Abstract
The ABCB1 gene encodes P-glycoprotein, an ATP-dependent drug efflux pump, which is responsible for drug transport across extra- and intra-cellular membranes. The variability in the expression of ABCB1 may contribute to variable plasma efavirenz concentration which results in variability in the levels of suppression of the human immunodeficiency syndrome virus (HIV). The aim of the study was to evaluate the role of polymorphisms in ABCB1 gene on plasma efavirenz levels and treatment response in the form of change in viral load and CD-4 cell count in HIV/AIDS patients receiving efavirenz-containing highly active antiretroviral treatment regimens. Two hundred and eighty-two HIV-infected patients were recruited from Themba Lethu Clinic in Johannesburg and plasma efavirenz drug concentration levels were measured using LC-MS/MS. SNaPshot was used to genotype five known ABCB1 single nucleotide polymorphisms (SNPs). Genotype-phenotype correlations were computed. The ABCB1 4036A/G and 4036G/G genotypes were significantly associated with low plasma efavirenz concentrations (P = 0.0236), while the ABCB1 1236C/T and 1236T/T genotypes were associated with high efavirenz concentrations (P = 0.0282). A haplotype ABCB1 T-G-T-A is reported that is associated with significantly increased plasma efavirenz levels. This is the first report on 61A>G, 2677G>T/A, and 4036A>G SNPs in the South African population. ABCB1 plays a role in determining the plasma concentrations of efavirenz and should be taken into account in future design of assays for genotype-based dosing of efavirenz-containing regimens.
Keywords: ABCB1, efavirenz, HIV/AIDS, South Africa, pharmacogenetics
Introduction
Efavirenz provides the backbone to first-line highly active antiretroviral treatment (HAART) in South Africa. HAART effectively suppresses human immunodeficiency syndrome virus (HIV) replication in the majority of patients (Mocroft et al., 2003). Thus, many HIV-infected patients are now living longer compared to the pre-HAART period. However, long term antiretroviral (ARV) treatment has its own challenges such as drug–drug interactions and the development of adverse drug reactions (ADRs). Drug–drug interactions are a major problem in HIV/AIDS patients due to co-morbidities such as TB and malaria.
FDA-approved ARV drugs, including efavirenz, indinavir, nelfinavir, and ritonavir are affected by the activities of the multidrug transporter P-glycoprotein (P-gp), coded by the ABCB1 gene. ABCB1 forms part of the ATP-binding cassette gene family with about 50 members and is located on chromosome 7q21.12, spanning 209.6 kb, and containing 29 exons (Bodor et al., 2005). Genetic variation in the ABCB1 gene is known to alter mRNA stability or splicing activity (Fung and Gottesman, 2009). The three most common single nucleotide polymorphisms (SNPs) in the protein coding region of ABCB1 are 1236C>T (rs1128503), 3435C>T (rs1045642), and 2677G>T/A (rs2032582), where multiple alleles have been reported. The 1236C>T SNP occurring in exon 13, does not result in an amino acid change, but may affect ABCB1 expression through codon usage (Gu et al., 2004). The allele frequency of 1236T variant ranges from 10% among South Africans (Dandara et al., 2011) to 90% among Asians (Ambudkar et al., 1999). The 2677G>T/A SNP results in a change from serine to alanine or threonine at residue 893, but the effect of the changes on protein function is uncertain. The 3435T allele has been associated with reduced expression of P-gp, although it is synonymous (Meissner et al., 2004). Large inter-ethnic variability has been reported for the 3435C>T SNP with the ABCB1 3435C variant being the most frequent at 83, 58, 57, and 11% among Africans (Kenyans and Ghanaians), Asians (Chinese), Caucasians, and Yoruba individuals, respectively (Ameyaw et al., 2001). The ABCB1 3435T variant has been linked with good immune recovery in HIV/AIDS individuals, while the presence of the ABCB1 2677T variant has been strongly associated with virological failure (Motsinger et al., 2006). A few studies have suggested associations between ABCB1 gene polymorphisms and variability in plasma efavirenz concentrations (Fellay et al., 2002; Mukonzo et al., 2009), but all the studies lack adequate sample size. There are conflicting reports on the effects of these SNPs on efavirenz treatment response (Cascorbi et al., 2001; Fellay et al., 2002; Cascorbi, 2006). Replication studies are thus necessary to understand the contribution of ABCB1 gene variants to plasma efavirenz levels. Dandara et al. (2011) showed that genetic variants in ABCB1 are frequent in the South African population, and this study is a continuation further evaluating the clinical significance of these SNPs. Therefore, the aim of this study was to investigate the role of genetic polymorphisms in ABCB1 on plasma efavirenz levels in HIV/AIDS patients in the South African population.
Results
The mean age of the HIV/AIDS patients was 41.3 years, and more than 75% (n = 227) were female. Of the patients, 7 and 10% smoked and consumed alcohol. The clinical characteristic of the patients included viral load and CD-4 cell count (Table 1).
Table 1.
Clinical characteristics of the South African HIV/AIDS patients.
| Clinical characteristics | HIV/AIDS patients (n = 301) |
|---|---|
| Median HIV-RNA at baseline, copies/mL ± SD (range) |
26917.71 ± 27133.50 (25–98400) |
| Median HIV-RNA at 6 months post-initiation of HAART, copies/mL ± SD (range) |
1518.52 ± 9004.10 (0–75000) |
| Average CD-4 cell count at baseline, cells/μL ± SD (range) |
136.09 ± 113.24 (2–605) |
| Average CD-4 cell count at 6 months post-initiation of HAART, cells/μL ± SD (range) |
261.76 ± 137.68 (28–775) |
| ARV regimens | |
| 3TC_TDF_EFV | 9 |
| AZT_3TV_EFV | 11 |
| d4T_3TC_EFV | 222 |
| d4T_3TC_LPVr | 18 |
| d4T_3TC_NVP | 22 |
| Average plasma efavirenz concentration, μg/mL (range) |
4.64 (0.6–22) |
Comparison of allele frequencies among world populations
The genotypes for all the SNPs were observed in the HIV/AIDS patients, except the ABCB1 61G/G (rs9282564) genotype. All ABCB1 SNPs were in Hardy–Weinberg Equilibrium (HWE). The ABCB1 61A/G (rs9282564), 3435T/T (rs1045642), 4036G/G (rs3842), 1236T/T (rs1128503), 2677T/A, and 2677G/A (rs2032582) genotypes were present at frequencies of 0.006, 0.024, 0.036, 0.015, 0.004, and 0.004, respectively, among the South Africans. No individuals with an ABCB1 3435A allele was observed in the South African cohort (Table 2), and this is similar to what was reported by Dandara et al. (2011). The allele frequencies of SNPs in the South Africans were compared to the allele frequencies reported previously in other populations (Table 2), available from previous studies or the HapMap project (http://hapmap.ncbi.nlm.nih.gov/).
Table 2.
Allele frequencies in the South Africans compared to other populations.
| Population | Reference | N | 61G | 1236T | 3435T | 2677T | 2677A | 4036G |
|---|---|---|---|---|---|---|---|---|
| Black South Africans# | (Dandara et al., 2011)/This study | 979 | 0.003 | 0.090 | 0.120 | 0.040 | 0.004 | 0.202 |
| Sotho/Tswana South Africans | This study | 127 | 0.000 | 0.106 | 0.110 | 0.012 | 0.004 | 0.165 |
| Xhosa South Africans | This study | 107 | 0.014 | 0.178* | 0.210* | 0.098* | 0.005 | 0.224 |
| Zulu South Africans | This study | 139 | 0.000 | 0.119 | 0.140 | 0.022 | 0.000 | 0.209 |
| Malawi | Brown et al. (2011) | 30 | N/A | N/A | 0.210 | 0.000 | 0.000 | N/A |
| Yoruba | Hapmap | 226 | 0.000 | 0.124 | 0.111 | 0.000* | 0.000 | 0.142 |
| Luhya | Ikediobi et al. (2011) | 89 | N/A | 0.110 | N/A | N/A | N/A | N/A |
| Maasai | Ikediobi et al. (2011) | 143 | N/A | 0.140 | 0.840* | N/A | N/A | N/A |
| African-American | Hapmap | 46 | 0.000 | 0.136 | 0.071 | 0.077 | 0.000 | 0.000* |
| Caucasian | Hapmap | 226 | 0.100* | 0.451* | 0.571* | 0.340* | 0.042* | 0.142 |
| Gujarati Indian | Hapmap | 176 | 0.017 | 0.597* | 0.597* | 0.653* | 0.000 | 0.163 |
| Mexican | Hapmap | 96 | 0.052* | 0.460* | 0.460* | 0.430* | 0.000 | 0.230 |
| Toscan | Hapmap | 176 | 0.062* | 0.426* | 0.466* | 0.438* | 0.000 | 0.138 |
| Chinese | Ikediobi et al. (2011) | 45 | 0.000 | 0.680* | 0.580* | N/A | N/A | N/A |
| Japanese | Hapmap | 90 | 0.000 | 0.587* | 0.459* | 0.552* | 0.000 | 0.320* |
N/A, not available, *statistically significant difference from the frequencies among the Black South African group#.
Correlation of genetic variation with plasma efavirenz concentration
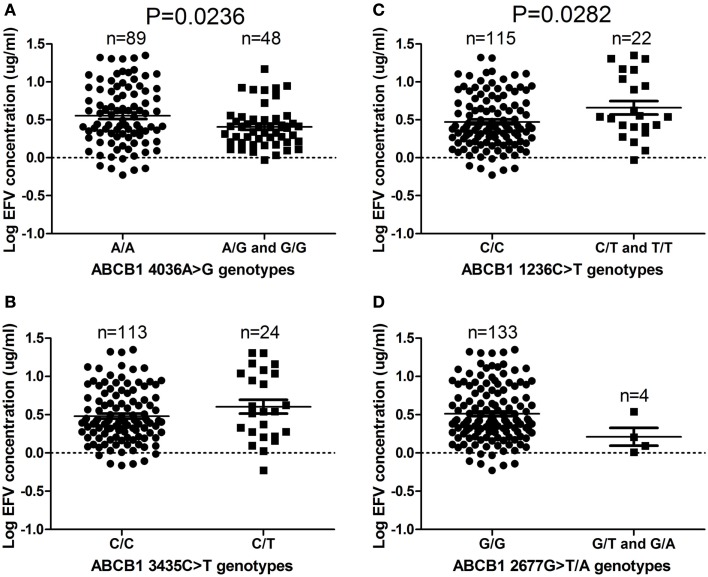
The ABCB1 4036A/G and 4036G/G genotypes were associated with significantly decreased efavirenz levels (P = 0.0236), compared to the 4036A/A genotype (Figure 1A). Fewer individuals with the ABCB1 4036G/G genotype changed treatment compared to the individuals with the 4036A/A genotype (Table 3). The ABCB1 1236C/T and 1236T/T genotypes were associated with significantly higher plasma efavirenz concentrations, compared to the 1236C/C genotype (P = 0.0282; Figure 1C). Compared to the ABCB1 1236C/C genotype, more individuals with the 1236T/T genotype changed antiretroviral regimens 1 year post treatment initiation (Table 3). No difference was observed when comparing individuals with efavirenz concentration above 4 μg/mL to those with concentrations below 4 μg/mL, with respect to change in treatment regimens (P = 0.571). No significant differences were observed in efavirenz concentrations between the ABCB1 2677G>T/A and ABCB1 3435C>T genotypes (Figures 1B,D).
Figure 1.
(A–D) ABCB1 4036A/G and G/G genotypes (P = 0.0236) are associated with reduced efavirenz levels, while 1236C/T and T/T genotypes (P = 0.0282) are associated with increased efavirenz levels in South African HIV/AIDS patients. Only significant P-values (without Bonferroni correction), using the dominant genetic model, are shown.
Table 3.
Frequency of HIV/AIDS patients changing ART regimens within 3 months, 6 months and 1 year post-initiation of treatment*.
| Genotype | Treatment initiation (n) | 3 months | P-value | 6 months | P-value | 1 year | P-value |
|---|---|---|---|---|---|---|---|
| EFV-CONTAINING ARV REGIMEN | |||||||
| 3TC_TDF_EFV | 9 | 0.11 | 0.00 | 0.11 | |||
| AZT_3TC_EFV | 11 | 0.00 | 0.182 | 0.09 | 0.993 | 0.18 | 0.898 |
| d4T_3TC_EFV | 222 | 0.03 | 0.05 | 0.14 | |||
| 1236C/C | 192 | 0.04 | 0.05 | 0.12 | |||
| 1236C/T | 44 | 0.02 | 0.399 | 0.04 | 0.636 | 0.18 | 0.089 |
| 1236T/T | 3 | 0.00 | 0.00 | 0.50 | |||
| 3435C/C | 192 | 0.04 | 0.05 | 0.13 | |||
| 3435C/T | 44 | 0.00 | 0.387 | 0.05 | 0.962 | 0.16 | 0.653 |
| 3435T/T | 3 | 0.33 | 0.00 | 0.00 | |||
| 2677G/G | 229 | 0.03 | 0.06 | 0.13 | |||
| 2677G/T | 7 | 0.00 | 0.563 | 0.11 | 0.666 | 0.00 | 0.706 |
| 2677T/A and G/A | 2 | 0.00 | 0.00 | 0.50 | |||
| 4036A/A | 153 | 0.03 | 0.05 | 0.14 | |||
| 4036A/G | 78 | 0.03 | 0.987 | 0.06 | 0.834 | 0.14 | 0.852 |
| 4036G/G | 8 | 0.00 | 0.00 | 0.11 | |||
*Only 282 patients had information on treatment regimens. ABCB1 61A>G was excluded based on being monomorphic.
Haplotype analysis
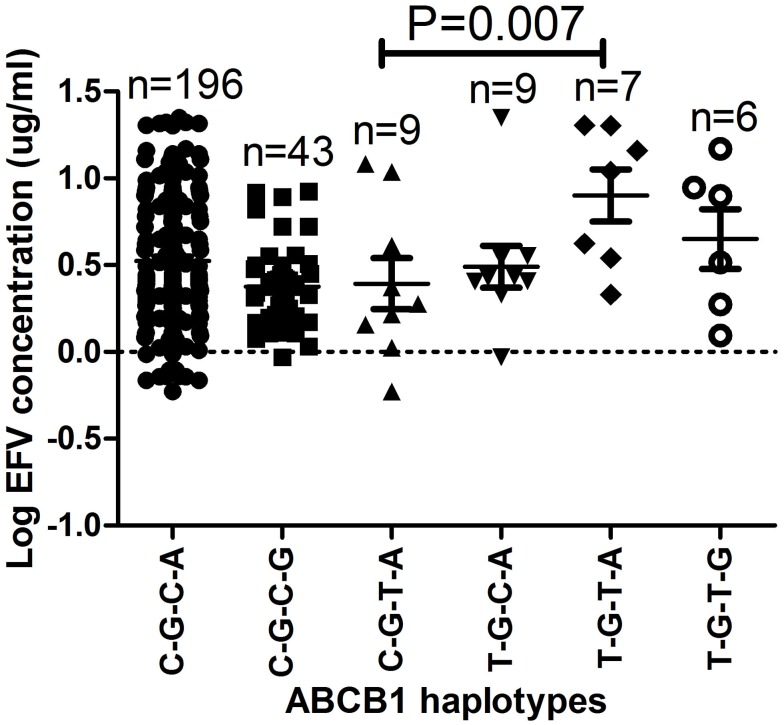
Haplotype and efavirenz plasma levels for each patient are presented in supplementary Table A1. The haplotypes with respect to 1236C>T, 2677G>T/A, 3435C>T, and 4036A>G SNPs C-G-C-A, C-G-C-G, C-G-T-G, T-G-C-A, T-G-T-A, T-G-T-G, T-T-T-A, and T-T-T-G had the following frequencies in the HIV/AIDS patients; 0.67, 0.17, 0.04, 0.03, 0.06, 0.01, 0.001, and 0.01, respectively. The ABCB1 T-G-T-A haplotype had the highest mean plasma log10 efavirenz concentrations (0.90 μg/mL) compared to 0.49 and 0.65 μg/mL among patients with the T-G-C-A or T-G-T-G haplotypes, respectively (Figure 2). The efavirenz concentrations differed significantly between individuals with the ABCB1 C-G-C-G and T-G-T-A haplotypes (P = 0.007) and were still significant after Bonferroni’s correction for multiple testing (cut-off significant P < 0.01).
Figure 2.
The effect of ABCB1 haplotypes on plasma efavirenz concentrations. Only significant P-values after Bonferroni’s correction for multiple testing at significance P < 0.01 are shown. The ABCB1 C-A-C-A, C-T-C-A, and T-T-T-G haplotypes were only observed once and thus excluded from the analysis.
Univariate and multivariate regression analysis of efavirenz concentration
Univariate regression analysis was performed to determine the effect of age, gender, tobacco smoking, alcohol use, BMI at baseline, CD-4 cell count, log10 HIV-RNA levels, ABCB1 haplotypes, ABCB1 1236C>T, 4036A>G, 3435C>T, and 2677G>T/A genotypes on plasma efavirenz concentrations (Table 4). The genotypes of the four SNPs 1236C>T, 2677G>T/A, 3435C>T, and 4036A>G significantly predicted efavirenz concentration individually and were, thus, included in the multivariate analysis together with age, gender, tobacco smoking, alcohol use and BMI at baseline. Stepwise backward regression analysis was then performed to identify the minimum set of independent variables that are predictive of plasma efavirenz levels and to determine the relative contribution of each variable to efavirenz concentration variability. Only three independent variables remained in the final model with P < 0.05, including ABCB1 1236C>T (standardized regression coefficient = 0.24; P = 0.004), ABCB1 4036A>G (standardized regression coefficient = 0.17; P = 0.009), and ABCB1 2677G>T/A (standardized regression coefficient = 0.36; P = 0.047). The adjusted coefficient of determination (R2) for the regression was 0.16, indicating that 16% of the total variance in efavirenz concentrations was explained by the model. ABCB1 1236C>T, 4036A>G, and 2677G>T/A genotypes accounted for 29, 23, and 17% (respectively) of the total variance in plasma efavirenz concentrations. When repeating the multivariate analysis among female patients only, the adjusted coefficient of determination (R2) for the regression was 0.18, indicating that 18% of the total variance in efavirenz concentrations was explained by the model compared to the 16% explained when males and females were combined. However, the majority of HIV/AIDS patients in our study were female. Further statistical analysis comparing only the genetic- and non-genetic factors in the multivariate analysis, showed that the genetic factors alone explained 11% of variance in efavirenz levels, while the non-genetic factors only explained 3% compared to the 16% explained by the combined multivariate analysis.
Table 4.
Univariate and multivariate regression analysis of efavirenz concentration.
| Independent variable |
Log10 efavirenz, % (95%CI) |
P | Contribution in model (%) |
|---|---|---|---|
| UNIVARIATE | |||
| Age | 0.01 (−0.06 to 0.08) | 0.738 | 1.36 |
| Gender | −7.34 (−21.5 to 6.81) | 0.307 | 0.12 |
| Tobacco smoking | −0.75 (−27.2 to 25.7) | 0.956 | 2.60 |
| Alcohol use | −12.4 (−34.3 to 9.45) | 0.263 | 11.9 |
| BMI at baseline | −0.70 (−2.16 to 0.76) | 0.344 | 15.2 |
| CD-4 cell count at baseline |
0.01 (−0.06 to 0.07) | 0.793 | – |
| CD-4 cell count at 6 months |
−0.04 (−0.09 to 0.02) | 0.186 | – |
| Log10 HIV-RNA at baseline |
−11.9 (−21.3 to −2.44) | 0.015 | – |
| Log10 HIV-RNA at 6 months |
10.4 (−7.26 to 28.0) | 0.245 | – |
| Genotype (dominant model) |
|||
| WT/WT | Ref | ||
| ABCB1 1236C>T | 18.6 (2.02 to 35.2) | 0.028 | 28.9 |
| ABCB1 4036A>G | −14.8 (−27.5 to −2.01) | 0.024 | 23.2 |
| ABCB1 3435C>T | 12.3 (−3.90 to 28.4) | 0.136 | 0.12 |
| ABCB1 2677G>T/A | −30.0 (−66.4 to 6.47) | 0.106 | 16.6 |
| ABCB1 haplotypes | 0.19 (−1.98 to 2.37) | 0.862 | – |
| MULTIVARIATE# | |||
| ABCB1 1236C>T | 24.2 (7.81 to 40.6) | 0.004 | – |
| ABCB1 4036A>G | −16.6 (−29.1 to −4.15) | 0.009 | – |
| ABCB1 2677G>T/A | −35.9 (−71.3 to −0.43) | 0.047 | – |
#Only significant covariates in the multivariate regression analysis are shown.
Correlation of genetic variation with clinical parameters
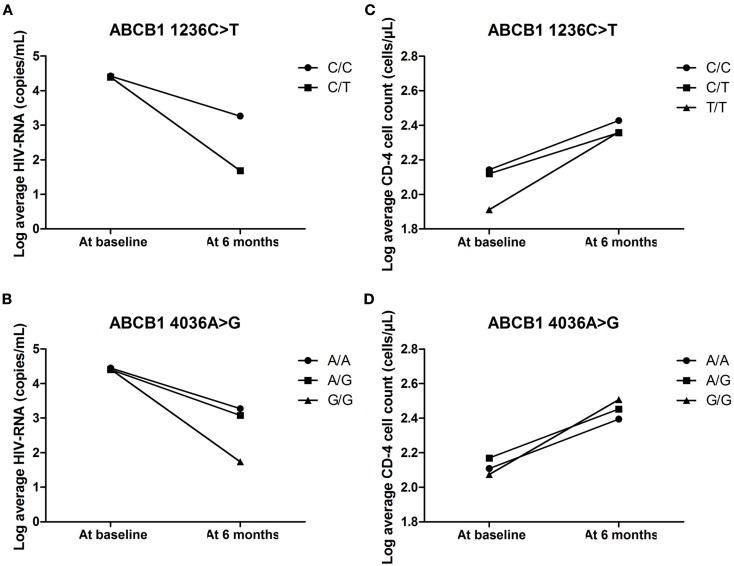
The average CD-4 cell count and viral load of individuals with the different genotypes for ABCB1 1236C>T and 4036A>G were compared at baseline and also 6 months post-initiation of ARV therapy. The ABCB1 1236C/T and 4036G/G genotypes were associated with the biggest decreases in viral load at 6 months (as shown in Figures 3A,B). None of the individuals with the ABCB1 1236T/T genotype had information on viral load at baseline or 6 months post-initiation of treatment. The ABCB1 1236T allele is associated with decreased expression of P-gp (Gow et al., 2008) and the ABCB1 4036G allele is associated with increased expression of P-gp. However, there were no major genotype associated differences in the recovery of the CD-4 cells with all genotypes showing a positive response (Figures 3C,D).
Figure 3.
(A–D) The effect of ABCB1 4036A>G and 1236C>T on average HIV-RNA and CD-4 cell count at baseline and 6 months post-initiation of treatment.
Discussion
Heterogeneity of african populations in terms of genetics
To our knowledge, the present study is the first to report on the allele and genotype frequencies of the ABCB1 61A>G, 2677G>T/A, and 4036A>G polymorphisms in the South African population. However, the other SNPs have been previously reported by Dandara et al. (2011). This data contributes to the accumulation of information on genetic variants of pharmacogenetics relevance among Africans. The allele frequencies of the genetic polymorphisms in South Africans were also compared to the frequencies among other African, African-American, Asian, and Caucasian populations (Table 2). As expected there are significant differences in the allele frequencies between African populations and Caucasians, for example, the allele frequency of the ABCB1 3435T allele in African populations ranges from 0.07 to 0.12, but is present at a frequency of almost 0.6 in Caucasian individuals. Differences in allele frequencies between the South African population compared with other African populations were also observed. The allele frequencies of the ABCB1 4036G, 2677T, and 2677A alleles were different to the frequencies reported in the Yoruba individuals (P < 0.0001). The differences in allele frequencies between the African and Caucasian individuals show that therapeutic drugs, including efavirenz, may not have similar effectiveness in different populations when given at standard dosages. Similarly, fine scale genetic structure exists within the African population which, therefore, should not be treated as one population.
Implications for disease or drug treatment and possible development of diagnostic tools
We observed lower plasma efavirenz concentrations among individuals with the ABCB1 4036A/G and 4036G/G genotypes and this could perhaps be as a result of the disruption of a miRNA binding site in the 3′UTR of ABCB1. Five poorly conserved miRNAs namely; miR-129, miR-491, miR-4795, miR-561, and miR-4717 have been predicted to target the 3′UTR region surrounding the ABCB1 4036A>G SNP using the TargetScanHuman 6.1 miRNA target prediction software. Disruption of these sites could potentially cause reduced transport of efavirenz by ABCB1 resulting in lower plasma efavirenz levels. In a different study among Ugandans, the ABCB1 4036A/G and 4036G/G genotypes were associated with higher efavirenz bioavailability (Mukonzo et al., 2009). In the current study, ABCB1 1236C/T and 1236T/T genotypes were associated with high plasma efavirenz concentrations, but there are conflicting reports on the effect of ABCB1 1236C>T genotypes in tacrolimus, cyclosporine, and sirolimus drug responses (Kuzuya et al., 2003; Anglicheau et al., 2004; Haufroid et al., 2004), making it difficult to draw conclusions. There are conflicting reports as well for the role or effects of ABCB1 2677G>T/A and 3435C>T polymorphisms (Schwab et al., 2003; Leschziner et al., 2007). Haas et al. (2005) reported an association between the ABCB1 3435T/T genotype and a decreased likelihood of virologic failure and decreased resistance to efavirenz, but not with plasma efavirenz exposure.
Clinical parameters such as CD-4 cell count, viral load, disease stage, hemoglobin, AST, and ALT levels were used as indicators of ARV treatment efficacy, underlying liver disease and disease progression in the HIV/AIDS patients. As expected, efavirenz-containing HAART led to a general (48%) increase in CD-4 cell count (cells/μL) and a 94% decrease in viral load (copies/mL) when baseline levels where compared to levels at 6 months post-initiation of treatment. Failure of reduction in viral load and emergence of opportunistic infections after 6 months led to ARV switching. Sustained viral load after 6 months and the presence of opportunistic infections are indications of possible treatment failure or non-adherence. On the other hand, other studies have shown that high plasma efavirenz concentrations are associated with development of adverse drug events leading to drug discontinuation (Marzolini et al., 2001; Lubomirov et al., 2011; Wyen et al., 2011).
Conclusion
The current study showed that the drug transporter ABCB1 contributes in predicting response to efavirenz treatment in the South African HIV/AIDS population. The ABCB1 4036A>G and 1236C>T polymorphisms were significantly correlated with low and high plasma efavirenz concentration levels, respectively. However, this data should be taken together with the variation in CYP2B6 which has a profound effect on efavirenz metabolism. The CYP2B6 516G>T SNP is known to be associated with high plasma efavirenz levels and the combined effect of CYP2B6 together with ABCB1 SNPs will be more informative in predicting response to efavirenz treatment. This strongly supports the development of a pharmacogenetic suite of gene variants to assist in deciding the HAART regimen for HIV/AIDS treatment in a clinical setting as well as the starting ARV dosage.
Materials and Methods
Research participants
All participants provided written informed consent and study approval was obtained from the University of Cape Town Health Science Faculty Research Ethics Committee, Cape Town, South Africa and the University of Witwatersrand Human Research Ethics Committee, Gauteng, South Africa. The research was performed in accordance with the guidelines of the Helsinki Declaration of 2008. Two hundred and eighty-two (n = 282) South African HIV/AIDS patients receiving efavirenz-based treatment for at least 6 months, were recruited to participate in this study. All subjects were of Bantu origin and comprised of Sotho/Tswana from Gauteng and Xhosa subjects from the Western Cape Province, South Africa. All subjects gave information on their ethnicity, age, health status (including self-reported adherence to treatment or pill counts), dietary, and smoking habits.
A 5 mL whole blood sample was obtained from each subject, and used for plasma sample collection as well as DNA extraction. DNA was isolated using a salting-out method adapted from Gustafson et al. (1987) or the GenElute™ Blood Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA). Steady-state plasma samples were available for 137 HIV/AIDS patients 12–16 h post dose with efavirenz. Plasma efavirenz concentrations were determined by LC/MS/MS (API 4000 triple quadrupole MS/MS Applied Biosystems, South Africa) according to the method by Chi et al. (2002).
Selection of SNPs and genotyping methods used
Five previously reported SNPs in ABCB1 [GenBank accession: NM_000927.4], namely 61A>G, 1236C>T, 2677G>T/A, 3435C>T, and 4036A>G were selected for investigation based on having minor allele frequencies above 10% in the African-Americans, South African, or other African populations and were genotyped using SNaPshot mini-sequencing (Table 5).
Table 5.
PCR and SNaPshot amplification conditions for ABCB1 SNPs (GenBank accession: NM_000927.4).
| SNP | Primer sequence (5′-3′) | Ta (°C) | PCR product | References |
|---|---|---|---|---|
| 4036A>G | F: CCTCAGTCAAGTTCAGAGTCTTCA | 54°C | 297 | Designed |
| R: TCACAGGCAGTTGGACAAG | ||||
| SNaPshot primer: TCTTGGCAGAAACTGCAAAAGGAGATTGAT | ||||
| 3435C>T | F: ACTCTTGTTTTCAGCTGCTTG | 54°C | 230 | Rhodes et al. (2007) |
| R: AGAGACTTACATTAGGCAGTGACTC | ||||
| SNaPshot primer: ACTCGTCCTGGTAGATCTTGAAGGG | ||||
| 2677G>T/A | F: ATGGTTGGCAACTAACACTGTTA | 54°C | 206 | Rhodes et al. (2007) |
| R: AGCAGTAGGGAGTAACAAAATAACA | ||||
| SNaPshot primer: CTTCGACCTAAGTGGAGAATGAGTTATTCTAAGGA | ||||
| 1236C>T | F: TGTGTCTGTGAATTGCCTTGAAG | 51°C | 228 | Rhodes et al. (2007) |
| R: CCTCTGCATCAGCTGGACTGT | ||||
| SNaPshot primer: TTAATTAATCAATCATATTTAGTTTGACTCACCTTCCCAG | ||||
| 61A>G | F: CTGCGGTTTCTCTTCAGGTC | 51°C | 149 | Designed |
| R: GATTCCAAAGGCTAGCTTGC | ||||
| SNaPshot primer: CTCCTTTGCTGCCCTCAC |
Each PCR reaction contained, 50 ng of genomic DNA, 1X Green GoTaq Flexi Reaction Buffer (Promega Corporation, Madison, WI, USA), 0.2 mM of each of the deoxynucleotide triphosphates (dNTPs; Bioline, London, UK); 1.5 mM MgCl2 (Promega Corporation, Madison, WI, USA); 40 pmol of the forward and reverse primers (Integrated DNA Technologies, Inc., Coralville, IA, USA); 1 U of GoTaq Flexi DNA Polymerase (Promega Corporation, Madison, WI, USA). The PCR reactions were carried out using a “MyCycler Thermal cycler” from Bio-Rad. PCR conditions were as follows: 3 min at 94°C; 40 cycles of 94°C for 30 s, the annealing temperature specific to each SNP for 30 s, 72°C for 50 s; and 10 min at 72°C for final extension.
Five microliters of each PCR product was pooled and 10 μL of the combined PCR products were cleaned using 1.5 U shrimp alkaline phosphatase (Fermentas Life Sciences, Burlington, Canada) and 2 U ExoI (Fermentas Life Sciences, Burlington, Canada) in a total reaction volume of 20 μL. The shrimp alkaline phosphatase and ExoI reaction was incubated at 37°C for 1 h and the enzymes were inactivated at 75°C for 15 min. SNaPshot single base extension of the genetic polymorphisms was performed on the “GeneAmp® PCR System 9700 version 3.08′′ (Applied Biosystems, Carlsbad, CA, USA) using the SNaPshot cycling programme as 96°C for 10 s, and then repeated for 25 cycles at 50°C for 5 s and 60°C for 30 s. The SNaPshot reaction (10 μL) contained 1 μL ABI Prism® SNaPshot™ Multiplex Kit (Applied Biosystems, CA, USA) and the pooled internal SNaPshot primers (Integrated DNA Technologies, Inc., Coralville, IA, USA). Following the SNaPshot reaction, the clean-up reaction was repeated using 1 U shrimp alkaline phosphatase using cycling conditions as mentioned before, and capillary electrophoresis was performed using a ABI 3130xl Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA). SNaPshot results were analyzed on the GeneMapper © Software version 4.1 (Applied Biosystems, Carlsbad, CA, USA).
Statistical analysis
Statistical analysis was performed using the Graphpad Prism (Version 5, GraphPad Software Inc., San Diego, CA) and STATA (Version 11, StatSoft, USA) statistical programs. ABCB1 haplotypes were inferred using Phase v2.1 (Stephens et al., 2001; Stephens and Donnelly, 2003; Stephens and Scheet, 2005). Pearson’s χ2-test and Fisher’s exact test were used to compare the allele frequencies to results previously published in populations of different ethnicity. Fisher’s exact test was also used to compare change in treatment regimen between the ABCB1 genotypes. One-way analysis of variance, followed by Bonferroni’s multiple comparison tests, was used to determine the effect of ABCB1 haplotypes on plasma log10 efavirenz levels. Genotypes were dichotomized according to the dominant genetic model (wildtype = 0 and heterozygote/homozygote variants = 1). Univariate regression analysis was applied to log10 efavirenz concentrations as dependant variable and the percentage change in efavirenz levels, with the 95% CI, was calculated as 100 × regression coefficient. Multivariate regression analysis was performed by including covariates from the univariate analysis, followed by stepwise backward removal. TargetScanHuman 6.1 miRNA target prediction software were used to predict miRNA binding to the ABCB1 3′UTR. The following equation was used to calculate the sample size required to achieve a 99% confidence-interval: N = [DEFF * Np(1−p)]/[(d2/Z21−α/2 * (N−1) + p * (1−p)]. DEFF is defined as the design effect, Z is the value for 99% confidence, d is an α-value = 0.05, while p is the frequency of the variant allele. A DEFF-value of 1 was used for random sampling, a Z-value of 1.96 and allele frequency of 0.1 was used to calculate the sample size of N = 239 samples. All statistical tests were performed two tailed, and statistical significance was defined as P < 0.05.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Patrick McPhail for facilitating sample collection from the Clinical HIV Research Unit at Themba Lethu Clinic, Helen Joseph Hospital, Johannesburg, South Africa. Special thanks to the South African Medical Research Council (MRC), the National Research Foundation of South Africa (NRF), and the University of Cape Town for funding.
Appendix
Table A1.
ABCB1 haplotype composition for each HIV/AIDS patient where steady-state plasma efavirenz levels were available.
| HIV/AIDS patients |
ABCB1 haplotype combination* |
Log10 efavirenz levels (μg/mL) |
|---|---|---|
| 1 | C-G-C-A/C-G-C-A | 0.111 |
| 2 | C-G-C-A/C-G-C-A | 0.838 |
| 3 | C-G-C-A/C-G-T-A | 0.023 |
| 4 | C-G-C-A/C-G-C-A | −0.013 |
| 5 | C-G-C-A/C-G-C-A | 0.676 |
| 6 | C-G-C-A/C-G-C-G | 0.507 |
| 7 | C-G-C-A/C-G-T-A | 0.275 |
| 8 | C-G-C-A/C-G-C-G | 0.199 |
| 9 | C-G-C-A/C-G-C-G | 0.428 |
| 10 | C-G-C-A/C-G-C-G | 0.327 |
| 11 | C-G-C-A/C-G-T-A | −0.229 |
| 12 | C-G-C-A/C-G-C-G | 0.917 |
| 13 | C-G-C-A/C-G-C-G | 0.208 |
| 14 | C-G-C-A/C-G-C-G | 0.415 |
| 15 | C-G-C-A/C-G-C-G | 0.478 |
| 16 | C-G-C-A/C-G-C-A | 0.321 |
| 17 | C-G-C-A/C-G-C-G | 0.276 |
| 18 | C-G-C-A/T-G-T-A | 0.624 |
| 19 | C-G-C-A/C-G-T-A | 1.083 |
| 20 | C-G-C-A/C-G-C-G | 0.170 |
| 21 | C-G-C-A/C-G-C-G | 0.445 |
| 22 | C-G-C-A/C-G-T-A | 0.611 |
| 23 | C-G-C-A/C-G-C-G | 0.419 |
| 24 | C-G-C-A/C-G-C-G | 0.558 |
| 25 | C-G-C-A/C-G-C-A | 0.622 |
| 26 | C-G-C-A/C-G-C-A | 0.267 |
| 27 | C-G-C-G/C-G-C-G | 0.107 |
| 28 | C-G-C-A/T-G-T-G | 1.170 |
| 29 | C-G-C-A/C-G-C-A | 0.083 |
| 30 | C-G-C-A/C-G-C-A | 0.390 |
| 31 | C-G-C-A/C-G-C-A | 0.408 |
| 32 | C-G-C-A/C-G-C-A | 0.720 |
| 33 | C-G-C-A/C-G-C-A | 0.378 |
| 34 | C-G-C-A/C-G-T-A | 0.157 |
| 35 | C-G-C-A/C-G-C-G | 0.172 |
| 36 | C-G-C-A/C-G-C-A | 0.029 |
| 37 | C-G-C-A/C-G-C-A | 0.902 |
| 38 | C-G-C-A/C-G-C-A | 0.380 |
| 39 | C-G-C-A/C-G-C-A | 1.016 |
| 40 | C-G-C-A/C-G-C-A | 0.880 |
| 41 | C-G-C-A/T-G-C-A | 0.407 |
| 42 | C-G-C-A/C-G-C-G | 0.819 |
| 43 | C-G-C-A/C-G-C-A | 0.780 |
| 44 | C-G-C-A/C-G-C-A | 1.316 |
| 45 | C-G-C-A/C-G-C-G | 0.112 |
| 46 | C-G-C-G/T-G-C-A | 0.554 |
| 47 | C-G-C-A/C-G-C-G | 0.419 |
| 48 | C-G-C-A/C-G-C-A | 0.352 |
| 49 | C-G-C-A/C-G-C-A | 1.141 |
| 50 | C-G-C-A/T-T-T-G | 0.539 |
| 51 | C-G-C-A/T-G-T-A | 1.303 |
| 52 | C-G-C-A/C-G-C-A | 0.927 |
| 53 | C-G-C-A/C-G-C-G | 0.298 |
| 54 | C-G-C-A/C-G-C-G | 0.892 |
| 55 | C-G-C-A/C-G-C-A | 0.422 |
| 56 | C-G-C-A/T-G-T-G | 0.899 |
| 57 | C-G-C-A/T-G-C-A | 0.428 |
| 58 | C-G-C-G/C-G-C-G | 0.723 |
| 59 | C-G-C-A/T-T-T-G | 0.205 |
| 60 | C-G-C-A/C-G-C-A | 0.353 |
| 61 | C-G-C-A/C-G-C-G | 0.252 |
| 62 | C-G-C-G/C-G-C-G | 0.499 |
| 63 | C-G-C-A/C-G-C-A | 0.442 |
| 64 | C-G-C-A/T-G-T-G | 0.095 |
| 65 | C-G-C-A/C-G-C-A | 0.649 |
| 66 | C-G-C-G/T-G-C-A | 0.405 |
| 67 | C-G-C-A/C-G-C-A | 0.495 |
| 68 | C-G-C-A/C-G-C-G | 0.413 |
| 69 | C-G-C-A/C-G-C-A | 0.619 |
| 70 | C-G-C-A/C-G-C-A | 0.196 |
| 71 | C-G-C-A/T-G-T-A | 1.306 |
| 72 | C-G-C-A/C-G-C-G | 0.328 |
| 73 | C-G-C-A/C-G-C-A | 0.932 |
| 74 | C-G-C-A/C-G-C-A | 0.315 |
| 75 | C-G-C-A/C-G-C-A | 1.322 |
| 76 | C-G-C-A/C-G-C-A | 0.585 |
| 77 | C-G-C-A/C-G-C-A | 0.623 |
| 78 | C-G-C-G/C-G-T-A | 0.215 |
| 79 | C-G-C-A/C-G-C-A | 1.125 |
| 80 | C-G-C-A/C-G-C-A | 0.294 |
| 81 | C-G-C-A/T-G-T-G | 0.947 |
| 82 | C-G-C-A/T-G-T-A | 1.160 |
| 83 | C-G-C-A/C-G-C-A | 0.989 |
| 84 | C-G-C-A/C-G-C-A | 0.072 |
| 85 | C-G-C-A/C-G-C-A | 0.415 |
| 86 | C-G-C-A/C-G-C-A | 0.214 |
| 87 | C-G-C-A/C-G-C-A | 0.874 |
| 88 | C-G-C-A/C-G-C-G | 0.450 |
| 89 | C-G-C-A/C-G-C-A | −0.140 |
| 90 | C-G-C-A/C-G-T-A | 1.037 |
| 91 | C-G-C-A/C-G-C-A | 0.381 |
| 92 | C-G-C-A/C-G-C-A | −0.143 |
| 93 | C-G-C-A/C-G-C-G | 0.076 |
| 94 | C-G-C-A/C-G-C-G | 0.924 |
| 95 | C-G-C-A/C-G-C-G | 0.033 |
| 96 | C-G-C-A/C-G-C-A | 0.946 |
| 97 | C-G-C-A/C-G-C-A | −0.164 |
| 98 | C-G-C-G/T-G-C-A | −0.029 |
| 99 | C-G-C-A/C-G-C-A | −0.105 |
| 100 | C-G-C-A/C-G-C-A | 0.645 |
| 101 | C-G-C-A/C-G-C-A | 0.911 |
| 102 | C-G-C-A/C-G-C-G | 0.394 |
| 103 | C-G-C-A/T-G-T-G | 0.274 |
| 104 | C-G-C-A/C-G-C-G | 0.313 |
| 105 | C-G-C-A/C-G-C-G | 0.164 |
| 106 | C-G-C-A/C-G-T-A | 0.371 |
| 107 | C-G-C-A/C-G-C-A | 0.264 |
| 108 | C-G-C-A/C-T-C-A | 0.010 |
| 109 | C-G-C-A/C-G-C-G | 0.111 |
| 110 | C-G-C-A/C-G-C-A | 1.109 |
| 111 | C-G-C-A/C-G-C-A | 0.671 |
| 112 | C-G-C-A/C-G-C-A | 0.324 |
| 113 | C-G-C-A/C-G-C-A | 0.431 |
| 114 | C-G-C-A/T-G-C-A | 0.431 |
| 115 | C-G-C-A/C-G-C-A | 0.566 |
| 116 | C-G-C-A/C-G-C-A | 0.780 |
| 117 | C-G-C-A/T-G-T-G | 0.516 |
| 118 | C-G-C-A/C-G-C-G | 0.276 |
| 119 | T-G-C-A/T-G-T-A | 0.541 |
| 120 | C-G-C-A/C-G-C-A | 0.821 |
| 121 | C-G-C-A/T-G-T-A | 1.038 |
| 122 | C-G-C-A/C-G-C-A | 0.364 |
| 123 | C-G-C-A/C-G-C-A | 1.106 |
| 124 | C-G-C-A/C-G-C-A | 0.584 |
| 125 | C-G-C-A/C-G-C-A | 1.093 |
| 126 | C-G-C-A/C-G-C-A | 0.692 |
| 127 | C-G-C-A/C-G-C-G | 0.340 |
| 128 | C-G-C-A/C-G-C-A | 0.751 |
| 129 | C-G-C-A/C-G-C-A | 0.192 |
| 130 | C-A-C-A/C-G-C-A | 0.092 |
| 131 | C-G-C-A/C-G-C-A | 0.192 |
| 132 | C-G-C-A/C-G-C-A | 0.106 |
| 133 | C-G-C-A/C-G-C-A | 0.941 |
| 134 | C-G-C-A/C-G-C-A | 0.204 |
| 135 | C-G-C-A/T-G-C-A | 1.348 |
| 136 | T-G-C-A/T-G-T-A | 0.329 |
| 137 | C-G-C-A/C-G-C-G | 0.271 |
*ABCB1 haplotypes with respect to 1236C>T-2677G>T/A-3435C>T-4036A>G, 61A>G was excluded from the haplotype based on being monomorphic.
Authors’ Contributions
Marelize Swart carried out all of the molecular genetic studies and drafted the manuscript. Yuan Ren and Peter Smith both carried out the LC/MS/MS analysis of plasma efavirenz concentration. Collet Dandara conceived of the study, designed, coordinated the study, collected all the samples, assisted with statistical data analysis, helped to draft the manuscript and approved the final version. All authors read and approved the final manuscript.
References
- Ambudkar S. V., Dey S., Hrycyna C. A., Ramachandra M., Pastan I., Gottesman M. M. (1999). Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 39, 361–398 10.1146/annurev.pharmtox.39.1.361 [DOI] [PubMed] [Google Scholar]
- Ameyaw M. M., Regateiro F., Li T., Liu X., Tariq M., Mobarek A., et al. (2001). MDR1 pharmacogenetics: frequency of the C3435T mutation in exon 26 is significantly influenced by ethnicity. Pharmacogenetics 11, 217–221 10.1097/00008571-200104000-00005 [DOI] [PubMed] [Google Scholar]
- Anglicheau D., Thervet E., Etienne I., Hurault De Ligny B., Le Meur Y., Touchard G., et al. (2004). CYP3A5 and MDR1 genetic polymorphisms and cyclosporine pharmacokinetics after renal transplantation. Clin. Pharmacol. Ther. 75, 422–433 10.1016/j.clpt.2004.01.009 [DOI] [PubMed] [Google Scholar]
- Bodor M., Kelly E. J., Ho R. J. (2005). Characterization of the human MDR1 gene. AAPS J. 7, E1–E5 10.1208/aapsj070479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. C., Hosseinipour M. C., Hoskins J. M., Thirumaran R. K., Tien H. C., Weigel R., et al. (2011). Exploration of CYP450 and drug transporter genotypes and correlations with nevirapine exposure in Malawians. Pharmacogenomics 13, 113–121 10.2217/pgs.11.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascorbi I. (2006). Role of pharmacogenetics of ATP-binding cassette transporters in the pharmacokinetics of drugs. Pharmacol. Ther. 112, 457–473 10.1016/j.pharmthera.2006.04.009 [DOI] [PubMed] [Google Scholar]
- Cascorbi I., Gerloff T., Johne A., Meisel C., Hoffmeyer S., Schwab M., et al. (2001). Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin. Pharmacol. Ther. 69, 169–174 10.1067/mcp.2001.114164 [DOI] [PubMed] [Google Scholar]
- Chi J., Jayewardene A. L., Stone J. A., Motoya T., Aweeka F. T. (2002). Simultaneous determination of five HIV protease inhibitors nelfinavir, indinavir, ritonavir, saquinavir, and amprenavir in human plasma by LC/MS/MS. J. Pharm. Biomed. Anal. 30, 675–684 10.1016/S0731-7085(02)00357-6 [DOI] [PubMed] [Google Scholar]
- Dandara C., Lombard Z., Du Plooy I., Mclellan T., Norris S. A., Ramsay M. (2011). Genetic variants in CYP (−1A2, −2C9, −2C19, −3A4, and −3A5), VKORC1 and ABCB1 genes in a black South African population: a window into diversity. Pharmacogenomics 12, 1663–1670 10.2217/pgs.11.106 [DOI] [PubMed] [Google Scholar]
- Fellay J., Marzolini C., Meaden E. R., Back D. J., Buclin T., Chave J. P., et al. (2002). Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet 359, 30–36 10.1016/S0140-6736(02)07276-8 [DOI] [PubMed] [Google Scholar]
- Fung K. L., Gottesman M. M. (2009). A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim. Biophys. Acta 1794, 860–871 10.1016/j.bbapap.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow J. M., Hodges L. M., Chinn L. W., Kroetz D. L. (2008). Substrate-dependent effects of human ABCB1 coding polymorphisms. J. Pharmacol. Exp. Ther. 325, 435–442 10.1124/jpet.107.135194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Zhou T., Ma J., Sun X., Lu Z. (2004). The relationship between synonymous codon usage and protein structure in Escherichia coli and Homo sapiens. BioSystems 73, 89–97 10.1016/j.biosystems.2003.10.001 [DOI] [PubMed] [Google Scholar]
- Gustafson S., Proper J. A., Bowie E. J., Sommer S. S. (1987). Parameters affecting the yield of DNA from human blood. Anal. Biochem. 165, 294–299 10.1016/0003-2697(87)90272-7 [DOI] [PubMed] [Google Scholar]
- Haas D. W., Smeaton L. M., Shafer R. W., Robbins G. K., Morse G. D., Labbe L., et al. (2005). Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an adult aids clinical trials group study. J. Infect. Dis. 192, 1931–1942 10.1086/497610 [DOI] [PubMed] [Google Scholar]
- Haufroid V., Mourad M., Van Kerckhove V., Wawrzyniak J., De Meyer M., Eddour D. C., et al. (2004). The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics 14, 147–154 10.1097/00008571-200403000-00002 [DOI] [PubMed] [Google Scholar]
- Ikediobi O., Aouizerat B., Xiao Y., Gandhi M., Gebhardt S., Warnich L. (2011). Analysis of pharmacogenetic traits in two distinct South African populations. Hum. Genomics 5, 265–282 10.1186/1479-7364-5-4-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuya T., Kobayashi T., Moriyama N., Nagasaka T., Yokoyama I., Uchida K., et al. (2003). Amlodipine, but not MDR1 polymorphisms, alters the pharmacokinetics of cyclosporine A in Japanese kidney transplant recipients. Transplantation 76, 865–868 10.1097/01.TP.0000084873.20157.67 [DOI] [PubMed] [Google Scholar]
- Leschziner G. D., Andrew T., Pirmohamed M., Johnson M. R. (2007). ABCB1 genotype and PGP expression, function and therapeutic drug response: a critical review and recommendations for future research. Pharmacogenomics J. 7, 154–179 10.1038/sj.tpj.6500413 [DOI] [PubMed] [Google Scholar]
- Lubomirov R., Colombo S., Di Iulio J., Ledergerber B., Martinez R., Cavassini M., et al. (2011). Association of pharmacogenetic markers with premature discontinuation of first-line anti-HIV therapy: an observational cohort study. J. Infect. Dis. 203, 246–257 10.1093/infdis/jiq043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzolini C., Telenti A., Decosterd L. A., Greub G., Biollaz J., Buclin T. (2001). Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV-1-infected patients. AIDS 15, 71–75 10.1097/00002030-200106150-00023 [DOI] [PubMed] [Google Scholar]
- Meissner K., Jedlitschky G., Meyer Zu Schwabedissen H., Dazert P., Eckel L., Vogelgesang S., et al. (2004). Modulation of multidrug resistance P-glycoprotein 1 (ABCB1) expression in human heart by hereditary polymorphisms. Pharmacogenetics 14, 381–385 10.1097/00008571-200406000-00007 [DOI] [PubMed] [Google Scholar]
- Mocroft A., Ledergerber B., Katlama C., Kirk O., Reiss P., d’Arminio Monforte A., et al. (2003). Decline in the AIDS and death rates in the EuroSIDA study: an observational study. Lancet 362, 22–29 10.1016/S0140-6736(03)15062-3 [DOI] [PubMed] [Google Scholar]
- Motsinger A. A., Ritchie M. D., Shafer R. W., Robbins G. K., Morse G. D., Labbe L., et al. (2006). Multilocus genetic interactions and response to efavirenz-containing regimens: an adult AIDS clinical trials group study. Pharmacogenet. Genomics 16, 837–845 10.1097/01.fpc.0000230413.97596.fa [DOI] [PubMed] [Google Scholar]
- Mukonzo J. K., Roshammar D., Waako P., Andersson M., Fukasawa T., Milani L., et al. (2009). A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single-dose efavirenz population pharmacokinetics in Ugandans. Br. J. Clin. Pharmacol. 68, 690–699 10.1111/j.1365-2125.2009.03516.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes K. E., Zhang W., Yang D., Press O. A., Gordon M., Vallbohmer D., et al. (2007). ABCB1, SLCO1B1 and UGT1A1 gene polymorphisms are associated with toxicity in metastatic colorectal cancer patients treated with first-line irinotecan. Drug Metab. Lett. 1, 23–30 10.2174/187231207779814328 [DOI] [PubMed] [Google Scholar]
- Schwab M., Eichelbaum M., Fromm M. F. (2003). Genetic polymorphisms of the human MDR1 drug transporter. Annu. Rev. Pharmacol. Toxicol. 43, 285–307 10.1146/annurev.pharmtox.43.100901.140233 [DOI] [PubMed] [Google Scholar]
- Stephens M., Donnelly P. (2003). A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 10.1086/379378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Scheet P. (2005). Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 76, 449–462 10.1086/428594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Smith N. J., Donnelly P. (2001). A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 10.1086/319501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyen C., Hendra H., Siccardi M., Platten M., Jaeger H., Harrer T., et al. (2011). Cytochrome P450 2B6 (CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. J. Antimicrob. Chemother. 66, 2092–2098 10.1093/jac/dkr272 [DOI] [PubMed] [Google Scholar]