SUMMARY
Although uridine-rich small nuclear RNAs (U-snRNAs) are essential for pre-mRNA splicing, little is known regarding their function in the regulation of alternative splicing or of the biological consequences of their dysfunction in mammals. Here, we demonstrate that mutation of Rnu2–8, one of the mouse multicopy U2 snRNA genes, causes ataxia and neurodegeneration. Coincident with the observed pathology, the level of mutant U2 RNAs was highest in the cerebellum and increased after granule neuron maturation. Furthermore, neuron loss was strongly dependent on the dosage of mutant and wild type snRNA genes. Comprehensive transcriptome analysis identified a group of alternative splicing events, including the splicing of small introns, which were disrupted in the mutant cerebellum. Our results suggest that the expression of mammalian U2 snRNA genes, previously presumed to be ubiquitious, is spatially and temporally regulated, and dysfunction of a single U2 snRNA causes neuron degeneration through distortion of pre-mRNA splicing.
INTRODUCTION
Splicing of pre-mRNAs is performed by the spliceosome machinery consisting of numerous proteins and 5 small nuclear RNAs (snRNAs) (Wahl et al., 2009). Assembly of the major spliceosome, responsible for splicing of over 90% of human pre-mRNAs, begins with the recruitment of U1 snRNPs (small nuclear ribonucleoproteins) to the pre-mRNA 5' exon/intron junction via basepairing of the U1 snRNA with the splice site. U2 snRNPs subsequently bind to the pre-mRNA branchpoint sequence (BPS) near the 3' intron boundary, also in part via basepairing interactions. Further remodeling of the spliceosome results in the recruitment of additional snRNPs, including U6 snRNPs that will replace U1 snRNPs at 5' pre-mRNA splice sites. Basepairing of U6 with U2 snRNAs juxtaposes the 5' splice site and the BPS, the reactants of the first transesterification reaction. Thus recognition and removal of introns and subsequent exon ligation depend in part on the dynamic basepairing of snRNAs with pre-mRNAs and with each other.
Given the critical importance of pre-mRNA splicing to the regulation of gene expression and proteome diversity, it follows that mutations in RNA-processing proteins that may influence pre-mRNA splicing cause human disease (Cooper et al., 2009; Licatalosi and Darnell, 2006). Indeed, dominant mutations in multiple ubiquitously expressed protein components of the U4/U5/U6 tri-snRNP cause retinitis pigmentosa (Mordes et al., 2006). Mutations in the SMN1 (survival of motor neuron) gene, which encodes a protein essential for the U snRNP biogenesis, cause motor neuron degeneration in spinal muscular atrophy (SMA) patients (Lefebvre et al., 1995). Although SMN is ubiquitous expressed, alterations in the levels of individual snRNAs and numerous aberrant transcripts were observed in a tissue-specific fashion in SMN-deficient mice (Zhang et al., 2008). Mutations in two other ubiquitously expressed DNA/RNA-binding proteins, TDP-43 and FUS/TLS, are associated with some cases of familial and sporadic amyotrophic lateral sclerosis and frontotemporal dementia (Lagier-Tourenne et al., 2010; Lemmens et al., 2010). Recent studies demonstrated that decreased expression or altered subcellular localization of TDP-43 resulted in reduced expression and altered splicing of multiple mRNAs (Polymenidou et al., 2011; Tollervey et al., 2011). These findings strongly implicate alterations in RNA processing as a key event in several neurodegenerative disorders. However, given the multiple functions of these proteins, the pathogenic basis of these diseases remains unclear.
Recently, mutations in the gene encoding U4atac snRNA, a component of the minor spliceosome that splices a restricted (U12-dependent) class of introns found in less than 1% of human genes, have been linked to the developmental disorder microcephalic osteodysplastic primordial dwarfism (Edery et al., 2011; He et al., 2011). However to date, mutations in the major spliceosomal snRNA genes have not been associated with disease, perhaps in part due to their ubiquitous expression and essential function. Furthermore, unlike the U4atac gene, the snRNAs of the major spliceosome are encoded by multiple genes and pseudogenes in metazoans (Marz et al., 2008), suggesting these genes may be highly redundant. Although differing in organization (e.g., U1 and U2 genes exist in large chromosomal clusters in the human genome, whereas other snRNA genes are not clustered (Manser and Gesteland, 1982; Van Arsdell and Weiner, 1984; Westin et al., 1984)), sequence analysis of snRNA genes suggests they have apparently undergone concerted evolution, i.e., members of gene families are identical or nearly so within a species (Marz et al., 2008; Pavelitz et al., 1995). However, the effect of polymorphisms in individual members of these repetitive gene families on pre-mRNA splicing and the extent to which the expression of single genes is independently regulated are largely unknown.
Here, we report that a 5-nucleotide deletion in one member of a cluster of mouse U2 snRNA genes causes neurodegeneration and alternative splicing defects, including the retention of small introns. Our study provides definitive evidence for the causative role of splicing dysfunction in neurodegeneration and a model to dissect the role of major spliceosomal snRNAs on the regulation of mammalian pre-mRNA splicing.
RESULTS
Progressive neurodegeneration in NMF291 mutant mice
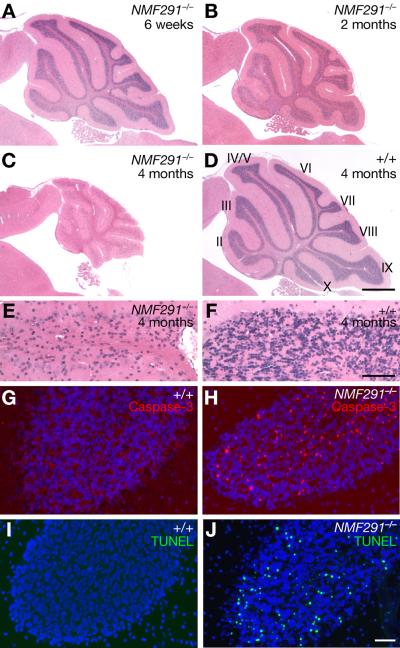
The NMF (Neuroscience Mutagenesis Facility) 291 mutation was identified in a chemical mutagenesis screen for recessive mutations that result in neurological phenotypes (Bult et al., 2004). Mice homozygous for this mutation developed mild tremors at 8 weeks of age, which progressed to overt truncal ataxia by 12 weeks (Movie 1). At 4 weeks of age, the brains of mutant mice appeared grossly normal (Figure S1A–S1D). However, histological analysis revealed granule cells with pyknotic nuclei in the mutant cerebellum beginning at 4 weeks of age (data not shown). Neuron loss was progressive, and by 4 months of age most granule cells had degenerated (Figure 1A–1F). However, other cerebellar neurons, including Purkinje cells, did not degenerate, even in aged mice (data not shown). Although neuron death was most severe in the cerebellum, later-onset degeneration of granule cells in the dentate gyrus region of the hippocampus was also observed (Figure S1E–S1H).
Figure 1. Progressive Granule Cell Degeneration in the NMF291−/− Cerebellum.
(A–F) Hematoxylin and eosin stained sagittal sections of wild type (+/+) and NMF291−/− cerebella. Cerebellar lobules are indicated by Roman numerals (D). (E, F) Higher magnification images of lobule IV/V from (C) and (D).
(G–J) Cleaved caspase 3 immunostaining (G, H) and TUNEL analysis (I, J) of sections from 6-week-old cerebella. Sections were counterstained with Hoechst 33342. Scale bars: (D), 500 μm; (F), 50 μm; (J), 10 μm. See also Figure S1 and Movie 1.
To determine the nature of neuron death, immunostaining with activated caspase-3 antibodies was performed on cerebellar sections from 6-week-old mice. Many immunopositive granule cells were observed in the mutant, but not the wild type cerebellum, suggesting mutant neurons undergo apoptosis (Figure 1G and 1H). Granule cell apoptosis was confirmed by TUNEL (TdT-mediated dUTP nick end labeling) analysis (Figure 1I and 1J).
The NMF291 mutation deletes 5 nucleotides in the Rnu2–8 U2 snRNA
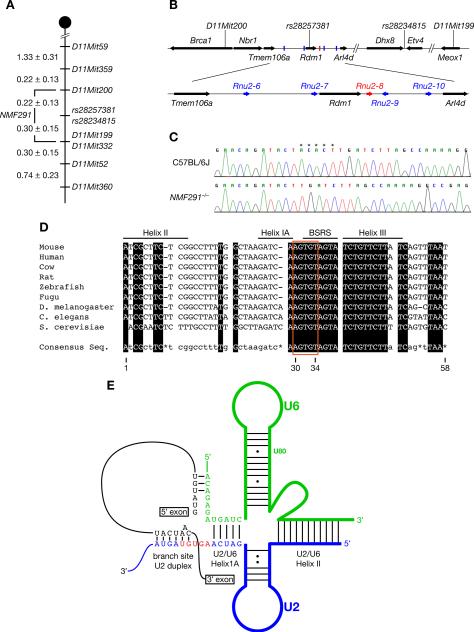
The NMF291 mutation was initially localized to distal Chromosome 11 by a genome scan analysis of F2 mice using polymorphic microsatellite markers. Additional recombination mapping narrowed the mutation interval to a 0.52 centimorgan (0.34 megabase) region between D11Mit200 and D11Mit199 containing 8 protein-coding genes (Figure 2A and 2B, and Figure S2A). No coding sequence polymorphisms between mutant and wild type DNA were found in these genes, nor did we observe differences in the expression of the cerebellar transcripts of these genes between genotypes (data not shown).
Figure 2. The NMF291 Mutation is a 5-nucleotide Deletion in a U2 Small Nuclear RNA Gene.
(A) The NMF291 mutation was mapped to Chromosome 11 between D11Mit200 and D11Mit199 (values are in centimorgans (cM) ± SEM).
(B) The NMF291 critical interval contains 8 protein-coding genes (arrows), and 5 U2 snRNA genes (Rnu2–6 – Rnu2–10; bars).
(C) Sequence chromatograms of wild type (C57BL6/J) and mutant (NMF291−/−) Rnu2–8 genomic DNA amplified using unique primers outside of the transcription unit. *, nucleotides deleted in the mutant genome.
(D) Deleted nucleotides are evolutionarily conserved. Conserved nucleotides are boxed in black and indicated in capitals in the consensus sequence. Nucleotides with one or more mismatches or absent across species are indicated in the consensus sequence in lower case or with an asterisk, respectively. The U2 branch site recognition sequence (BSRS), and the nucleotides in helices IA, II, and III, which basepair with U6 snRNA, are indicated. Nucleotides are numbered according to the mouse sequence.
(E) The schematic of RNA:RNA interactions contributing to the first step of splicing. The 5-nucleotide deletion in Rnu2–8 is highlighted in red. See also Figure S2.
In addition to the protein-coding genes, a cluster of five U2 snRNA (Rnu2–6 to Rnu2–10) genes also resides in the NMF291 critical interval (Figure 2B). The RNAs encoded by these genes are identical, except for a single nucleotide polymorphism in Rnu2–6 (Figure S2B). Sequencing of mutant genomic DNA revealed a 5 base-pair deletion between nucleotides 30 and 34 (30AGUGU34) in a highly conserved region of the Rnu2–8 transcription unit (Figure 2C and 2D). This deletion removes the first 2-nucleotides of the U2 consensus branch site recognition sequence (BSRS) (33GUAGUA38) and the 3-nucleotide linker region (30AGU33) between the BSRS and U2/U6 helix IA (Figure 2D and 2E) (Wahl et al., 2009). The deletion was not observed in genomic DNA from several other inbred stains, including C57BL6/J and 129S4/SvJae, from which the F1 ES cells used for mutagenesis were derived (data not shown).
Rnu2–8 is spatially and temporally regulated
To confirm that the U2 gene disrupted by the NMF291 deletion is indeed expressed, we performed northern blot analysis and RNase protection assays (RPA) using cerebellar RNA isolated from P30 wild type and mutant mice. Total U2 snRNA levels were similar between the wild type and the NMF291−/− cerebellum (Figure 3A) and both wild type and the deletion-containing Rnu2–8 RNAs were detected in the mutant cerebellum (Figure 3B and 3C), with the mutant transcript found at 78.9±6.3% (n=6) of the level of the wild-type U2 snRNA (or ~45% of total U2 levels). Similar analysis of highly pure granule cell cultures demonstrated that both wild type and mutant U2 RNAs were expressed within the same cell type (data not shown). Lastly, immunoprecipitation experiments using Y12 antibody, which precipitates U snRNPs (Lerner et al., 1981), demonstrated that the mutant U2 snRNAs were assembled into mutant cerebellar U2 snRNPs (Figure S3A and S3B), consistent with previous reports that the free snRNA pool is relatively very small and unstable (Sauterer et al., 1988).
Figure 3. The Expression of Rnu2–8 is Temporally and Spatially Regulated.
(A and B) Total RNA from wild type and NMF291−/− (−/−) P30 cerebella was separated by denaturing acrylamide gels for shorter (A) or longer (B) periods and subsequently analyzed by northern blot analysis using an U2-specific oligonucleotide probe. For loading controls, the gel was stained with ethidium bromide (A, lower panel) or the membrane was re-hybridized with a probe for 5s rRNA (B).
(C) Ribonuclease protection assay (RPA) using increasing amounts of cerebellar RNA (0.2–1 μg) from P30 wild type and NMF291−/− mice. For loading controls, RPA was also performed using a ß-actin RNA probe.
(D) RPA analysis of U2 snRNAs in various tissues taken from P30 wild type and NMF291−/− mice.
(E) RPA analysis of U2 snRNAs in wild type, NMF291+/− (+/−), and NMF291−/− cerebella at different postnatal (P) ages (days after birth). Cere., cerebellum. S.C., spinal cord. See also Figure S3.
Mammalian U2 snRNAs, like other U snRNAs, are believed to be ubiquitously and highly expressed (Egloff et al., 2008; Hernandez, 2001). However, neurodegeneration in NMF291−/− mice was very distinct, with profound neuron loss occurring within the cerebellum beginning at P30. To determine if expression of mutant Rnu2–8 correlates with the specificity of pathological changes, we performed RPA and northern blot assays using neuronal and non-neuronal tissues from the P30 wild type and homozygous mutant mice (Figure 3D and Figure S3C). Surprisingly, mutant Rnu2–8 was differentially expressed between tissues, with highest levels observed in the cerebellum. Furthermore, mutant Rnu2–8 expression was also temporally regulated in the postnatal cerebellum. Mutant U2 RNAs comprised ~20–25% of the total U2 RNA levels in the P10 and P20 NMF291−/− cerebellum, but rose to ~45% of the total level at P30 (Figure 3E, Figure S3D and S3E). This increase was maintained at P40 but dropped slightly at P60, likely due to neuron loss in the mutant cerebellum. This temporal regulation of expression was also observed in heterozygous mice, where amounts of mutant U2 RNA were approximately half of that observed in homozygous mice (Figure 3E and Figure S3E). Together, these data suggest that mammalian U2 snRNAs display previously unsuspected temporal and spatial variation with the expression of mutant Rnu2–8 correlating with the onset and specificity of neurodegeneration in the NMF291−/− mouse.
Mutant Rnu2–8 induces neurodegeneration in a dosage-dependent manner
Although mice homozygous for the NMF291 mutation were originally ascertained in our screen, the site of the Rnu2–8 deletion, the multicopy nature of the U2 genes, and the expression of wild type U2 snRNAs in the mutant cerebellum suggested that this mutation might act in a gene dosage-dependent fashion. To investigate this possibility, we performed histological analysis of the cerebellum of NMF291+/− mice. Apoptotic granule cells were observed in the cerebellum of 1-month-old and older NMF291+/− mice, but the number of dying neurons was considerably reduced compared to that observed in homozygous NMF291 mice (data not shown). Differences in the overall size of the NMF291+/− cerebellum were not obvious until mice were close to 2 years of age, when mild tremors were apparent and histological analysis revealed that many granule cells had degenerated (Figure 4A–4F).
Figure 4. Mutant Rnu2–8 Acts in a Dosage-dependent Fashion on Neuron Survival.
(A–F) Hematoxylin and eosin stained sagittal sections of NMF291+/− and wild type (+/+) cerebella. (E, F) Higher magnification images of lobule IV/V from (C) and (D).
(G) The 1.5kb DNA fragment, containing the transcription unit of the mutant Rnu2–8 and its 5' and 3' flanking sequences, used for pronuclear injection.
(H) RPA analysis of cerebellar RNA from P30 wild type, NMF291+/−, NMF291−/− mice and mice hemizygous for the mutant Rnu2–8 transgene (Tg).
(I) RPA analysis of cerebellar RNA from P20, P30, and P40 NMF291−/− mice and mice hemizygous for the transgene (Tg).
(J) RPA analysis of tissues from P30 NMF291−/−and hemizygous Tg-MuU2 mice. Note the NMF291−/− cerebellar lane was run on a separate gel.
(K and L) Hematoxylin and eosin stained sagittal sections of Tg-MuU2 cerebella. DES, distal sequence element; PES, proximal sequence element. Scale bars: (D and L) 500 μm; (F) 50 μm. See also Figure S4.
To further test the mechanism of the Rnu2–8 mutation, we generated mice transgenic for a 1.5kb genomic DNA fragment containing the mutant Rnu2–8 transcriptional unit and the basal regulatory elements necessary for U2 transcription and processing (Figure 4G) (Egloff et al., 2008; Hernandez, 2001). Mice from one transgenic line (Tg-MuU2) expressed mutant Rnu2–8 in the cerebellum, and expression was temporally and spatially regulated similar to that observed in NMF291−/− mice (Figure 4H–4J and Figure S4A). Like NMF291−/− mice, hemizygous mice from this line developed pronounced ataxia by 12 weeks of age (data not shown). Histological analysis revealed dying granule cells in the cerebellum of 1-month-old transgenic mice, and by 3 months of age most granule cells had died, similar to the time course of neuron death in NMF291−/− mice (Figure 4K and 4L, and data not shown). As observed in NMF291−/− mice, neuron loss in the dentate gyrus was observed in 3-month-old transgenic mice (Figure S4B and S4C).
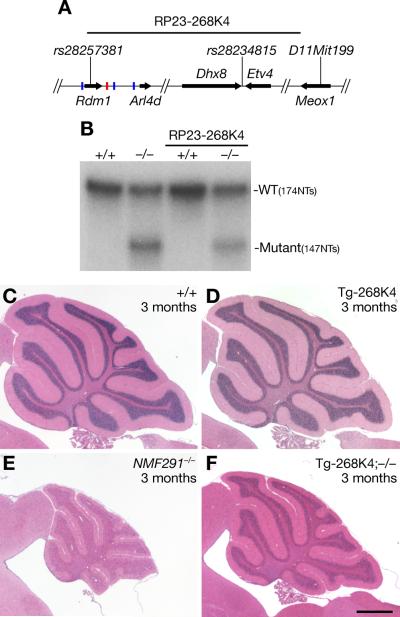
If the severity of neurodegeneration is indeed dependent on the ratio of mutant to wild type snRNAs, then the complementary experiment in which an increase in expression of wild type U2 RNA on the NMF291−/− background should lead to attenuation of pathology. To test this hypothesis we generated a transgenic mouse line carrying a BAC (bacterial artificial chromosome RP23-268K4) containing Rnu2–8 and two other U2 genes (Rnu2–9 and −10; Figure 5A). This line was crossed to NMF291 mice to generate NMF291−/− mice carrying the BAC transgene. The percentage of mutant/total U2 snRNA was reduced in the cerebellum of Tg-268K4; NMF291−/− and Tg-268K4; NMF291+/− mice relative to that observed in the cerebellum of age-matched NMF291−/− and NMF291+/− mice (Figure 5B and Figure S5A and S5B). Neurodegeneration was also decreased in the cerebellum of 3-month-old Tg-268K4; NMF291−/− mice relative to that of age-matched NMF291−/− mice (Figure 5C–5F). Surprisingly, when mutant U2 RNAs represented ~17% of total U2 levels, little granule cell loss was observed in aged Tg-268K4; NMF291+/− mice (Figure S5C–S5F). When the percentage of mutant/total U2 RNA increased to ~25% as observed in the NMF291+/− cerebellum, neurodegeneration was slowly progressive (Figure 4A–4F). However, when this percentage reached ~45% (as in the P30 NMF291−/− and Tg-MuU2 cerebellum), rapid granule neuron loss was observed (Figure 1C and 4L). Taken together, our data demonstrate that the expression of the mutant Rnu2–8 gene is sufficient to induce granule cell death even in the presence of wild type U2 expression, and the extent of neuron loss is dependent on the expression level of the mutant U2 snRNAs relative to that of the wild type U2 snRNA.
Figure 5. Transgenic Expression of Wild Type U2 snRNAs Partially Rescues the NMF291−/− Phenotype.
(A) Schematic of the RP23-268K4 BAC, containing three snRNA genes (Rnu2–8 red bar, Rnu2–9, −10 blue bars), used for transgenesis.
(B) RPA of cerebellar RNA from 1-month-old wild type (+/+) or NMF291−/− (−/−) mice with or without the BAC transgene (RP23-268K4).
(C–F) Hematoxylin and eosin stained sagittal sections of wild type (+/+), RP23-268K4 (Tg-268K4), NMF291−/− and Tg-268K4; −1/− cerebella. Scale bars: (D) 500 μm. See also Figure S5.
Expression of the mutant U2 snRNA decreases splicing efficiency
The central role of U2 snRNA in pre-mRNA splicing suggested that abnormalities in this process underlie neurodegeneration in NMF291−/− mice. To study the impact of the mutant Rnu2–8 on pre-mRNA splicing, we employed a previously reported splicing reporter construct, TN24 (ISS+), which encodes both ß-galactosidase and luciferase separated by an exon-intron-exon cassette (Kollmus et al., 1996; Nasim et al., 2002). Although ß-galactosidase is constitutively expressed from this plasmid, luciferase is only expressed when accurate splicing occurs between the adenovirus 5' exon and the alternatively spliced SK exon of human tropomyosin 3 (see diagram in Figure 6C).
Figure 6. Mutant Rnu2–8 Selectively Decreases Splicing Efficiency of Introns.
(A) The sequences of wild type and deletion (Δ) U2 constructs used for cell transfection experiments.
(B) RPA analysis of RNA from mock-transfected HEK293T cells or cells transfected with wild type (WT) or ΔU2 plasmids.
(C and D) HEK293T cells were transfected with the ISS+ splicing reporter (C) or the ISS− splicing reporter (D), or co-transfected with WT or ΔU2 constructs as indicated. The splicing of pBPLUGA parental vector is shown in (C) as a control, and the splicing of the ISS+ reporter is shown in (D) as a reference. The splicing efficiency of the reporters was measured by luciferase/ beta-gal activity (upper panels; C and D) and confirmed by RT-PCR (lower panels). Values represent mean ± SEM, n = 3; *p<0.01; one-way ANOVA. * and arrows in reporter diagrams denote stop codons and RT-PCR primers, respectively.
(E) A diagram of the 3' region of the L1CAM gene. *, stop codon. Arrows, RT-PCR primers.
(F) RT-PCR was performed on wild type (+/+) and NMF291−/− (−/−) cerebellar cDNA at indicated ages.
(G and H). HEK293T cells were transfected with L1CAM exon27-intron27-exon28 (G) and exon28-intron28-exon29 (H) splicing reporters with or without wild type or ΔU2 constructs as indicated. Splicing of these reporters was analyzed as described above in C and D. As references, the spliced RT-PCR products of exon27-intron27-exon28 reporter and the unspliced PCR product amplified from the exon28-intron28-exon29 reporter plasmid (DNA) are also shown in (H). Values represent mean ± SEM, n = 3; *p<0.01; one-way ANOVA. * in reporter diagrams, stop codons.
HEK293T cells were transfected with ISS+ alone, or co-transfected with the reporter and a 1.5kb genomic DNA fragment containing the wild type (WT) or the mutant (Δ5nt) U2 transcription unit (Figure 6A–6C). As controls, cells were co-transfected with the reporter and a plasmid encoding the RNA-binding protein hnRNPG, which has been shown to silence splicing of this exon-intron-exon cassette via binding of the 25 bp intronic splicing silencer sequence (ISS) (Nasim et al., 2002).
Transfection of wild type Rnu2–8 did not have a significant effect on reporter splicing as determined by the ratio of luciferase/ ß-galactosidase activity or by RT-PCR (Figure 6C). However, transfection of mutant Rnu2–8 led to a significant decrease (28%; p<0.01) in the splicing efficiency of the reporter compared to that of cells transfected with reporter alone.
Similar experiments were performed using the same reporter lacking the 25bp ISS (ISS-) (Nasim et al., 2002) (Figure 6D). In agreement with the role of the ISS sequence in splicing silencing, we observed a 4–5 fold increase in splicing of this reporter over that of the reporter containing the ISS sequence. As previously shown (Nasim et al., 2002), the inhibitory effect of hnRNPG expression on ISS- reporter splicing was greatly reduced (Figure 6D). As observed for the ISS+ reporter, splicing of the ISS- reporter was not changed by expression of the wild type U2. However, in contrast to the attenuation of ISS+ reporter splicing by expression of the mutant U2 construct, expression of the mutant U2 did not significantly affect splicing of the ISS- reporter. These results suggest that alterations in splicing efficiency mediated by the expression of the mutant U2 snRNAs may be dependent on the presence of splicing regulatory sequences.
The presence of intronic or exonic splicing regulatory sequences often accompany alternative splicing of pre-mRNA (Black, 2003), thus we initially analyzed several alternative-splicing events that were previously reported to occur in the human cerebellum (Wang et al., 2008). Four out of the twenty alternative splicing events, including one in the neural cell adhesion molecule L1 (L1CAM), displayed obvious changes in relative isoform expression between the wild type and NMF291−/− cerebellum (data not shown). PCR amplification of the distal exons of the L1CAM transcript from wild type cerebellar RNA generated two isoforms: the major form containing exons 27–28–29, and a minor isoform that retains the 95-bp intron located between exons 27 and 28 (Figure 6E and 6F). Coincident with the decrease of the fully spliced isoform, the abundance of the intron 27-containing isoform was increased in mutant cerebellar RNA. Interestingly, these alterations in splicing were most dramatic in the P30 and P40 cerebellum demonstrating that the change in the ratio of splicing variants was temporally regulated in a manner coincident with expression of the mutant U2 snRNA.
To directly evaluate the effects of the mutant U2 on splicing of intron 27, we generated L1CAM splicing reporters. A cassette containing exon 27, the alternatively spliced intron 27, and exon 28, or a cassette with exon 28, the constitutively spliced intron 28, and exon 29 were cloned into the pBPLUGA plasmid (Figure 6G and 6H). These plasmids were co-transfected into HEK293T cells with the wild type or mutant Rnu2–8 plasmid. Transfection of the intron27-containing reporter resulted in both spliced and unspliced transcripts as evidenced by RT-PCR (Figure 6G). In contrast, only spliced transcripts were observed in cells transfected with the intron28-containing reporter, and luciferase/ß-galactosidase activity was increased accordingly (Figure 6H). Overexpression of the wild type U2 had no effect on splicing of either reporter. However, in agreement with our in vivo data, expression of mutant Rnu2–8 caused a decrease in splicing efficiency (34%; p<0.01) of intron 27, but not of intron 28, the splicing efficiency of which was increased for unknown reasons (Figure 6G and 6H). These reporter data confirm that expression of the mutant U2 disrupts splicing of select introns.
Deletion of the U2/U6 helix IA linker is sufficient to disrupt splicing
The 5-nucleotide deletion in Rnu2–8 removes 2 nucleotides of the BSRS, which recognizes and forms a duplex with the intronic branch site (BS) of the pre-mRNA that is important for the spliceosome assembly and the first catalytic step of splicing (Wahl et al., 2009). In addition, the NMF291 deletion removes the 3-nucleotide linker between the BSRS and the sequence that forms the U2/U6 helix IA. The distance between these two sequences has been shown to change splicing kinetics in yeast (McGrail et al., 2006; Ryan and Abelson, 2002; Smith et al., 2009). To further understand the underlying mechanism of mutant Rnu2–8-induced splicing defects, we created a series of U2 deletion plasmids: Δ6, in which the entire BSRS was deleted; Δ3, in which the linker sequence between BSRS and U2/U6 helix IA was deleted; and Δ9, in which both the entire BSRS and the linker was deleted (Figure 6A).
Experiments in yeast demonstrated that mutations in U2 snRNA which disrupt the duplex formation between the BSRS and the branch site, inhibit spliceosome assembly and result in the degradation of these RNAs (Smith et al., 2009). In agreement with these studies, we consistently observed lower expression levels of Δ6 and Δ9 Rnu2–8 RNAs, both of which lack the BSRS, compared to that observed for the wild type, Δ5 (the NMF291 mutant form), and Δ3 Rnu2–8 RNAs in transfected cells (Figure 6B). Co-transfection of the Δ6 or the Δ9 plasmid with the ISS+ reporter did not affect reporter splicing (Figure 6C). However, expression of the Δ3 construct significantly decreased (24%; p<0.01) splicing of the ISS+ reporter, closely mimicking the repression of splicing that was observed when Δ5 Rnu2–8 was co-transfected with the reporter. Also as observed with co-transfection of Δ5 Rnu2–8, expression of Δ3 construct had no effect on splicing of ISS- splicing reporter (Figure 6D).
Similarly, expression of the Δ3 Rnu2–8 plasmid was sufficient to reduce splicing (13%; p<0.01) of intron 27 from the L1CAM intron27-containing splicing reporter, although not as efficiently as co-transfection of the reporter with Δ5 Rnu2–8, suggesting that the deletion of two nucleotides of the BSRS may also contribute to the Δ5 effect on splicing (Figure 6G). Co-transfection of the Δ3 plasmid with the intron28-containing reporter again led to an increase, rather than decrease of splicing, as did expression of the Δ6 and Δ9 plasmids (Figure 6H). Our data suggests that expression of a U2 snRNA with either the NMF291-associated 5nt deletion or lacking the linker sequence between the BSRS and the U2/U6 helix IA disrupts select intron splicing.
Abnormal alternative splicing in the NMF291 mutant cerebellum
To analyze alternative splicing in the NMF291 mutant cerebellum, exon expression was examined by Affymetrix mouse exon 1.0 ST microarrays. Microarrays were hybridized with RNA from three wild type and three NMF291−/− cerebella isolated at P30, a time when the mutant U2 RNA is expressed at peak levels but few neurons have degenerated (Figure S1A–S1D, Figure 3E, and Figure S3E). 123 genes were differentially expressed between the wild type and mutant cerebellum as demonstrated by analysis of signal across all probe clusters for a gene (fold-change ≥1.5; p<0.05) (Table S1A). Differential expression of most (78%) of these genes was relatively low (1.5–2 fold), suggesting that overall gene expression was not dramatically altered in the P30 mutant cerebellum.
Paired analysis of the exon splicing index (SI), which was calculated by normalizing exon expression to gene level expression (Clark et al., 2007; de la Grange et al., 2010), revealed that the expression of 206 exons in 178 genes were differentially expressed between the wild type and mutant cerebellum (SI fold-change ≥1.5; p<0.05) (Table S1B). These exons were compared to the comprehensive list of known human and mouse splice variants in the FAST DB database (de la Grange et al., 2007) and 146 (71%) events, representing all major types of alternative splicing patterns, were previously annotated as alternatively spliced (Table S1B and Figure 7A). To validate our exon array results, RT-PCR was performed using cerebellar cDNA from four independent mice of each genotype. 92% (12/13) of these events were validated as differentially spliced between the wild type and mutant cerebellum (Figure S6A and data not shown).
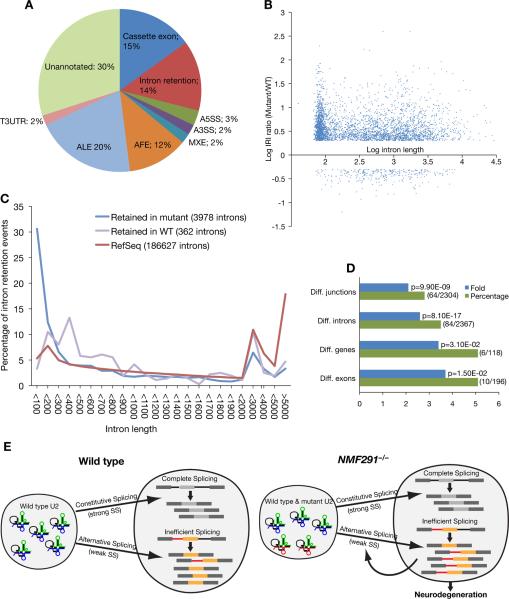
Figure 7. Global Splicing Abnormalities in the NMF291−/− Cerebellum.
(A) Types of alternative splicing events identified by exon array analysis as differentially spliced between the wild type and the NMF291−/− cerebellum. A3SS, alternative 3' splice site; A5SS; alternative 5' splice site; AFE, alternative first exon; ALE, alternative last exon; MXE, mutally exclusive exon; T3UTR, truncated 3'UTR.
(B) The log of the mutant/WT Relative Intron retention Index (IRI) generated by RNA-Seq analysis plotted against the log of the intron length (bp).
(C) Significantly retained introns in the cerebellum of NMF291−/− or wild type mice, or RefSeq introns were grouped by length and plotted as a percentage of the total introns.
(D) Gene Ontology (GO) analysis of genes annotated to the pre-mRNA splicing GO term were significantly enriched in the differentially spliced exons (Diff. junctions) and introns (Diff. introns) identified by RNA-Seq, and differentially spliced exons (Diff. exons) and differentially expressed genes (Diff. genes) identified by exon-array. Percentage, the number of genes (the numerator in parentheses) annotated to the GO term divided by the total number of inputted genes (the denominator in parentheses). Fold enrichment, the magnitude of enrichment of input genes relative to all mouse genes annotated to pre-mRNA splicing. p-value, the significance of gene-term enrichment with a modified Fisher's exact test.
(E) The working model for our findings. The splicing status and biological consequences of expression of wild type (blue) and NMF291 mutant (red) U2 snRNAs in wild type and NMF291−/− neurons are shown. Our in vivo and in vitro data suggest that expression of mutant U2 snRNA disrupts splicing at many suboptimal or weaker splice sites, including some of those used in alternative splicing. These changes in alternative splicing may be further amplified by altered splicing of RNA processing factors caused directly by mutant U2 snRNAs or by autoregulation. See also Figure S6 and Table S1A–H.
Interestingly, 14% of the differentially spliced events detected by exon array analysis were annotated as alternatively spliced introns, although introns normally represent a very minor (1–3%) class of alternative splicing events (Chacko and Ranganathan, 2009; de la Grange et al., 2010; Wang et al., 2008). To examine this more closely and to expand our exon array results, we performed RNA-Seq analysis. Libraries were prepared from cerebellar mRNA from three P30 mutant and two P30 wild-type mice and sequenced. The resulting reads were aligned to the mouse genome reference sequence via SpliceMap (http://www.stanford.edu/group/wonglab/SpliceMap/), an algorithm that maps exon-exon junctions without relying on previous exon annotations for recovering the splicing junctions (Au et al., 2010). A total of 53.48 ± 6.35 × 106 reads per sample were uniquely mapped to the mouse genome, and 61.23 ± 1.18%, 20.80 ± 1.54%, and 6.20 ± 0.19% of these reads uniquely mapped to the exons, introns, and splice junctions, respectively (Table S1C).
To identify alternative splicing pattern changes between the wild type and NMF291−/− cerebellum, splicing junction reads were extracted and normalized for isoform expression to generate the Relative Junction Index (RJI) (Figure S6). Comparison of the RJI between the two genotypes identified 1636 junctions that were differentially expressed (RJI ratio≥ 2.0; p< 0.05, Table S1D). In addition, 52 and 1137 junction reads that were present only in the wild type and mutant datasets, respectively, indicating that many splicing variants in the mutant cerebellum were not present, or present at a very low levels, in the wild-type cerebellum (Table S1E). As described above, RT-PCR using wild type and mutant cerebellar cDNA was performed to test differential splicing of junctions. 20/20 junctions that were originally identified in both genotypes and 20/21 (95%) of the junctions identified only in one genotype were validated (Figure S6B and S6C and data not shown).
To examine global abnormalities in splicing of introns in the mutant cerebellum, we calculated the Relative Intron retention Index (RII) from our wild type and mutant RNA-Seq datasets (Figure S6). Comparison of wild type and mutant RIIs indicated that 3978 introns were retained at higher levels in the mutant cerebellum (RII ratio ≥ 2; p<0.05, Figure 7B and Table S1F), including intron 27 of L1CAM (RII ratio = 3.55; p = 0.0012). In contrast, we identified only 362 introns that were retained at higher levels in the wild-type cerebellum (RII ratio ≥ 2; p<0.05). To check the validity of our analysis, we performed RT-PCR on 23 introns that were present at higher levels in the mutant transcriptome by RNA-Seq and all were validated (Figure S6D, S6E, and data not shown). Furthermore, 19/23 (82.61%) of these introns, like intron 27 of LICAM, and 5/7 RT-PCR-validated introns identified by exon array, were also present in wild-type cerebellum as a minor isoform (Figure S6A and S6D). These results indicate that these introns represent alternative splicing events and are likely to be suboptimal pre-mRNA substrates even in presence of the wild-type spliceosome. Previous studies have shown that small introns are prone to being retained in human and mouse transcripts in physiological conditions (Sakabe and de Souza, 2007). These prompted us to check the size distribution of the intron retention events identified by RNA-Seq. Indeed, 30.62% of the retained introns in the mutant cerebellum were less than 100 bps, which largely differs from the size distribution of introns in the mouse genome (RefSeq; 5.31%<100 bps) or those significantly retained in the wild type cerebellum (3.31%<100 bps) (Figure 7C). As expected, differences in splicing of small introns removed by the U12-dependent minor spliceosome were not observed in the mutant cerebellum (Figure S6F). Thus, both exon array and RNA-Seq analysis demonstrated that a group of alternative splicing events, including the splicing of small introns, were altered in the NMF291−/− cerebellum.
To check the overlap of our exon array and RNA-Seq analysis, we compared intron retention events (Table S1H). 96.4% (27/28) of these events detected by exon array displayed differences in the same direction in RNA-Seq data, although only 46.4% (13/28) passed our stringent threshold set for RNA-Seq analysis. Although differential events in other splicing event categories ascertained by both analyses were clearly validated by RT-PCR, little overlap was observed in the datasets, likely due to the different experimental and analysis parameters used in the two methods (see Extended Experimental Procedures).
The role of U2 snRNA as a basal component of the spliceosome would predict that splicing of transcripts encoding proteins across many functional classes would be disrupted in the mutant cerebellum. Indeed, splicing alterations between the wild type and mutant cerebellum occur in genes with many predicted cellular roles (Table S1B and S1D–S1F). However, Gene Ontology (GO) analysis of the differentially expressed junctions and spliced introns detected by RNA-Seq and exon array analysis revealed that genes annotated to GO terms of nucleoplasm (GO:0005654) were significantly enriched, as were those annotated to ribonucleotide binding (GO:0032553) (Table S1G). Interestingly, after these two functional clusters, the most significantly enriched genes in the differentially spliced intron data set were annotated to mRNA processing (GO:0006397), including those annotated to the spliceosomal complex (GO:0005681) (Figure 7D and Table S1G). Differentially expressed exons and genes also showed significant enrichment for mRNA processing, although at higher p-values.
DISCUSSION
We demonstrate that a mutation in one of the multicopy mouse U2 snRNA genes causes defects in pre-mRNA splicing leading to neurodegeneration. U2 snRNAs play an essential role in formation of the catalytically active spliceosome by base pairing with both the intron branch point and the U6 snRNA (Wahl et al., 2009). Expression of the mutant U2 snRNA alters pre-mRNA splicing at selective splice sites that are often associated with alternative splicing demonstrating that U-snRNA dysfunction, like downregulation of core spliceosomal proteins, can influence splice site choice (Corioni et al., 2011; Park et al., 2004; Saltzman et al., 2011; Shaw et al., 2007).
One of the main pathological features of the NMF291−/− cerebellum is the increased retention of small introns, which likely represent a unique spliceosomal substrate. Unlike splicing of large introns, which is thought to first occur by pairing of splice sites across the exons (“exon definition”), splicing of short introns likely occurs by pairing of 5' and 3' splice sites across the intron (“intron definition”) (Berget, 1995). Furthermore, even under normal physiological conditions many of the highly retained introns in the mutant cerebellum are not fully spliced, consistent with the observation that weak splicing sites often flank small introns (Lim and Burge, 2001; Sakabe and de Souza, 2007). Given the disruption of short intron splicing and other alternative splicing events, it is plausible that mutant U2 snRNPs are fully functional on optimal, but not suboptimal, substrates (Figure 7E).
Interestingly, we found that mRNA-processing genes were significantly enriched among the alternative splicing events differentially expressed between the wild type and mutant cerebellum, suggesting that neurons may utilize alternative splicing to regulate the function of proteins involved in RNA splicing and processing in an effort to restore splicing homeostasis. Indeed, studies have shown that a number of splicing regulators autoregulate their expression and activity via transcriptional feedback loops and alternative splicing (Ni et al., 2007; Saltzman et al., 2011; Wollerton et al., 2004). However, rather than reestablishing homeostasis, these splicing alterations could in fact act to amplify the amount of abnormal splicing and ultimately prove deleterious. It will be intriguing to see whether the expression level and/or alternative splicing status of these mRNA-processing genes is also affected in SMA patients or ALS/FTD patients with mutations in TDP-43 or FUS/TLS.
The ultimate cause of neuron death in the mutant cerebellum is unclear. Cell death could be caused by the generation of proteins with altered function or by the production of RNAs containing abnormal sequences, which could themselves be toxic as previously reported for trinucleotide repeat expansion diseases (Li et al., 2008). Retained introns may also contribute to neuron death by sequestering splicing regulators and/or other RNA binding proteins. In addition, many of these introns likely harbor premature translation termination codons (PTCs) that would be predicted to trigger the nonsense-mediated mRNA-decay (NMD) pathway. In addition to its role in degradation of abnormal transcripts, NMD also regulates many natural PTC-containing transcripts, including those involved in synaptic physiology and cellular stress (Gardner, 2010; Giorgi et al., 2007). Enhanced and/or prolonged NMD activation could overwhelm the NMD pathway, causing dysregulation of natural NMD targets that might be essential for cell survival, or itself cause cellular stress.
Finally, our data demonstrate the potential for disease-causing mutations in multicopy genes. While a single U2 gene is present in yeast, multiple copies of these genes exist in higher organisms, each producing identical or nearly identical products. Indeed, the region on human Chromosome 17p21 homologous to the (Rnu2–6 to Rnu2–10-containing) region on mouse Chromosome 11 also contains a cluster of stably inherited U2 genes that vary in copy number from 5 to 25 (Van Arsdell and Weiner, 1984; Westin et al., 1984). In addition to our findings of temporal and spatial regulation of a mouse U2 gene, developmental regulation of other snRNA genes, including U2 snRNA genes, has been reported (Forbes et al., 1984; Lund et al., 1985; Sierra-Montes et al., 2005; Stefanovic et al., 1991). Furthermore, the developmental arrest associated with homozygosity for a mutation in one of twelve C. elegans U1 genes raises possibility of differences in expression and/or function between C. elegans U1 genes (Zahler et al., 2004). Whether individual human U2s (or other multicopy genes) are differentially expressed during development, in different cell types, or even as a result of pathogenic processes is unknown. However, the potential for discrete regulation of individual members of multigene families, combined with their potential for copy number variation, increases the prospect of uncovering novel disease-causing mutations in repetitive genes.
EXPERIMENTAL PROCEDURES
Mice and mouse U2 snRNA genes
The NMF291 mutant strain was derived from EMS-treated ES cells and a two-generation mating scheme, as described previously (Munroe and Schimenti, 2009). Genetic mapping of the NMF291 mutation was performed using an intersubspecific intercross (B6; 129 NMF291 × CAST/Ei). The NMF291 critical interval contains 5 mouse U2 snRNA genes. GenBank accession numbers for these genes are: Rnu2–6 (JN863957), Rnu2–7 (JN863958), Rnu2–8 (JN863956), Rnu2–9 (JN863959), and Rnu2–10 (JN863960).
Histology Analysis
Hematoxylin and eosin staining was performed on Bouin's fixed tissue. Cleaved Caspase-3 (Cell Signaling) immunostaining and TUNEL assays (Roche) were performed as described previously (Zhao et al., 2005). At least three mice of each genotype were used for all histological analyzes.
RNase Protection Assay, Northern Blot and RT-PCR assays
Total RNA was extracted by Trizol (Invitrogen) and treated with DNase I (Ambion). RPA analysis was performed as suggested by the manufacturer's instructions (Ambion). For northern blots, total RNA was separated by 10% denaturing polyacrylamide gel and the membrane was hybridized with endlabeled DNA oligo probes. RT-PCR was performed on random-primed cDNA (Invitrogen). Density of bands in scanned X-ray films was determined by ImageQuant 5.2 software (Amersham).
Constructs, Cell Culture, Transfection and Luciferase assays
The mutant Rnu2–8 DNA fragment used for generating transgenic mice and the corresponding wild type DNA fragment were cloned into pCR2.1-TOPO for expression of wild type and mutant U2 snRNA in cultured cells. The Δ3, Δ6, and Δ9 U2 snRNA and ISS- expression constructs were generated by sitedirected mutagenesis (Stratagene). HEK293T cellswere used for transient transfection (Lipofectamine 2000, Invitrogen). Luciferase and β-galactosidase activities (Promega) were measured in a multilabel counter 48 hours after transfection. Statistical significance was determined by ANOVA analysis (SPSS).
Exon Array, RNA-Seq and Functional GO Analysis
Data treatment of Mouse Exon 1.0 ST Arrays (Affymetrix) was done by the EASANA® analysis and the interface visualization system (GenoSplice Technology, www.genosplice.com), which is based on the FAST DB® annotations (de la Grange et al., 2007). Probe selection and statistical analysis were preformed as previously described (Clark et al., 2007; de la Grange et al., 2010). The GEO (Gene Expression Omnibus) accession number for our exon array data is GSE33069. For RNA-Seq, mRNA purification and DNA library preparation were performed according to the manufacturer's protocol (Illumina). The library was sequenced on Illumina GAIIX using paired-end read strategy. For de novo identification of junctions, the reads were inputted to SpliceMap 3.3.6 (Au et al., 2010) and aligned to the mouse reference genome. A workflow chart of our RNA-Seq analysis is shown in Figure S6G. RefSeq intron length information was extracted from UCSC. The Gene Ontology analysis was conducted by using DAVID bioinformatics resources 6.7 (Huang da et al., 2009).
Supplementary Material
Acknowledgements
We thank The Jackson Laboratory sequencing, microchemistry, histology, microinjection, and multimedia services for their assistance. We also thank W. H. Wong for discussions on RNA-Seq analysis, J. Blake and C. Bult for discussion on GO analysis, L. Beverly-Staggs and J. Cook for mouse husbandry assistance, P. de la Grange for exon array analysis, Jesse Hammer and L. Zhao for graphics assistance, John E. G. McCarthy and M. Talat Nasim for plasmids, and R. Burgess, G. Cox and M. Hibbs for comments on the manuscript. This work was partially supported by an institutional National Cancer Institute core grant (JAX) and the work of John C. Mu was partially supported by NIH grants R01HG004634 and R01HG005717. S.L.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Au KF, Jiang H, Lin L, Xing Y, Wong WH. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38:4570–4578. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Bult C, Kibbe WA, Snoddy J, Vitaterna M, Swanson D, Pretel S, Li Y, Hohman MM, Rinchik E, Takahashi JS, et al. A genome end-game: understanding gene function in the nervous system. Nat Neurosci. 2004;7:484–485. doi: 10.1038/nn0504-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko E, Ranganathan S. Comprehensive splicing graph analysis of alternative splicing patterns in chicken, compared to human and mouse. BMC Genomics. 2009;10(Suppl 1):S5. doi: 10.1186/1471-2164-10-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corioni M, Antih N, Tanackovic G, Zavolan M, Kramer A. Analysis of in situ pre-mRNA targets of human splicing factor SF1 reveals a function in alternative splicing. Nucleic Acids Res. 2011;39:1868–1879. doi: 10.1093/nar/gkq1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Grange P, Dutertre M, Correa M, Auboeuf D. A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC Bioinformatics. 2007;8:180. doi: 10.1186/1471-2105-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues. Nucleic Acids Res. 2010;38:2825–2838. doi: 10.1093/nar/gkq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332:240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochem Soc Trans. 2008;36:590–594. doi: 10.1042/BST0360590. [DOI] [PubMed] [Google Scholar]
- Forbes DJ, Kirschner MW, Caput D, Dahlberg JE, Lund E. Differential expression of multiple U1 small nuclear RNAs in oocytes and embryos of Xenopus laevis. Cell. 1984;38:681–689. doi: 10.1016/0092-8674(84)90263-0. [DOI] [PubMed] [Google Scholar]
- Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332:238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kollmus H, Flohe L, McCarthy JE. Analysis of eukaryotic mRNA structures directing cotranslational incorporation of selenocysteine. Nucleic Acids Res. 1996;24:1195–1201. doi: 10.1093/nar/24.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lemmens R, Moore MJ, Al-Chalabi A, Brown RH, Jr., Robberecht W. RNA metabolism and the pathogenesis of motor neuron diseases. Trends Neurosci. 2010;33:249–258. doi: 10.1016/j.tins.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr., Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci U S A. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Kahan B, Dahlberg JE. Differential control of U1 small nuclear RNA expression during mouse development. Science. 1985;229:1271–1274. doi: 10.1126/science.2412294. [DOI] [PubMed] [Google Scholar]
- Manser T, Gesteland RF. Human U1 loci: genes for human U1 RNA have dramatically similar genomic environments. Cell. 1982;29:257–264. doi: 10.1016/0092-8674(82)90110-6. [DOI] [PubMed] [Google Scholar]
- Marz M, Kirsten T, Stadler PF. Evolution of spliceosomal snRNA genes in metazoan animals. J Mol Evol. 2008;67:594–607. doi: 10.1007/s00239-008-9149-6. [DOI] [PubMed] [Google Scholar]
- McGrail JC, Tatum EM, O'Keefe RT. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J. 2006;25:3813–3822. doi: 10.1038/sj.emboj.7601258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P, Jr., Wu JY. Pre-mRNA splicing and retinitis pigmentosa. Mol Vis. 2006;12:1259–1271. [PMC free article] [PubMed] [Google Scholar]
- Munroe R, Schimenti J. Mutagenesis of mouse embryonic stem cells with ethylmethanesulfonate. Methods Mol Biol. 2009;530:131–138. doi: 10.1007/978-1-59745-471-1_7. [DOI] [PubMed] [Google Scholar]
- Nasim MT, Chowdhury HM, Eperon IC. A double reporter assay for detecting changes in the ratio of spliced and unspliced mRNA in mammalian cells. Nucleic Acids Res. 2002;30:e109. doi: 10.1093/nar/gnf108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci U S A. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelitz T, Rusche L, Matera AG, Scharf JM, Weiner AM. Concerted evolution of the tandem array encoding primate U2 snRNA occurs in situ, without changing the cytological context of the RNU2 locus. The EMBO journal. 1995;14:169–177. doi: 10.1002/j.1460-2075.1995.tb06987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DE, Abelson J. The conserved central domain of yeast U6 snRNA: importance of U2-U6 helix Ia in spliceosome assembly. Rna. 2002;8:997–1010. doi: 10.1017/s1355838202025013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 2011;25:373–384. doi: 10.1101/gad.2004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauterer RA, Feeney RJ, Zieve GW. Cytoplasmic assembly of snRNP particles from stored proteins and newly transcribed snRNA's in L929 mouse fibroblasts. Exp Cell Res. 1988;176:344–359. doi: 10.1016/0014-4827(88)90336-9. [DOI] [PubMed] [Google Scholar]
- Shaw SD, Chakrabarti S, Ghosh G, Krainer AR. Deletion of the N-terminus of SF2/ASF permits RS-domain-independent pre-mRNA splicing. PLoS One. 2007;2:e854. doi: 10.1371/journal.pone.0000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra-Montes JM, Pereira-Simon S, Smail SS, Herrera RJ. The silk moth Bombyx mori U1 and U2 snRNA variants are differentially expressed. Gene. 2005;352:127–136. doi: 10.1016/j.gene.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Konarska MM, Query CC. Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast. Mol Cell. 2009;34:333–343. doi: 10.1016/j.molcel.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B, Li JM, Sakallah S, Marzluff WF. Isolation and characterization of developmentally regulated sea urchin U2 snRNA genes. Dev Biol. 1991;148:284–294. doi: 10.1016/0012-1606(91)90337-3. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arsdell SW, Weiner AM. Human genes for U2 small nuclear RNA are tandemly repeated. Mol Cell Biol. 1984;4:492–499. doi: 10.1128/mcb.4.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G, Zabielski J, Hammarstrom K, Monstein HJ, Bark C, Pettersson U. Clustered genes for human U2 RNA. Proc Natl Acad Sci U S A. 1984;81:3811–3815. doi: 10.1073/pnas.81.12.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Tuttle JD, Chisholm AD. Genetic suppression of intronic +1G mutations by compensatory U1 snRNA changes in Caenorhabditis elegans. Genetics. 2004;167:1689–1696. doi: 10.1534/genetics.104.028746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet. 2005;37:974–979. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.