Abstract
The cluster of microcystin synthetase genes from Anabaena strain 90 was sequenced and characterized. The total size of the region is 55.4 kb, and the genes are organized in three putative operons. The first operon (mcyA-mcyB-mcyC) is transcribed in the opposite direction from the second operon (mcyG-mcyD-mcyJ-mcyE-mcyF-mcyI) and the third operon (mcyH). The genes mcyA, mcyB, and mcyC encode nonribosomal peptide synthetases (NRPS), while mcyD codes for a polyketide synthase (PKS), and mcyG and mcyE are mixed NRPS-PKS genes. The genes mcyJ, mcyF, and mcyI are similar to genes coding for a methyltransferase, an aspartate racemase, and a d-3-phosphoglycerate dehydrogenase, respectively. The region in the first module of mcyB coding for the adenylation domain was found to be 96% identical with the corresponding part of mcyC, suggesting a recent duplication of this fragment and a replacement in mcyB. In Anabaena strain 90, the order of the domains encoded by the genes in the two sets (from mcyG to mcyI and from mcyA to mcyC) is colinear with the hypothetical order of the enzymatic reactions for microcystin biosynthesis. The order of the microcystin synthetase genes in Anabaena strain 90 differs from the arrangement found in two other cyanobacterial species, Microcystis aeruginosa and Planktothrix agardhii. The average sequence match between the microcystin synthetase genes of Anabaena strain 90 and the corresponding genes of the other species is 74%. The identity of the individual proteins varies from 67 to 81%. The genes of microcystin biosynthesis from three major producers of this toxin are now known. This makes it possible to design probes and primers to identify the toxin producers in the environment.
Cyanobacteria produce a wide variety of bioactive compounds. Many of these are potent toxins, which cause health problems for animals and humans when producer organisms occur in masses in lakes and water reservoirs (26). Cyanobacteria are also found to be a rich source of pharmaceutical compounds (17). Most well known of the cyanobacterial toxins are the hepatotoxic heptapeptides known as microcystins. The general structure of microcystins is cyclo(-d-Ala-X-d-MeAsp-Z-Adda-d-Glu-Mdha-), where X and Z are variable l-amino acids, d-MeAsp is d-erythro-β-methylaspartic acid, Mdha is N-methyldehydroalanine, and Adda is 3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid. More than 65 structurally different microcystins are known (26). The most common variant has l-leucine and l-arginine in the positions of X and Z, respectively. Demethylated microcystins are also frequently found. Toxicity of microcystins is caused by the inhibition of protein phosphatases 1 and 2A (13). The level of inhibition varies depending on the structure, but the Adda and d-Glu moieties, which are almost invariable in microcystins, are essential for the inhibition (7) and hence for the toxicity. This activity makes the microcystins powerful tools for cell biological investigations.
Microcystins have been found principally in cyanobacteria of three planktonic, bloom-forming genera, Anabaena, Microcystis, and Planktothrix (26). All members of these genera do not make microcystins, and both toxic and nontoxic strains occur in the same species. Toxic and nontoxic strains of Anabaena, Microcystis, or Planktothrix cannot be separated based on the classical morphological taxonomy or ribosomal gene sequences (12). On the other hand, one strain may produce different microcystins and also other peptides simultaneously (5, 6, 25). Thus, the sequences of the microcystin synthetase genes are important for the recognition of toxic cyanobacteria by molecular methods.
Recently, the gene clusters encoding microcystin synthetase were sequenced and characterized from the unicellular Microcystis aeruginosa (18, 29) and from the filamentous Planktothrix agardhii (2). It was demonstrated that the biosynthesis requires a combination of polyketide and nonribosomal peptide synthesis (18, 29).
The bioactive peptides produced by Anabaena strain 90 have been characterized: three microcystins (MCYST-LR, MCYST-RR, and d-Asp-MCYST-LR) (25), two seven-residue depsipeptides (anabaenopeptilide 90A and 90B), and three six-residue peptides having a ureido linkage (anabaenopeptins A, B, and C) (6). Previously we have described the biosynthetic genes of the depsipeptides, anabenopeptilides 90A and 90 B, and constructed a mutant that lacks these anbaenopeptilides but still makes other peptides (21). Here we report an analysis of the complete microcystin synthetase gene cluster from Anabaena strain 90, show a comparison with the corresponding genes from other cyanobacteria, present a genetic basis for the production of variable microcystins, and propose a model for microcystin biosynthesis in Anabaena strain 90.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The cyanobacterial strain Anabaena strain 90 was isolated from Lake Vesijärvi, Finland, and purified axenic (20, 25). It was shown to produce three microcystins (MCYST-LR, MCYST-RR, and d-Asp-MCYST-LR (25). Anabaena strain 90 was grown in Z8 medium (11) without nitrate at ∼22°C with continuous illumination of 20 to 25 μmol m−2 s−1. Escherichia coli strain DH5α, which was used as a host for DNA cloning and sequencing, was cultured in Luria broth at 37°C.
DNA manipulations, screening, mapping, and sequencing of cosmids.
Extraction of cyanobacterial DNA and the preparation of genomic library has been described earlier (21). The genomic library was screened by colony hybridization (22). The probe labeled with [32P]dCTP was a 2.5-kb fragment from mcyA of M. aeruginosa provided by Elke Dittmann (Humboldt University, Berlin, Germany). A total of about 6,000 colonies were tested. The insert DNA of 29 positive cosmid clones was mapped with HindIII, EcoRI, and SpeI. The ends of 18 inserts were sequenced with SP6 and T7 primers, and the cosmid clones for sequencing the microcystin synthetase genes were selected. DNA of the cosmid clones was digested with restriction enzymes BstEII, HindIII, EcoRI, ScaI, SpeI, or XbaI and ligated to pBluescript SK(+). Nested deletions and other DNA manipulations were performed according to the method of Sambrook et al. (22). Sequencing was carried out mainly by the University of Chicago Cancer Research Center DNA Sequencing Facility. Gaps were filled, and the verifications were done by amplifying chromosomal DNA in PCR with DyNAzyme EXT Polymerase (Finnzymes), the sequencing reactions were done with the BigDye Terminator Cycle Sequencing kit (Applied Biosystems) and analyzed on the ABI 310 Genetic Analyzer. The standard T3 and T7 primers and oligonucleotides derived from already determined sequences were employed.
Sequence analysis.
Analysis and comparisons of sequences were performed with the Sequence analysis software package (version 8.0; University of Wisconsin Genetics Computer Group) and with EMBOSS (European Molecular Biology Open Software Suite). The CAP program (http://bioweb.pasteur.fr/seqanal/interfaces/cap.html) was used for sequence assembly. Sequence similarity searches in databases were done with Blast through the Web site of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). Searches for conserved domains and motifs were accomplished with the CD-Search program (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and with the Motif Scan program (http://hits.isb-sib.ch/cgi-bin/PFSCAN?). Clustal W was applied for multiple sequence alignments (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html).
Nucleotide sequence accession number.
The sequences reported in this paper have been deposited in the EMBL data bank under the accession number AJ536156.
RESULTS
Organization of the microcystin synthetase genes.
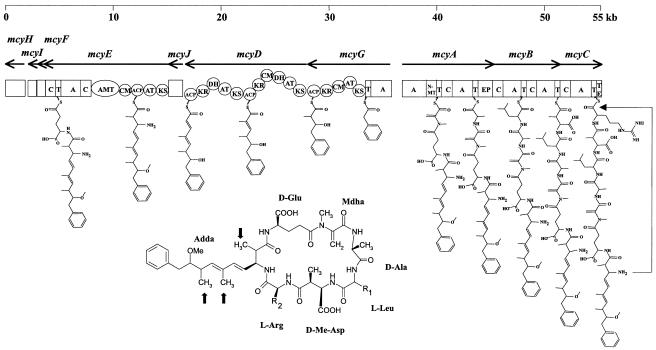
Microcystin synthetase genes in Anabaena strain 90 (mcyABCGDJEFIH) are organized in three putative operons (Fig. 1) with a total size of 55.4 kb. The first operon (mcyA-mcyB-mcyC) is transcribed in the opposite direction from the second (mcyG-mcyD-mcyJ-mcyE-mcyF-mcyI) and the third operon (mcyH). Putative promoter regions were identified in front of mcyA, mcyG, and mcyH. Transcriptional starts prior to mcyD (93 bp from mcyG), mcyE (37 or 95 bp from mcyJ), mcyF (42 bp from mcyE), and before mcyI (51 bp from mcyF) cannot be ruled out, although no transcription stop loops were identified following the preceding genes, and no Pribnow box could be identified in front of mcyD.
FIG. 1.
Microcystin synthetase gene cluster of Anabaena strain 90, a linear model for the assembly and the structure of microcystin-LR. Abbreviations for domains: A, adenylation; C, condensation; T, thiolation; NMT, N-methyltransferase; EP, epimerase; TE, thioesterase; KR, β-ketoacyl reductase. Adda is 3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid, Mdha is N-methyldehydroalanine, and d-Me-Asp is d-erythro-β-methylaspartic acid. The thick arrows point to three methyl groups, which are putatively introduced by the CM domains. The way of cyclization of the microcystin precursor is shown with an arrow on the right of the picture.
Characterization of the peptide synthetase genes.
In the first operon there are three open reading frames (ORFs) named mcyA, mcyB, and mcyC. The translations of mcyA, mcyB and mcyC start with ATG codons, and potential ribosome binding sites were recognized preceding the ORFs. mcyB is separated by 17 bp from the previous stop codon and overlaps mcyC with 1 bp. The lengths of mcyA, mcyB, and mcyC are 8364, 6399, and 3852 bp, and they encode polypeptides with predicted masses of 315,663, 243,072, and 146,877 Da, respectively. Sequence analysis of mcyA, mcyB, and mcyC revealed a typical modular structure for nonribosomal peptide synthetase (NRPS) genes (14) (Fig. 1). McyA contains two adenylation and thiolation domains, a condensation, an N-methyltransferase, and an epimerization domain. In McyB there are two modules, each including one condensation, adenylation, and thiolation domain. McyC is composed of one module, containing a condensation, an adenylation, a thiolation, and a thioesterase domain (Fig. 1).
Identification of the PKS genes.
The second operon contains six ORFs, mcyG-mcyD-mcyJ-mcyE-mcyF-mcyI. There are two possible translation start codons (ATG) for mcyG separated by 75 bp, giving ORFs which are 7,827 and 7,905 bp long. They can code for proteins of 2,609 and 2,635 amino acids (aa) with predicted masses of 289,859 and 292,851 Da. The next ORF, mcyD, is separated from mcyG by 96 bp. The size of this large gene is 11,607 bp, and it encodes a polypeptide of 3,869 aa with the predicted mass of 430,216 Da. Two alternative starts (ATG codons) for mcyE were identified 57 bp apart from each other. These versions of mcyE (10,446 bp and 10,386 bp) can code for polypeptides of 3,482 and 3,462 aa with the masses of 388,755 and 386,501 Da, respectively. mcyD was identified as a PKS gene, whereas mcyG and mcyE have a combined NRPS-PKS gene structure (Fig. 1).
Auxiliary genes.
The ORF mcyJ is suggested to initiate with a GTG codon 59 bp downstream of the stop codon (TAA) of mcyD and 5 bp from a putative Shine-Dalgarno sequence AGGAGAG. There is no ATG codon located nearby. Accordingly, mcyJ is predicted to be 930 bp long. The small, 756-bp ORF mcyF follows 42 bp after the stop codon (TAG) of mcyE. The distance between mcyF and the next 1,011-bp ORF mcyI is 54 bp. An alleged ribosome binding site and the designated start codon (ATG) were found upstream from both mcyF and mcyI. Downstream (295 bp) from the stop codon (TAA) of mcyI the ORF mcyH (1,776 bp) was found. mcyJ, mcyF, mcyI, and mcyH encode polypeptides of 310, 252, 337, and 592 aa with the predicted masses of 35,812, 28,426, 36,750, and 67,731 Da, respectively. McyF is similar to aspartate racemases, McyJ belongs to the family of methyltransferases, and McyI is related to d-3-phosphoglycerate dehydrogenases. McyH contains a membrane-spanning and an ATP-binding domain of ABC transporters. A Blast search of McyH found 75% identity (in 589 aa) to NosG from Nostoc sp. strain GSV224 (AF204805) and 39% identity (in 543 aa) to the hypothetical ABC transporter ATP-binding protein SLL0182 of Synechocystis sp. strain PCC6803 (Q55774).
Comparison of microcystin synthetase genes.
The microcystin synthetase genes were previously sequenced from M. aeruginosa strains PCC7806 (mcyA-mcyJ) (29), K-139 (mcyA-mcyI) (18), and UV027 (mcyA-mcyC; Raps et al. unpublished data [GenBank accession no. AF458094]), and from P. agardhii CYA126 (2). When Anabaena strain 90 sequences were compared to M. aeruginosa sequences, they revealed identities of 65 to 75% (mcyJ, 80%) at the amino acid level and identities of 69 to 75% (mcyJ, 79%) at the nucleotide level (Table 1).
TABLE 1.
Comparison of microcystin synthetase genes and polypeptides from Anabaena strain 90 with those sequenced from other cyanobacteriaa
| Strain | % Identity of DNA/% identity of amino acids (mol% G + C)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | |
| M. aeruginosa PCC7806 | 69/68 (41) | 72/69 (39) | 74/73 (37) | 72/69 (40) | 75/74 (39) | 71/65 (38) | 74/71 (38) | 74/70 (35) | 74/71 (40) | 79/80 (39) |
| M. aeruginosa K-139 | 69/68 (41) | 71/69 (39) | 74/73 (37) | 72/69 (40) | 75/75 (39) | 71/65 (37) | 74/71 (38) | 74/70 (36) | 74/72 (39) | |
| M. aeruginosa UV027 | 69/68 (41) | 73/71 (39) | 74/73 (37) | |||||||
| P. agardhii CYA126/8 | 67/66 (45) | 72/70 (39) | 80/79 (35) | 77/73 (38) | 78/77 (38) | 77/74 (38) | 78/75 (35) | 81/82 (37) | ||
| Anabaena strain 90 (mol% G+C) | 41 | 38 | 37 | 40 | 38 | 34 | 39 | 36 | 38 | 39 |
Substrate specificity of the adenylation domains.
The substrate specificity-conferring amino acids in the adenylation domains of the microcystin synthetases of Anabaena strain 90, P. agardhii CYA126, M. aeruginosa PCC7806, K-139, and UV027 were determined according to Stachelhaus et al. (27) (Table 2). The specificity codes of both modules of McyA, the second module of McyB and the NRPS modules in McyG and McyE are identical or nearly identical in all the sequenced microcystin synthetases (Table 2). There are, however, more differences in the specificity codes of the first module of McyB (McyB-1) and the single module of McyC, which activate variable amino acids. The substrate specificity regions of the adenylation domains (corresponding to aa 235 to 331 of GrsA [27]) in McyA, McyB, and in McyC from Anabaena strain 90, from P. agardhii, and from M. aeruginosa were compared by using the algorithm of Smith and Waterman in the EMBOSS program package. The specificity regions of McyA, McyC, and the second module of McyB (McyB-2) are highly conserved.
TABLE 2.
Specificity-conferring amino acids (signature sequences) of adenylation domains in the microcystin synthetases from different cyanobacterial strains
| Module | Strain | Signature sequencea | Precedent SS | Activated amino acid | Reference templateb |
|---|---|---|---|---|---|
| McyA | Anabaena sp. strain 90 | DVWHISLID | DVWHLSLID | Ser | SyrE (1) |
| M. aeruginosa 7806 | DVWHFSLID | DVWHFSLVD | Ser | EntF, MycC (1, 2) | |
| M. aeruginosa K-139 | DVWHFSLID | Ser | |||
| M. aeruginosa UV027 | DVWHFSLID | Ser | |||
| P. agardhii CYA 126/8 | DVWHISLID | Ser | |||
| McyA 2 | Anabaena sp. strain 90 | DLFNNALTY | Ala | ||
| M. aeruginosa 7806 | DLFNNALTY | Ala | |||
| M. aeruginosa K-139 | DLFNNALTY | DLFNNALTY | Ala | BlmIX, MxA (3, 4) | |
| M. aeruginosa UV027 | DLFNNALTY | Ala | |||
| P. agardhii CYA 126/8 | DLFNNALSY | Ala | |||
| McyB 1 | Anabaena sp. strain 90 | DVWFFGLVD | (Leu/Arg) | ||
| M. aeruginosa 7806 | DAWFLGNVV | DAWFLGNVV | Leu | BacA, LicA, LicB, SrFA (1) | |
| M. aeruginosa K-139 | DAWFLGNVV | Leu | |||
| M. aeruginosa UV027 | DVWTIGAVE | (Arg) | |||
| P. agardhii CYA 126/8 | DALFFGLVD | (Leu/Arg) | |||
| McyB 2 | Anabaena sp. strain 90 | DARHVGIFV | (Asp/MeAsp) | ||
| M. aeruginosa 7806 | DARHVGIFV | (Asp/MeAsp) | |||
| M. aeruginosa K-139 | DARHVGIFV | None | (Asp/MeAsp) | ||
| M. aeruginosa UV027 | DARHVGIFV | (Asp/MeAsp) | |||
| P. agardhii CYA 126/8 | DPRHVGIFI | (Asp/MeAsp) | |||
| McyC | Anabaena sp. strain 90 | DVWCFGLVD | (Arg) | ||
| M. aeruginosa 7806 | DVWTIGAVD | (Arg) | |||
| M. aeruginosa K-139 | DVWTIGAVE | None | (Arg) | ||
| M. aeruginosa UV027 | DVWTIGAVD | (Arg) | |||
| P. agardhii CYA 126/8 | DPWGFGLVD | (Arg) | |||
| McyG | Anabaena sp. strain 90 | GAFWVAASG | |||
| M. aeruginosa 7806 | GAFWVAASG | None | |||
| M. aeruginosa K-139 | GAFWVAASG | ||||
| P. agardhii CYA 126/8 | GAFWVAASG | ||||
| McyE | Anabaena sp. strain 90 | DPRHSGVVG | (Glu) | ||
| M. aeruginosa 7806 | DPRHSGVVG | None | (Glu) | ||
| M. aeruginosa K-139 | DPRHSGVVG | (Glu) | |||
| P. agardhii CYA 126/8 | DPRHSGVVG | (Glu) |
Nine variable amino acids of the signature sequences determined as described by Stachelhaus et al. (27). Letters in boldface type indicate the residues identical with the amino acids of the signature sequence from Anabaena sp. strain 90.
High similarity of mcyC and the first module of mcyB.
The alignment of mcyC and the first module of mcyB revealed high similarity, 96%, in the 1,617-bp fragments (nucleotides 1404 to 3020 in mcyC and 1416 to 3032 in mcyB). The sequences differ only in 67 positions. This region codes for the entire adenylation domain. Comparable but shorter sections were found in mcyB (nucleotides 1861 to 2706) and mcyC (nucleotides 1846 to 2688) of M. aeruginosa UV027 (AF458094), the identity between the sequences is 94.6%. This part encodes the adenylation domain core motifs from A3 to A8 (14) and thus the major part of the putative substrate binding pocket (27).
Activities encoded by mcyG, mcyD, and mcyE of Anabaena strain 90.
Motif Scan at Prosite (database of protein families and domains) and at Pfam (Protein families) database (http://hits.isb-sib.ch/cgi-bin/PFSCAN) and Conserved Domain (CD) search at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) were used to discover the putative functions of McyG, McyD, and McyE. In the N-terminal part of McyG an NRPS module was identified which contains an adenylation domain and a thiolation (phosphopantetheine carrier) domain. Next to this, toward the C terminus there are four PKS domains: β-ketoacyl synthase (KS), acyl transferase (AT), ketoreductase (KR), and acyl carrier protein (ACP), in this order. Between AT and KR domains there is a C-methyltransferase (CM) domain (Fig. 1 and 2). McyD contains two modules of type I PKSs. The first module consists of KS, AT, dehydratase (DH), methyltranferase (CM) (Fig. 1 and 2), KR, and ACP domains, and the second module has KS, AT, DH, KR, and ACP domains, in the presented orders. McyE is another mixed PKS-NRPS, including KS, AT, and ACP domains and a methyltranferase (CM) domain (Fig. 1 and 2). These are followed by a unique aminotransferase (AMT) domain (Fig. 1) found also in other microcystin synthetases (2, 18, 29), which is 52% identical and 68% similar to the AMT domains of mycosubtilin (4) and iturin synthetase (30). At the N-terminal region there is an NRPS module comprising two condensation domains, an adenylation domain, and a thiolation domain (Fig. 1).
FIG. 2.
Comparison of the putative CM domains in McyG, McyD, and McyE of Anabaena strain 90 with three bacterial CM domains in the region of the conserved motifs: 1, (VIL)(LV)(DE)(VI)G(GC)G(TP)G; 2, (PG)(QT)(FYA)DA(IVY)(FI)(CVL); and 3, LL(RK)PGG(RIL)(LI)(LFIV)(IL) (9). EpoE is the PKS in epothilone biosynthesis of Sorangium cellulosum (AF217189); HMWP1 is the high-molecular-weight-protein in yersiniabactin biosynthesis encoded by irp1 of Yersinia enterocolitica (Y12527); and ECUbiE is E. coli CM, UbiE (P27851). Residues in boldface type (in the boxed areas) are identical to the consensus amino acids of the motifs. Amino acids, which are identical in at least five of the six sequences, are marked with the asterisks. Strongly similar amino acids are pointed out by two dots.
KR and DH domains.
The activity of the KR domains of McyG (one) and McyD (two) can be predicted from the microcystin synthetase structure, and these domains carry the NAD cofactor binding motifs, GXGXX(G/A)(X)3(G/A)(X)6G, common to oxidoreductases (23). Two DH domains of McyD contain the following active site motifs: H(X)3D(X)4P and H(X)3G(X)4P. The latter motif is identical to the consensus sequence of DH domains (1). The motif H(X)3D(X)4P, where Gly is replaced by Asp, is also found in the active DH domain of module 10 in rifamycin synthase (28).
Specificity of the AT domains.
From the structure of the microcystins it is possible to conclude that the single AT domains of McyG and McyE and the first AT domain of McyD load methylmalonyl-coenzyme A (CoA), but the presence of methyltransferase domains in McyG, McyD, and McyE (Fig. 1 and 2) suggests that the loading unit can be malonyl-CoA. Regions have been identified in AT domains, where the sequences are different depending on the specificity for either malonyl-CoA or methylmalonyl-CoA (8). By analyzing the sequences of the acyltransferase domains (Fig. 3) and comparing them with the AT domains of soraphen and rapamycin synthases, which utilize malonyl subunits, we conclude that all the AT domains of microcystin synthetase load malonyl units. The methyltranferase domains of McyG, McyD, and McyE would then carry out three methylations (Fig. 1).
FIG. 3.
Comparison of the AT domains of the microcystin synthetases with the consensus sequences of malonyl and methylmalonyl loading AT domains described by Ikeda et al. (8). AT domains (AT1 to AT4) are from Anabaena strain 90 (AMcyG, AMcyD, and AmcyE), from M. aeruginosa (AF183408) (MMcyG, MMcyD, and MMcyE), and from P. agardhii (AJ441056) (PMcyG, PMcyD, and PmcyE). Letters in boldface type indicate the amino acids which are significantly specific to malonyl loading domains, and underlined letters in boldface type point out the residues, which are specific to methylmalonyl loading domains. Serines of the active site are marked with an asterisk.
Ketosynthase and ACP domains.
The active-site cysteine and the two histidine residues, which are present in PKSs (1), were identified in the KS domains of McyG, McyD, and McyE. The active sites of the single ACP domain of McyG and the first ACP domain of McyD have the sequence MGXDS, where methionine replaces the commonly identified leucine residue. There is also variation in this position of rifamycin synthase (28). The ACP domain in the second module of McyD has the active site motif LGLNS, where Asn takes the place of the generally found Asp as in module 11 of the rapamycin synthase (1). The consensus motif LGXDS is found in the single ACP domain of McyE of Anabaena strain 90.
DISCUSSION
The arrangements of the gene clusters of microcystin synthetases from three species are different (Fig. 4). In Anabaena strain 90, M. aeruginosa (18, 29), and P. agardhii CYA126 (2) the NRPS genes mcyA, mcyB, and mcyC have the same order, but the organization of the other genes is different. In Anabaena strain 90 and in M. aeruginosa the mcy genes are in two clusters, which are transcribed in opposite directions, whereas in P. agardhii they are in one cluster transcribed in the same direction (except mcyT, which was not found in Anabaena and Microcystis). The organization of the genes from mcyD to mcyH in Microcystis corresponds the order in Planktothrix with the exception that mcyF and mcyI were not found in Planktothrix and mcyJ is located after mcyC (2) (Fig. 4). In Anabaena strain 90 the arrangement of the genes from mcyG to mcyI and then from mcyA to mcyC is colinear with the structure of microcystin (Fig. 1). Most NRPSs follow the colinearity rule, although several exceptions are known (16). It can be speculated that the order of the microcystin synthetase genes in Anabaena is more original than the one in Microcystis and Planktothrix, in which reorganizations of these genes have taken place.
FIG. 4.
Comparison of Anabaena (A), Microcystis (B), and Planktothrix (C) microcystin synthetase gene clusters. Letters refer to the corresponding mcy genes. Figure (not to scale) shows the order of genes and direction of the gene transcription.
In Anabaena, the order of the domains encoded by the genes in the two sets is colinear with the hypothetical sequence of the enzymatic reactions for microcystin biosynthesis (Fig. 1). The progression of the biosynthetic reactions is consistent with the order of the functions encoded first by mcyG and continuing with the activities encoded by mcyD, mcyJ, mcyE, mcyF, mcyI, mcyA, mcyB, and mcyC. Phenyl acetate is the assumed starting unit in the biosynthesis of Adda (15). It is activated by the adenylating domain identified in the N terminus of McyG and transferred onto the subsequent thiolation site. Polyketide synthesis reactions are monitored (Fig. 1). All four-extension units are malonyl-CoA molecules according to the substrate specificity of the AT domains (Fig. 3). In McyG there is one KS domain to catalyze the first condensation reaction between phenylacetate and malonyl-CoA. The reductive reactions needed to fashion the polyketide chain are putatively catalyzed by KR and DH domains of McyD and McyE. The KR domain of McyG is in the right position to reduce the carbonyl group of the putative starter molecule. The methyltransferase domains of McyG, McyD, and McyE (Fig. 1 and 2) are the obvious candidates to introduce three methyl groups into the carbon frame of Adda. It was recently verified with a knockout mutant (2) that the incorporation of the fourth methyl, which is seen in the methoxy group of Adda, is catalyzed by McyJ. The AMT domain of McyE most likely adds the amino group, which participates in the final peptide bond with the arginine residue.
There are two condensation domains of peptide synthetases in McyE. The first one logically catalyzes the peptide bond between Adda and glutamate, which is activated by the adenylation domain of McyE. The signature sequence, which was also determined as DPRHSGVVG for McyE of both M. aeruginosa and P. agardhii, has no precedents in the databases (Table 2). The synthetases of other peptides, which contain glutamyl residues, are known for bacitracin, fengycin, and surfactin (accession numbers AF007865, AF023464, AF087452, and D13262). In these compounds the standard α-carboxyl of glutamate is part of the peptide bond, while in microcystins it is the γ-carboxyl. This is analogous to the activation of aspartate/methylaspartate by the second adenylation domain of McyB, which results in the β-carboxyl of aspartate/methylaspartate instead of the α-carboxyl being engaged in the peptide bond. This difference must have an impact on the compositions of the glutamate and aspartate/methylaspartate binding pockets in the adenylation domains. McyA has two adenylation domains for the activation of serine and alanine, respectively. The signature sequences of these domains have models in the databases and are almost identical in Anabaena strain 90, M. aeruginosa, and P. agardhii (Table 2). The dehydration of serine supposedly takes place after the activation by adenylation and is probably catalyzed by McyI, which is similar to phosphoglycerate dehydrogenases. There is only one, internal, condensation domain in McyA, which most likely links dehydroserine and d-alanine. The C-terminal condensation domain of McyE putatively catalyzes the bond between glutamate and dehydroserine. There is a methyltransferase domain in the first module of McyA for N-methylation of dehydroserine. The epimerase domain at the C terminus of McyA converts l-alanine to the d form.
Two modules of McyB and one module of McyC logically activate and then add three residues to the nascent peptide chain: l-leucine or l-arginine, methylaspartate or aspartate, and l-arginine, respectively (Fig. 1). The amino acids activated by the adenylation domains of McyC and by the first module of McyB (McyB-1) vary most frequently in microcystins. M. aeruginosa PCC7806 and M. aeruginosa K-139 produce mainly MCYST-LR, and the substrate specificity conferring sequences in McyB-1 of these strains are identical with the signature sequence for leucine (Table 2). M. aeruginosa UV027 and P. agardhii CYA126 produce mostly MCYST-RR, which is also produced by Anabaena strain 90 together with MCYST-LR. Their signature sequences in McyB-1 are different and have no precedents in the databases (Table 2). In M. aeruginosa UV027 the specificity codes of McyB-1 and McyC are almost identical [DVWTIGAV(E/D)WTIGAVD] and match with the codes of McyC from M. aeruginosa K-139 and M. aeruginosa PCC7806, respectively (Table 2). Accordingly McyB-1 of M. aeruginosa UV027 and McyC activate arginine.
There is no epimerase domain in McyB of Anabaena strain 90 or in the other sequenced versions of McyB, though in microcystins, the aspartyl or methylaspartyl moiety is in the d form. The epimerization in this position and in the glutamyl residue is putatively catalyzed by McyF, which is similar to aspartate racemases, and was shown by Nishizawa et al. (19) to complement a d-glutamate-deficient mutant of E. coli. The C-terminal thiosterase domain of McyC, as generally in bacterial nonribosomal peptide synthesis (10), catalyzes the final step in microcystin biosynthesis, the cyclization of the linear peptide (Fig. 1). McyH is probably not needed for the synthesis of microcystins but it may participate in the transport.
The first module of mcyB from Anabaena strain 90 contains a 1,617-bp fragment, which is almost identical to the corresponding part in mcyC. This similarity suggests a duplication of the part of mcyC, which codes for the adenylation domain, and the replacement of the corresponding section in mcyB. The adenylation domain mainly determines the substrate specificity in NRPSs (27). This substitution would explain why Anabaena strain 90 produces the arginine variant of microcystin, MCYST-RR. Because Anabaena strain 90 also produces MCYST-LR, the first module of mcyB has a somewhat-relaxed specificity, while mcyC is specific to arginine.
The genes coding for the microcystin synthesis in three major producers, Anabaena, Microcystis, and Planktothrix have been sequenced. There were several differences between the microcystin synthetase genes of these three producers: (i) gene order was different (Fig. 4), (ii) certain genes were lacking from some producers (Fig. 4), and (iii) the gene identities were rather low (Table 1). This all shows that it was necessary to characterize these genes from each organism. This research has now made it possible to design primers and probes to specifically detect and identify the toxin-producing species in natural samples even when the quantities are low (31). These early warning methods might become effective monitoring systems and valuable tools for protecting water users.
Acknowledgments
This study was supported by Research Center of Excellence funding, grants of the Academy of Finland (40978 and 46812), and grants from the European Union (CYANOTOX grant ENV4-CT98-0802 and TOPIC grant FMRX-CT98-0246) to K.S.
REFERENCES
- 1.Aparicio, J. F., I. Molnár, T. Schwecke, A. König, S. F. Haydock, L. E. Khaw, J. Staunton, and P. F. Leadlay. 1996. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169:9-16. [DOI] [PubMed] [Google Scholar]
- 2.Christiansen, G., J. Fastner, M. Erhard, T. Börner, and E. Dittmann. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 185:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du, L., C. Sanchez, M. Chen, D. J. Edwards, and B. Shen. 2000. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthesis and a polyketide synthase. Chem. Biol. 7:623-642. [DOI] [PubMed] [Google Scholar]
- 4.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fastner, J., M. Erhard, and H. von Döhren. 2001. Determination of oligopeptide diversity within a natural population of Microcystis spp. (cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 67:5069-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii, K., K.-I. Harada, M. Suzuki, F. Kondo, Y. Ikai, H. Oka, W. W. Carmichael, and K. Sivonen. 1996. Occurrence of novel cyclic peptides together with microcystins from toxic cyanobacteria, Anabaena species, p. 559-562 In T. Yasumoto, Y Oshima, and Y. Fukuyo (ed.), Harmful and toxic algal blooms. Intergovermental Oceanographic Commission of UNESCO, Paris, France.
- 7.Goldberg, J., H. B. Huang, Y. G. Kwon, P. Greengard, A. C. Nairn, and J. Kuriyan. 1995. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376:745-753. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda, H., T. Nonomiya, M. Usami, T. Ohta, and S. Omura. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 96:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kagan, R. M., and S. Clarke. 1994. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 310:417-427. [DOI] [PubMed] [Google Scholar]
- 10.Kohli, R. M., J. W. Trauger, D. Schwarzer, M. A. Marahiel, and C. T. Walsh. 2001. Generality of peptide cyclization catalyzed by isolated thioesterase domains of nonribosomal peptide. Biochemistry 40:7099-7108. [DOI] [PubMed] [Google Scholar]
- 11.Kotai, J. 1972. Instructions for preparation of modified nutrient solution Z8 for algae, publication B-11/69, Norwegian Institute for Water Research. Blindern, Oslo, Norway.
- 12.Lyra, C., S. Suomalainen, M. Gugger, C. Vezie, P. Sundman, L. Paulin, and K. Sivonen. 2001. Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis and Planktothrix genera. Int. J. Syst. Evol. Microbiol. 51:513-526. [DOI] [PubMed] [Google Scholar]
- 13.MacKintosh, C., K. A. Beattie, S. Klumpp, P. Cohen, and G. A. Codd. 1990. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatase 1 and 2A from both mammals and higher plants. FEBS Lett. 264:187-192. [DOI] [PubMed] [Google Scholar]
- 14.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 15.Moore, R. E., J. L. Chen, B. S. Moore, G. M. L. Patterson, and W. W. Charmichael. 1991. Biosynthesis of microcystin-LR. Origin of the carbons in the Adda and Masp units. J. Am. Chem. Soc. 113:5083-5084. [Google Scholar]
- 16.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem 3:490-504. [DOI] [PubMed] [Google Scholar]
- 17.Namikoshi, M., and K. L. Rinehart. 1996. Bioactive compounds produced by cyanobacteria. J. Ind. Microbiol. 17:373-384. [Google Scholar]
- 18.Nishizawa, T., A. Ueda, M. Asayama, K. Fujii, K.-I. Harada, K. Ochi, and M. Shirai. 2000. Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J. Biochem. 127:779-789. [DOI] [PubMed] [Google Scholar]
- 19.Nishizawa, T., M. Asayama, and M. Shirai. 2001. Cyclic heptapeptide microcystin biosynthesis requires the glutamate racemase gene. Microbiology 147:1235-1241. [DOI] [PubMed] [Google Scholar]
- 20.Rouhiainen, L., K. Sivonen, W. J. Buikema, and R. Haselkorn. 1995. Characterization of toxin-producing cyanobacteria by using an oligonucleotide probe containing a tandemly repeated heptamer. J. Bacteriol. 177:6021-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouhiainen, L., L. Paulin, S. Suomalainen, H. Hyytiäinen, W. Buikema, R. Haselkorn, and K. Sivonen. 2000. Genes encoding synthetases of cyclic depsipeptides, anabaenopeptilides, in Anabaena strain 90. Mol. Microbiol. 37:156-167. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 23.Scrutton, N. S., A. Berry, and R. N. Perham. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343:38-43. [DOI] [PubMed] [Google Scholar]
- 24.Silakowski, B., G. Nordsiek, B. Kunze, H. Blöcker, and R. Müller. 2001. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga 15. Chem. Biol. 8:59-69. [DOI] [PubMed] [Google Scholar]
- 25.Sivonen, K., M. Namikoshi, W. R. Evans, W. W. Carmichael, F. Sun, L. Rouhiainen, R. Luukkainen, and K. L. Rinehart. 1992. Isolation and characterization of a variety of microcystins from seven strains of the cyanobacterial genus Anabaena. Appl. Environ. Microbiol. 58:2495-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivonen, K., and G. Jones. 1999. Cyanobacterial toxins, p. 41-111. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management. E. & F. N. Spon, London, United Kingdom.
- 27.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 28.Tang, L., Y. J. Yoon, C.-Y. Choi, and C. R. Hutchinson. 1998. Characterization of the enzymatic domains in the modular polyketide synthase involved in rifamycin B biosynthesis by Amycolatopsis mediterranei. Gene 216:255-265. [DOI] [PubMed] [Google Scholar]
- 29.Tillett, D., E. Dittmann, M. Erhard, H. von Döhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 30.Tsuge, K., T. Akiyama, and M. Shoda. 2001. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaitomaa, J., A. Rantala, K. Halinen, L. Rouhiainen, P. Tallberg, L. Mokelke, and K. Sivonen. 2003. Quantitative real-time PCR for determination of microcystin synthetase E copy numbers for Microcystis and Anabaena in lakes. Appl. Environ. Microbiol. 69:7289-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]