densities were significantly positively correlated (rs = 0.59, n = 36, P < 0.0001).
Correlations between streptomycete density and antibiotic activity (mean inhibition and resistance zone size and the number of test isolates inhibited or resistant to) among locations and depths revealed no clear trends (results not presented). Furthermore, there were no distinct patterns between density and antibiotic activity on the smallest spatial scale. Specifically, among all 36 samples (three locations, three cores, four depths), there was no significant correlation between streptomycete density and the number of test isolates inhibited (r = −0.08, n = 36, P < 0.65) or resisted (r = 0.07, n = 36, P < 0.67). Further, mean inhibition zone size and streptomycete density (r = −0.06, n = 36, P < 0.74) and mean resistance zone size and streptomycete density (r = 0.04, n = 36, P < 0.81) were poorly correlated overall.
DISCUSSION
Streptomycetes from the CCNHA prairie soil were enormously variable in their antibiotic inhibition and resistance abilities. Overall, resistance was generally more common than inhibition. The majority of prairie soil streptomycetes could resist multiple test isolates, while isolates varied widely in the ability to inhibit the test isolates (Fig. 1). The greater frequency and stability of antibiotic resistance compared with antibiotic production ability (inhibition) within the soil community suggests that antibiotic resistance confers a greater relative fitness benefit than antibiotic production. Alternatively, resistance capabilities may be less costly energetically to maintain in the absence of direct selection pressure (35). Further, resistance may be more readily disseminated among individuals via horizontal gene transfer (26, 47, 71). These data are consistent with the expectation that resistance is an absolute requirement for survival in the presence of an antibiotic producer, while antibiotic production will confer only a relative benefit to the producer in the presence of sensitive strains.
There was substantial diversity in inhibition and resistance phenotypes (Fig. 2) within the prairie soil streptomycete community. The samples collected here, which identified 53 inhibition phenotypes and 23 resistance phenotypes in the spatially explicit sample of 153 individual streptomycete isolates, have likely uncovered only a fraction of the total phenotypic diversity within these soils. Indeed, other studies have revealed amazing levels of phenotypic and genetic diversity within soil microbial communities (e.g., see references 5, 29, 43, 44, and 45). Additional test isolates may uncover further diversity within the collection of prairie soil streptomycetes already analyzed. Larger sample sizes, including both field isolates and test isolates, are needed to fully characterize the diversity in antibiotic inhibitory and resistance phenotypes within this prairie soil.
The distinct differences in the frequency and intensity of antibiotic inhibition and resistance abilities among streptomycete communities in the different soil locations suggest that antibiotic interactions may vary in their significance to fitness among locations in soil. Competitive interactions can take a variety of forms, including resource and interference, or antibiotic, competition (59, 69). Locations where isolates have relatively little antibiotic inhibitory ability (e.g., location 3, Table 1) within the streptomycete community may be locations where interference competition is less important (i.e., where population density is low; location 3, Table 1). Alternatively, other competitive strategies, especially resource competition or niche differentiation, might be more important to fitness at locations where isolates have low antibiotic inhibitory activity. Overall, these data show that antibiotic activities, especially inhibition abilities, can be highly clustered in soil.
Spatial clustering of antibiotic phenotypes may reflect distinct selective environments among locations or repeated sampling of clonal individuals. Overlapping of clones on small spatial scales has been found for some soil microbes (56, 68), although for other soil microbes the same genotype has been found across wide geographic areas (29, 42, 45). In this work, genetic analyses with BOX-PCR genomic DNA fingerprints and 16S rRNA gene sequences of these prairie isolates revealed that in only 6 of 36 samples were more than 50% of the isolates from a single clone (K. Xiao, A. L. Davelos, and L. L. Kinkel, unpublished data). Thus, repeated sampling of clones does not account for the clustering of antibiotic activity in space observed here.
The structure of the soil microbial community may be altered by associated plant species (e.g., see references 23, 31, 33, 41, and 61). Within the 1-m2 plot sampled in this study, the percent cover and plant species composition did not vary significantly among the sampling locations (A. L. Davelos and L. L. Kinkel, unpublished data). While the effect of plant species is potentially important, the differences in antibiotic inhibitory activity in this study are not likely driven by differences in plant species among locations.
Population density (number of CFU per gram of soil) may provide an estimate of the potential significance of competitive interactions to microbial ecology in soil. In this study, we used counts from selective media as a crude estimate of the total density of the soil microbial community. Other work has shown that counts of microbial density from selective media and direct microscopic counts are highly correlated (64). We hypothesize that interference competition, or antibiotic interactions, has a greater effect on microbial fitness at high population densities than at low population densities. Certainly, the probability or frequency of microbial encounters will increase with increased population density, suggesting that the relative benefits of antibiotic inhibition or antibiotic resistance may be enhanced in a high-density community (70). Indeed, our findings of greater frequency and intensity of inhibition in locations with a greater density of total culturable bacteria (Table 1) are consistent with this idea. Further work should consider a wider range of population densities or more controlled experiments to elucidate any possible effects of population density on the frequency or intensity of antibiotic activities within the soil microflora.
The lack of correspondence between phenotype groups obtained on the basis of antibiotic inhibition profiles and those obtained on the basis of resistance profiles is intriguing. Specifically, the data indicate that streptomycete isolates having the same inhibition abilities, or the ability to inhibit the same collection of test isolates, generally had a wide variety of different resistance profiles. This pattern may be the result of the lateral gene transfer of resistance among the prairie isolates. Horizontal transfer of resistance genes among streptomycetes in soil has been reported (26, 71), as well as lateral transfer of antibiotic gene clusters (17, 18). The lack of correspondence between inhibition and resistance phenotypes suggests that antibiotic inhibitory and resistance abilities within the soil microbial community are selected for independently. More broadly, the distinct clustering of isolates into antibiotic inhibition and resistance phenotypes may reflect the diverse selection mosaic present in the soil habitat and the adaptation of streptomycete isolates to the precise physical and biological characteristics of their local habitat. Given the enormous diversity in antibiotic inhibition and resistance capabilities within the soil streptomycete community, it is not surprising that unique and presumably locally advantageous combinations of resistance and inhibition occur within individual isolates.
The vast diversity in antibiotic activities observed in this study likely reflects the complexity of interactions among members of the soil microbial community. The high diversity in antibiotic inhibition and resistance activities and the specificity of inhibition and resistance reactions among isolates are reminiscent of the huge diversity in host resistance and pathogen virulence genes noted in many agricultural and nonagricultural host pathosystems (e.g., see references 8 and 9; reviewed in reference 7). Coevolutionary models that predict the generation and maintenance of a high degree of diversity in host resistance and pathogen virulence genes and the establishment of polymorphic equilibria (3, 20, 27, 36, 65) demonstrate that an arms race between interacting organisms can promote genetic and phenotypic diversity. Particularly in a spatially structured environment, such as the soil, development of antibiotic production is especially likely and may provide fitness benefits under a wide range of conditions (11, 16). Results from a recent spatially explicit model showed that “chemical warfare” (i.e., antibiotic production) among interacting microbes can generate and maintain a diversity of phenotypes including antibiotic production, resistance, and even sensitivity (14). The work presented here offers the first empirical data quantifying the diversity of antibiotic activities in a spatially explicit sample of the soil microbial community and suggests that selection pressures and microbial responses vary across the soil landscape.
Acknowledgments
Funding for this research was provided by the National Science Foundation through Microbial Observatories project 9977907.
We thank Jennifer Flor for technical assistance and production of graphics and Jennifer Gilpin for assistance in performing the antibiotic assays.
REFERENCES
- 1.Agresti, A. 2002. Categorical data analysis, 2nd ed. Wiley, New York, N.Y.
- 2.Anderson, A. S., and E. M. H. Wellington. 2001. The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol. 51:797-814. [DOI] [PubMed] [Google Scholar]
- 3.Antonovics, J., and P. Thrall. 1994. The cost of resistance and the maintenance of genetic polymorphism in host-pathogen systems. Proc. R. Soc. Lond. B Biol. Sci. 257:105-110. [Google Scholar]
- 4.Bent, S. J., C. L. Gucker, Y. Oda, and L. J. Forney. 2003. Spatial distribution of Rhodopseudomonas palustris ecotypes on a local scale. Appl. Environ. Microbiol. 69:5192-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers, J. H., L. L. Kinkel, R. K. Jones, and N. A. Anderson. 1996. Influence of disease suppressive strains of Streptomyces on the native Streptomyces community in soil as determined by the analysis of cellular fatty acids. Can. J. Microbiol. 42:27-37. [DOI] [PubMed] [Google Scholar]
- 7.Burdon, J. J. 1987. Diseases and plant population biology. Cambridge University Press, Cambridge, United Kingdom.
- 8.Burdon, J. J., and A. M. Jarosz. 1992. Temporal variation in the racial structure of flax rust (Melampsora lini) populations growing on natural stands on wild flax (Linum marginale): local versus metapopulation dynamics. Plant Pathol. 41:165-179. [Google Scholar]
- 9.Burdon, J. J., and J. N. Thompson. 1995. Changed patterns of resistance in a population of Linum marginale attacked by the rust pathogen Melampsora lini. J. Ecol. 83:199-206. [Google Scholar]
- 10.Chamberlain, K., and D. L. Crawford. 1999. In vitro and vivo antagonism of pathogenic turfgrass fungi by Streptomyces hygroscopicus strains YCED9 and WYE53. J. Ind. Microbiol. Biotechnol. 23:641-646. [DOI] [PubMed] [Google Scholar]
- 11.Chao, L., and B. R. Levin. 1981. Structural habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. USA 78:6324-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colwell, R. K. 1997. EstimateS: statistical estimation of species richness and shared species from samples. Version 5. User's guide and application. [Online.] http://viceroy.eeb.uconn.edu/estimates.
- 13.Crawford, D. L., J. M. Lynch, J. M. Whipps, and M. A. Ousley. 1993. Isolation and characterization of actinomycete antagonists of a fungal root pathogen. Appl. Environ. Microbiol. 59:3899-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czárán, T. L., R. F. Hoekstra, and L. Pagie. 2002. Chemical warfare between microbes promotes biodiversity. Proc. Natl. Acad. Sci. USA 99:786-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doumbou, C. L., M. K. H. Salove, D. L. Crawford, and C. Beaulieu. 2001. Actinomycetes, promising tools to control plant diseases and to promote plant growth. Phytoprotection 82:85-102. [Google Scholar]
- 16.Durrett, R., and L. Simon. 1997. Allelopathy in spatially distributed populations. J. Theor. Biol. 185:165-171. [DOI] [PubMed] [Google Scholar]
- 17.Egan, S., P. Wiener, D. Kallifidas, and E. M. H. Wellington. 1998. Transfer of streptomycin biosynthesis gene clusters within streptomycetes isolated from soil. Appl. Environ. Microbiol. 64:5061-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egan, S., P. Wiener, D. Kallifidas, and E. M. H. Wellington. 2001. Phylogeny of Streptomyces species and evidence for horizontal transfer of entire and partial antibiotic gene clusters. Int. J. Gen. Mol. Microbiol. 79:127-133. [DOI] [PubMed] [Google Scholar]
- 19.El-Abyad, M. S., M. A. El-Sayed, A. R. El-Shanshoury, and S. M. El-Sabbagh. 1993. Towards the biological control of fungal and bacterial diseases of tomato using antagonistic Streptomyces spp. Plant Soil 149:185-195. [Google Scholar]
- 20.Frank, S. 1993. Coevolutionary genetics of plants and pathogens. Evol. Ecol. 7:45-75. [Google Scholar]
- 21.Franklin, R. B., and A. L. Mills. 2003. Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol. Ecol. 44:335-346. [DOI] [PubMed] [Google Scholar]
- 22.Fujisawa, Y., and B. Weisblum. 1981. A family of r-determinants in Streptomyces spp. that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 146:621-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grayston, S. J., S. Wang, C. D. Campbell, and A. C. Edwards. 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30:369-378. [Google Scholar]
- 24.Hopwood, D. A. 1988. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc. R. Soc. Lond. B Biol. Sci. 235:121-138. [DOI] [PubMed] [Google Scholar]
- 25.Hotta, K., and Y. Okami. 1996. Diversity in aminoglycoside antibiotic resistance of actinomycetes and its exploitation in the search for novel antibiotics. J. Ind. Microbiol. 17:352-358. [Google Scholar]
- 26.Huddleston, A. S., N. Cresswell, M. C. P. Neves, J. E. Beringer, S. Baumberg, D. I. Thomas, and E. M. H. Wellington. 1997. Molecular detection of streptomycin-producing streptomycetes in Brazilian soils. Appl. Environ. Microbiol. 63:1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayakar, S. D. 1970. A mathematical model for interaction of gene frequencies in a parasite and its host. Theor. Pop. Biol. 1:140-164. [DOI] [PubMed] [Google Scholar]
- 28.Jones, C. R., and D. A. Samac. 1996. Biological control of fungi causing alfalfa seedling damping-off with a disease-suppressive strain of Streptomyces. Biol. Control 7:196-204. [Google Scholar]
- 29.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl. Environ. Microbiol. 60:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khalad, A. E., G. E. S. J. Hardy, K. Sivasithamparam, A. M. Hussein, and D. I. Kurtboke. 1997. The potential for biological control of cavity-spot disease of carrots, caused by Pythium coloratum, by streptomycete and non-streptomycete actinomycetes. New Phytol. 137:495-507. [DOI] [PubMed] [Google Scholar]
- 31.Kowalchuk, G. A., D. S. Buma, W. de Boer, P. G. L. Klinkhamer, and J. A. van Veen. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Int. J. Gen. Mol. Microbiol. 81:509-520. [DOI] [PubMed] [Google Scholar]
- 32.Krechel, A., A. Faupel, J. Hallmann, A. Ulrich, and G. Berg. 2002. Potato-associated bacteria and their antagonistic potential towards plant-pathogenic fungi and the plant-parasitic nematode Meloidogyne incognita (Kofoid & White) Chitwood. Can. J. Microbiol. 48:772-786. [DOI] [PubMed] [Google Scholar]
- 33.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S. M. Burns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapointe, F.-J., and P. Legendre. 1992. Statistical significance of the matrix correlation coefficient for comparing independent phylogenetic trees. Syst. Biol. 41:378-384. [Google Scholar]
- 35.Lenski, R. E. 1997. The cost of antibiotic resistance—from the perspective of a bacterium. CIBA Found. Symp. 207:131-140. [DOI] [PubMed] [Google Scholar]
- 36.Leonard, K. J. 1977. Selection pressures and plant pathogens. Ann. N. Y. Acad. Sci. 287:207-222. [Google Scholar]
- 37.Liu, D. 1992. Biological control of Streptomyces scabies and other plant pathogens. Ph.D. thesis. University of Minnesota, St. Paul.
- 38.Liu, D., N. A. Anderson, and L. L. Kinkel. 1995. Biological control of potato scab in the field with antagonistic Streptomyces scabies. Phytopathology 85:827-831. [Google Scholar]
- 39.Liu, D., N. A. Anderson, and L. L. Kinkel. 1996. Selection and characterization of strains of Streptomyces suppressive to the potato scab pathogen. Can. J. Microbiol. 42:487-502. [Google Scholar]
- 40.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, N.J.
- 41.Marschner, P., C.-H. Yang, R. Lieberei, and D. E. Crowley. 2001. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33:1437-1445. [Google Scholar]
- 42.Mavrodi, O. V., B. B. McSpadden Gardener, D. V. Mavrodi, R. F. Bonsall, D. M. Weller, and L. S. Thomashow. 2001. Genetic diversity of phlD from 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. Phytopathology 91:35-43. [DOI] [PubMed] [Google Scholar]
- 43.Mazza, P., P. Monciardini, L. Cavaletti, M. Sosio, and S. Donadio. 2003. Diversity of Actinoplanes and related genera isolated from an Italian soil. Microb. Ecol. 45:362-372. [DOI] [PubMed] [Google Scholar]
- 44.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McSpadden Gardener, B. B., K. L. Schroeder, S. E. Kalloger, J. M. Raaijmakers, L. S. Thomashow, and D. M. Weller. 2000. Genotypic and phenotypic diversity of phlD-containing Pseudomonas strains isolated from the rhizosphere of wheat. Appl. Environ. Microbiol. 66:1939-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meinhardt, F., R. Schaffrath, and M. Larsen. 1997. Microbial linear plasmids. Appl. Microbiol. Biotechnol. 47:329-336. [DOI] [PubMed] [Google Scholar]
- 47.Nwosu, V. C. 2001. Antibiotic resistance with particular reference to soil microorganisms. Res. Microbiol. 152:421-430. [DOI] [PubMed] [Google Scholar]
- 48.Phillips, L., E. M. H. Wellington, and S. B. Rees. 1994. The distribution of antibiotic resistance patterns within streptomycetes and their use in secondary metabolite screening. J. Ind. Microbiol. 13:53-62. [Google Scholar]
- 49.Rohlf, F. J. 1998. NTSYSpc. Numerical taxonomy and multivariate analysis system, version 2. User guide. Applied Biostatistics, Inc., Setauket, N.Y.
- 50.Ryan, A. D., and L. L. Kinkel. 1997. Inoculum density and population dynamics of suppressive and pathogenic Streptomyces strains and their relationship to potato scab biocontrol. Biol. Control 10:180-186. [Google Scholar]
- 51.Samac, D. A., and L. L. Kinkel. 2001. Suppression of the root-lesion nematode (Pratylenchus penetrans) in alfalfa (Medicago sativa) by Streptomyces spp. Plant Soil 235:35-44. [Google Scholar]
- 52.Samac, D. A., A. M. Willert, M. J. McBride, and L. L. Kinkel. 2003. Effect of antibiotic-producing Streptomyces on nodulation and leaf spot in alfalfa. Appl. Soil Ecol. 22:55-66. [Google Scholar]
- 53.SAS Institute, Inc. 1988. SAS procedures guide, release 6.03 edition. SAS Institute, Inc., Cary, N.C.
- 54.SAS Institute, Inc. 1988. SAS/STAT user's guide, release 6.03 edition. SAS Institute, Inc., Cary, N.C.
- 55.SAS Institute, Inc. 1997. SAS/STAT software: changes and enhancements through release 6.12. SAS Institute, Inc., Cary, N.C.
- 56.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schabenberger, O., and F. J. Pierce. 2002. Contemporary statistical models for the plant and soil sciences. CRC Press, Inc., Boca Raton, Fla.
- 58.Séveno, N. A., D. Kallifidas, K. Smalla, J. D. van Elsas, J.-M. Collard, A. D. Karagouni, and E. M. H. Wellington. 2002. Occurrence and reservoirs of antibiotic resistance genes in the environment. Rev. Med. Microbiol. 13:15-27. [Google Scholar]
- 59.Slattery, M., I. Rajbhandari, and K. Wesson. 2001. Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb. Ecol. 41:90-96. [DOI] [PubMed] [Google Scholar]
- 60.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 61.Söderberg, K. H., P. A. Olsson, and E. Bååth. 2002. Structure and activity of the bacterial community in the rhizosphere of different plant species and the effect of arbuscular mycorrhizal colonisation. FEMS Microbiol. Ecol. 40:223-231. [DOI] [PubMed] [Google Scholar]
- 62.Sokal, R. R., and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. Kansas Univ. Sci. Bull. 38:1409-1438. [Google Scholar]
- 63.Tanaka, Y., and S. Omura. 1990. Metabolism and products of actinomycetes: an introduction. Actinomycetologica 4:13-14. [Google Scholar]
- 64.Taylor, J. P., B. Wilson, M. S. Mills, and R. G. Burns. 2002. Comparison of microbial numbers and enzymatic activities in surface soils and subsoils using various techniques. Soil Biol. Biochem. 34:387-401. [Google Scholar]
- 65.Thrall, P. H., and J. Antonovics. 1995. Theoretical and empirical studies of metapopulations: population and genetic dynamics of the Silene-Ustilago system. Can. J. Bot. 73:S1249-S1258. [Google Scholar]
- 66.Vidaver, A. K., M. L. Mathys, M. E. Thomas, and M. L. Schuster. 1972. Bacteriocins of the phytopathogens Pseudomonas syringae, P. glycinea, and P. phaseolicola. Can. J. Microbiol. 18:705-713. [DOI] [PubMed] [Google Scholar]
- 67.Vining, L. C. 1990. Functions of secondary metabolites. Annu. Rev. Microbiol. 44:395-427. [DOI] [PubMed] [Google Scholar]
- 68.Vogel, J., P. Normand, J. Thioulouse, X. Nesme, and G. L. Grundmann. 2003. Relationship between spatial and genetic distance in Agrobacterium spp. in 1 cubic centimeter of soil. Appl. Environ. Microbiol. 69:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiener, P. 1996. Experimental studies on the ecological role of antibiotic production in bacteria. Evol. Ecol. 10:405-421. [Google Scholar]
- 70.Wiener, P. 2000. Antibiotic production in a spatially structured environment. Ecol. Lett. 3:122-130. [Google Scholar]
- 71.Wiener, P., S. Egan, and E. M. H. Wellington. 1998. Evidence for transfer of antibiotic-resistance genes in soil populations of streptomycetes. Mol. Ecol. 7:1205-1216. [DOI] [PubMed] [Google Scholar]
- 72.Williams, S. T., M. Goodfellow, and G. Alderson. 1989. Genus Streptomyces Waksman and Henrici 1943, 339AL, p. 2453-2492. In S. T. Williams, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of determinative bacteriology, vol. 4. The Williams & Wilkins Co., Baltimore, Md.
- 73.Xiao, K., L. L. Kinkel, and D. A. Samac. 2002. Biological control of Phytophthora root rots on alfalfa and soybean with Streptomyces. Biol. Control 23:285-295. [Google Scholar]