Abstract
Three-week-old plants of two unrelated lines of maize (Zea mays L.) and their hybrid were submitted to progressive water stress for 10 d. Changes induced in leaf proteins were studied by two-dimensional electrophoresis and quantitatively analyzed using image analysis. Seventy-eight proteins out of a total of 413 showed a significant quantitative variation (increase or decrease), with 38 of them exhibiting a different expression in the two genotypes. Eleven proteins that increased by a factor of 1.3 to 5 in stressed plants and 8 proteins detected only in stressed plants were selected for internal amino acid microsequencing, and by similarity search 16 were found to be closely related to previously reported proteins. In addition to proteins already known to be involved in the response to water stress (e.g. RAB17 [Responsive to ABA]), several enzymes involved in basic metabolic cellular pathways such as glycolysis and the Krebs cycle (e.g. enolase and triose phosphate isomerase) were identified, as well as several others, including caffeate O-methyltransferase, the induction of which could be related to lignification.
Water availability is a major limiting factor for plant growth. Limited water availability leads to reduced growth of aerial parts and, to a lesser extent, of the root system. Several other responses have been described, such as stomatal closure and synthesis of osmolytes (e.g. betaine and Pro). These responses are at least partly controlled by ABA, a phytohormone that increases in concentration in plants subjected to water deficit (Zeevaart and Creelman, 1988).
Numerous genes expressed in response to water deficit and/or ABA in different species encode RAB (Responsive to ABA) proteins or dehydrins that exhibit high hydrophilicity and contain repeated domains. Most of these genes are also highly expressed during late embryogenesis (lea genes). Sequence features allowed their classification into different groups (Close et al., 1989; Dure et al., 1989), and tentative functions were proposed, according to the predicted protein structure (Dure, 1993; Lisse et al., 1996), to be sequestration of ions or water or preservation of membrane or protein structure (chaperone function). Other proteins with functions related to water deprivation were found to be induced by water deficit or saline stress: e.g. proteins showing domain or sequence similarities to transmembrane channel proteins (Guerrero et al., 1990; Yamaguchi-Shinozaki et al., 1992; Fray et al., 1994; Ruiter et al., 1997) and betaine aldehyde dehydrogenase, which catalyzes the last step of betaine synthesis (Weretilnyk and Hanson, 1990; Ishitani et al., 1995).
Water deficit also induces the expression of proteins not specifically related to this stress, but rather to reactions against cell damage. These include different classes of heat-shock protein genes or cognates (Heikkila et al., 1984; Almoguera and Jordano, 1992; Kiyosue et al., 1994), thiol proteases (Guerrero et al., 1990; Williams et al., 1994), proteinase inhibitors (Downing et al., 1992; Reviron et al., 1992), and osmotin (Kononowicz et al., 1993). In maize (Zea mays L.) a ferritin gene induced by iron stress is also induced by drought and ABA (Fobis-Loisy et al., 1995).
Finally, several genes encoding proteins with functions not directly related to stress were shown to be expressed at greater levels in response to drought or salinity stress: several enzymes involved in glycolysis (Umeda et al., 1994; Velasco et al., 1994) and in the synthesis of Met (Glaser et al., 1993), SAM (Espartero et al., 1994; Chang et al., 1995), peroxidases (Botella et al., 1994), nonspecific lipid transferases (Torres-Schumann et al., 1992; Ouvrard et al., 1996), and early light-induced proteins (Bartels et al., 1992; Ouvrard et al., 1996).
Although the simultaneous changes in gene expression and physiological responses strongly suggest that induced proteins play a role in these responses, the correlation between their expression and the level of stress tolerance in the different genotypes has been rarely studied (Ramagopal, 1987; Hurkman et al., 1989; Moons et al., 1995). As a first step in such a study, we have undertaken the characterization of protein responses of two maize lines and their F1 hybrid. These lines, which display contrasting behavior in response to water stress in the field, are the parents of a population of recombinant inbred lines, which will be used for analyzing the relationship between protein induction and stress tolerance.
In the vast majority of studies reporting the induction of gene expression upon water deprivation, osmotic stress, or ABA treatment, young seedlings were submitted to abrupt stress. However, in the field plants are submitted to more gradual stress because water availability in the soil does not change abruptly, and, therefore, responses might be different. For example, Leone et al. (1994) showed that different sets of polypeptides were synthesized in potato cells submitted to abrupt or gradual osmotic stress. In the present study we submitted autotrophic plants (approximately 3 weeks old) to a gradual dehydration. The protein response of the two maize lines and their hybrid was studied by 2DE, and computer-assisted quantitative analysis allowed the detection of proteins with accumulation altered by drought. Nineteen induced proteins were microsequenced.
MATERIALS AND METHODS
Plant Material
Two genetically distant lines of maize (Zea mays L.) and their hybrid were used: F2, a flint line from the Institut National de la Recherche Agronomique (France) referred to as Lc, and Io, an American dent line from the Iodent group. The parental lines were chosen for their differential response to water deprivation in the fields: the percentage of yield decrease in nonirrigated conditions was greater for Lc than for Io.
For the quantitative analysis the two parental lines and their hybrid were grown in four successive batches in a growth cabinet under controlled conditions (450 μmol m−2 s−1 irradiance during the 16 h-photoperiod, 25°C day and 20°C night temperatures, and 60% RH). Plants were grown in perlite (one plant per 14-cm-diameter pot) and watered with nutrient solution.
The plant material required for microsequencing was obtained from approximately 500 plants of the 2 parental lines in 5 successive batches. They were grown in a greenhouse (1 plant per 14-cm-diameter pot containing soil) at uncontrolled temperature and RH.
Both in the cabinet and in the greenhouse, a subset of plants was stressed by withholding watering at the fifth- leaf stage, corresponding approximately to 3-week-old plants. After 10 d of stress, 2 to 3 cm of the etiolated part of the blade of the sixth leaf was harvested and immediately frozen in liquid nitrogen for further protein analysis.
ABA Content and Water Potential Measurement
ABA content was determined according to the method of Quarrie et al. (1988). Leaf water potential was measured with a portable pressure chamber (PMS Intrument, Corvallis, OR).
Protein Extraction and Gel Electrophoresis
Analytical 2DE
Denaturing protein extraction was applied as described in Damerval et al. (1986), except that 60 μL of resolubilization solution was used to resuspend 1 mg of pellet. The isoelectric and SDS-PAGE dimensions were as described by Damerval et al. (1987) with a protein load of about 45 μg. Gels were silver stained according to the method of Damerval et al. (1987) as modified by Burstin et al. (1993).
Preparative 2DE for Microsequencing
To facilitate isolation of proteins of low abundance, enriched fractions were obtained by differential precipitation with acetone. Proteins were first solubilized in a Tris-HCl buffer, pH 8.7, according to the method of Zivy et al. (1983), and different fractions were obtained by gradually increasing the acetone concentration in the solution. The fractions were compared by analytical 2DE, and those showing the highest relative concentration for the protein to be microsequenced were used for preparative 2DE. Preparative 2DE was similar to analytical 2DE except for the following modifications: about 500 μg of protein was loaded on IEF gels (3 mm diameter) and the slab gels were 1.5 mm thick, with stacking gels. The gels were stained with Amido black according to a procedure adapted from Chen et al. (1993) prior to in situ gel digestion. Depending on the abundance of the protein, spots were taken from 5 to 30 preparative gels for microsequencing.
Protein Microsequencing and Search for Similarities in Amino Acid Sequences
Internal amino acid sequences of leaf proteins were obtained from Drs. J. d'Alayer and M. Davi at the Laboratoire de Microséquençage des Protéines, Institut Pasteur (Paris, France). The procedure is described in Touzet et al. (1996b). Amino acid sequences were compared with the sequences in the Non Redundant Peptide Sequences Database of the National Center for Biotechnology Information by using the BLAST program (Altschul et al., 1990).
2DE Quantitative Analysis
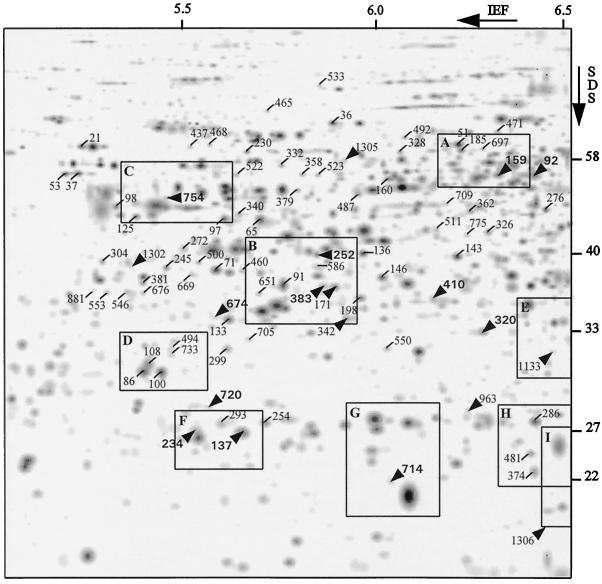
Wet, silver-stained gels were scanned (model 7899, Eikonix, MA) with a spatial resolution of 1 pixel/100 μm and an optical density range from 0.0 to 1.2. Image treatment, spot detection, and quantification were done using the Kepler package (LSB Corp., Rockville, MD). Spot detection followed a method developed by M. Zivy (unpublished data). Quantification was based on the modeling of spots by two-dimensional Gaussians, with parameters fitted to the image obtained after background subtraction. To compensate for staining variation between gels, spot intensities of each gel were scaled relative to the sum of spot intensities within a rectangle defined in the same way for each gel. The upper left and bottom right angles of this rectangle were at approximately the positions of spots 21 and 714, respectively (Fig. 1). As shown by Damerval (1994), the intensity of the large majority of proteins is linearly related to the protein quantity when this method of silver staining is used. Only the spots with responses not affine to the mean response would not be correctly scaled (Burstin et al., 1993). Most proteins that do not have a linear response to silver stain show a plateau; for these proteins, the computed induction ratio is a minimum value of induction.
Figure 1.
Computer image of a silver-stained 2DE gel of leaf proteins from the hybrid Io × Lc subjected to water stress in a growth cabinet. Numbered spots correspond to proteins in which quantity was altered during water stress (increased or decreased). Arrows indicate sequenced proteins induced in both growing conditions (bold type) or only in the greenhouse (normal type). Boxes correspond to gel regions enlarged in Figure 2. Molecular masses (in kilodaltons) are indicated.
One two-dimensional gel per plant was run. Considering the three genotypes (Io, Lc, and the hybrid) and the two treatments (irrigated and nonirrigated), six genotype × treatment combinations were studied. Fifty-six two-dimensional gels were used, approximately 10 gels per genotype × treatment combination. The four batches of cultivation counted 10, 16, 15, and 15 plants, and each genotype × treatment combination was represented in each batch. To take into account possible genotype × batch interactions, two models of analysis of variance were used:
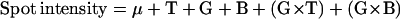 |
1 |
where T, G, B, G × T, and G × B stand for treatment, genotype, batch effect, genotype × treatment, and genotype × batch interactions, respectively. As only two spots (s143 and s254) showed a significant G × B interaction, a more simple model was used for the other spots:
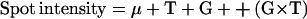 |
2 |
Spots were retained in the analysis when the treatment effect was significant at P < 0.01 or when the G × T interaction was significant at P < 0.05.
RESULTS
Plant Responses to Water Stress
Plants Grown in the Growth Cabinet
Watering was stopped when the fifth leaf was emerging. At this stage, plants from the two unrelated lines and their hybrid had three ligulated leaves. The height of the last ligule was on average 10.8 cm, although Io was 3 cm shorter than Lc and the hybrid (P < 0.0001). During the following 10 d, growth was substantially lower in stressed plants than in controls (P < 0.0001): height increase of the last ligule was 4.0 and 10.0 cm, respectively, for stressed and control plants. In the same time, stressed and control plants produced, respectively, 1.2 and 2.8 emerged leaves (P < 0.0001). In both control and stress conditions, the height increase was significantly less in Io than in the hybrid and Lc (difference of approximately 2 cm), whereas Lc plants produced 0.8 fewer new leaves than Io and the hybrid. A significant genotype × treatment interaction was found for the number of ligulated leaves: Lc plants produced 0.7 fewer ligulated leaves than Io or the hybrid in control conditions, but the same number in stress conditions.
The percentage of dry leaves was significantly smaller in stressed Lc plants (34%) than in the two other genotypes (46%), which is consistent with data from studies conducted in the field. ABA content in the 6th leaf was significantly higher in stressed plants (715 ng/g of dry matter) than in controls (227 ng/g). Water content ([fresh weight − dry weight]/[dry weight]) was also significantly lower in stressed plants (6.0) than in controls (12.4). ABA and water content showed no significant genotype effect.
Plants Grown in the Greenhouse
After 10 d of water deprivation, the average measures in the sixth leaf of stressed plants and controls were, respectively, −1.5 and −0.3 MPa for leaf water potential, 73.9% and 94.6% for relative water content, 7.0 and 14.0 for water content, and 1321 and 244 ng/g of dry matter for leaf ABA content. No significant genotype effect was observed.
Plants grown in the greenhouse were more stressed than those cultivated in the growth cabinet; not only was their ABA content almost doubled, but they also exhibited much more leaf rolling. Other experiments with the same genotypes cultivated in the same way confirmed that the 10 d of water stress was more severe in the greenhouse than in the growth cabinet: the water potential of stressed leaves was generally between −0.8 and −1.0 MPa in the growth cabinet compared with −1.5 MPa in the greenhouse, and plant growth was more quickly inhibited in the greenhouse than in the growth cabinet.
Quantitative Analysis of the Protein Responses
Out of 413 spots reproducibly detected, 78 were affected by drought (Table I); of these, 50 were increased, and of these, 10 were present only in stressed plants. Twenty-three other proteins were decreased, and 5 showed no treatment effect but did show a significant genotype × treatment interaction. Stress-affected proteins are shown in Figure 1 and several examples of protein induction are shown in Figure 2.
Table I.
Leaf proteins affected by 10 d of water stress
| Treatment Effecta | Genotype Effect | Interaction | Spot No. |
|---|---|---|---|
| ↑ P/A | Io and Lc | s159 s374 s383 s487 s720 | |
| ↑ P/A | Io only | s705 s714 s733 s775 | |
| ↑ P/A | Lc only | s494 | |
| ↑ **b | s21 s37 s86 s97 s100 s108 s133 s137 s143 s252 s272 s293 s304 s320 s340 s410 s492 s500 s553 s676 s709 s754 s881 | ||
| ↑ ** | * | s92 s136 s586 s674 | |
| ↑ ** | ** | s36 s53 s234 s358 s481 | |
| ↑ ** | * | s91 s245 s468 s533 | |
| ↑ ** | ** | * | s71 s198 |
| ↑ ** | ** | ** | s254 s550 |
| ↓ ** | s98 s230 s299 s328 s332 s362 s437 s460 s471 s523 s546 s669 | ||
| ↓ ** | * | s160 s185 s465 s522 | |
| ↓ ** | ** | s125 s697 | |
| ↓ ** | * | s65 | |
| ↓ ** | * | * | s146 |
| ↓ ** | ** | * | s511 s651 |
| ↓ ** | ** | ** | s276 |
| * | ** | s381 | |
| * | s51 s286 s326 s379 |
For proteins present in stressed plants, but absent in controls (P/A), the genotype column contains the parental lines in which they were observed.
Treatment effects are as follows: ↑, increased proteins in stressed plants; ↓, decreased protein in stressed plants; and P/A, present in stressed plants but absent in controls.
Statistical significance, P < 0.01 (**) and P < 0.05 (*) for treatment, genotype, and interaction effects in the analysis of variance.
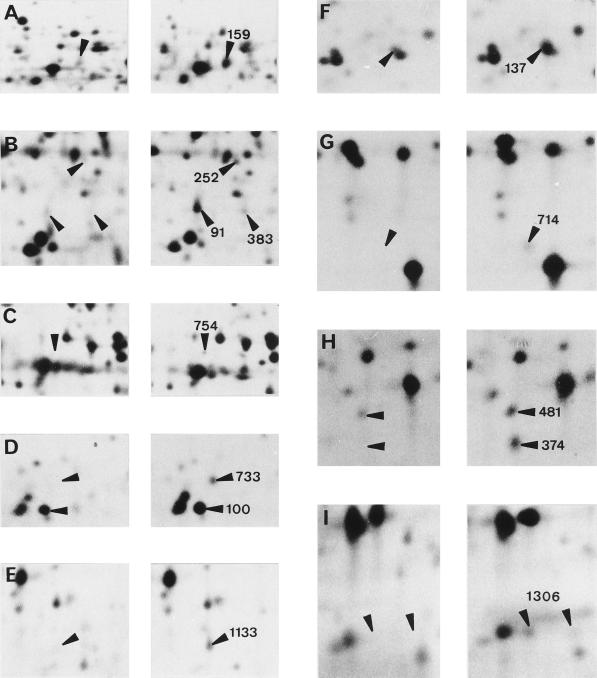
Figure 2.
Examples of protein induction in leaves of the maize line Io in response to 10 d of water deprivation in a control plant (A–E) and in a stressed plant (F–I). Letters correspond to the boxes in Figure 1. All plants except that shown in I were grown in a growth cabinet. Peptides from spots 159 (A), 252 (B), 383 (B), 754 (C), 1133 (E), 137 (F), 714 (G), and 1306 (I) were microsequenced (see Table II).
As shown in Table I, 38 proteins showed a different response according to the genotype: 5 proteins were synthesized by only 1 of the parental lines in response to water stress. The other 33 proteins showed a genotype effect in addition to the treatment effect and/or a genotype × treatment interaction. The quantities of proteins showing a significant genotype effect but no interaction were different between genotypes whatever the treatment, but showed the same increase (or decrease) in response to water stress. The existence of a significant interaction indicates that the response to the stress was not the same for the three genotypes. Tests of multiple comparisons of means (the Student-Newman-Keuls method) allowed us to study proteins showing significant genotype × treatment interaction. For all of them the interaction was due to a differential increase or decrease of protein by drought in the different genotypes. No protein was significantly increased in one genotype and significantly decreased in the other genotypes.
Identification of a Subset of the Induced Proteins
Two-dimensional analytical gels of the parental lines grown in the greenhouse were visually analyzed and compared with the one used for quantitative analysis. Among the proteins that reproducibly increased in quantity under water stress in both culture conditions, a subset of 12 was selected for amino acid sequencing. Four of them were detected only in stressed plants (s159, s383, s714, and s720), and in the others the induction factor (intensity in stressed plants/intensity in control plants) varied from 1.3 to 5.0 (Table II).
Table II.
Induction level and sequence similarity of proteins induced by water stress in leaves of maize lines Io and Lc
| Protein | Genotype | Induction Factora | Amino Acid Sequence | Description | Species | Amino Acid Identity | Reference |
|---|---|---|---|---|---|---|---|
| % | |||||||
| s92 | Io and Lc | 3.3 | KGENGXIGLAFDVMGR | β-Glucosidase, glu2 gene (EC 3.2.1.21) | Maize | 93 | Bandaranayake and Esen (1996) |
| KNWLTFNEPQTFTSFS | β-Glucosidase, glu2 gene (EC 3.2.1.21) | Maize | 100 | Bandaranayake and Esen (1996) | |||
| s137 | Io and Lc | 2.1 | KEVLSGVVFQPFEEIK | Ferritin | Maize | 100 | Lobreaux et al. (1992) |
| s159 | Io and Lc | Sb | KVCFDNFGDK | β-Glucosidase (EC 3.2.1.21) | Maize | 100 | Esen (1992) |
| s171 | Lc | – | KMELVDAAFPLLK | Putative cytoplasmic malate dehydrogenase (EC 1.1.1.37) | Maize | 100 | Keith et al. (1993) |
| s234 | Io and Lc | 1.3 | KDWSNVVLAYEPVWAI | Cytosolic triose phosphate isomerase (EC 5.3.1.1) | Maize | 93 | Marchionni and Gilbert (1986) |
| s252 | Io and Lc | 5.0 | KSVGGQPVVFDSVK | Glu 1-semialdehyde 2,1-aminotransferase (EC 5.4.3.8) | Barley | 92 | Grimm (1990) |
| s320 | Io and Lc | 1.7 | KHSLGQSHPVLLTRHN | ABA-induced protein | Rice | 68 | Moons et al. (1996) |
| s342 | Io and Lc | – | KVLEGAEERLQLLK | NIc | |||
| s383 | Io and Lc | S | KWILHDWSDAHXATLL | COMT (EC 2.1.1.6) | Maize | 93 | Collazo et al. (1992) |
| s410 | Io and Lc | 3.7 | KTIASPGRGILAMDES | Chloroplastic Fru bisphosphate aldolase (EC 4.1.2.13) | Pea | 100 | Razdan et al. (1992) |
| Chloroplastic Fru bisphosphate aldolase (EC 4.1.2.13) | Rice | 100 | Tsutsumi et al. (1994) | ||||
| s674 | Io and Lc | 1.7 | KGKVXIFIGGIGTGGT | Cys synthase, chloroplastic precursor (EC 4.2.99.8) | Spinach | 81 | Saito et al. (1993) |
| s714 | Io | S | KQHLGEAGAIAAGAFA | ASR 2 | Tomato | 60 | Amitai-Zeigerson et al. (1994) |
| Water-inducible protein DS2 | S. chacoense B. | 60 | Silhavy et al. (1995) | ||||
| cDNA clone similar to ASR protein (Expressed sequence tag) | Maize | 94 | Shen et al. (1994) | ||||
| s720 | Io and Lc | S | KRAPKLNERILSSLSR | Soluble inorganic pyrophosphatase (EC 3.6.1.1) | Potato | 81 | du Jardin et al. (1995) |
| s754 | Io and Lc | 1.4 | KYNQLLRIEEELGDAA | Enolase (EC 4.2.1.11) | Maize | 100 | Lal et al. (1991) |
| s963 | Lc | – | KSLEGAFVLNQHQPAE | NI | |||
| s1133 | Io | S | KIATVEPVTMK | NI | |||
| KAVHNLVEAVSQHGVA | NI | ||||||
| KYVHVVTVDSHDFWFM | NI | ||||||
| s1302 | Io and Lc | S | KASIEARKPDFDAFID | Phosphoribulokinase (EC 2.7.1.19) | Wheat | 100 | Raines et al. (1989) |
| s1305 | Io and Lc | – | KLAGSYNMLGLNYY | β-Glucosidase, root meristem precursor (EC 3.2.1.21) | Maize | 100 | Brzobohaty et al. (1993) |
| s1306 | Io and Lc | S | KDDQHATATTGGAYGQQGHTGSAYGQ | RAB17-dehydrin DHN1 | Maize | 100 | Vilardell et al. (1990) |
Average intensity in stressed plants/average intensity in control plants. Not calculated for proteins induced only in the greenhouse (visual scoring).
S, Protein only detected in stressed plants.
NI, No identification.
Seven other proteins (s171, s342, s963, s1133, s1302, s1305, and s1306) that were reproducibly increased in the greenhouse but not in the cabinet were microsequenced. Among them, s1133, s1302, s1305, and s1306 were observed only in stressed plants. Therefore, a total of 19 proteins was selected, for which 22 internal amino acid sequences were obtained, with a length of 10 to 26 residues (Table II).
Sequence comparisons showed nine similarities to proteins previously characterized in maize with 93% to 100% identity: ferritin, COMT (EC 2.1.1.6), β-glucosidase (three spots, EC 3.2.1.21), RAB17, enolase (EC 4.2.1.11), cytosolic triose phosphate isomerase (EC 5.3.1.1), and a putative cytoplasmic NAD-malate dehydrogenase (EC 1.1.1.37). An ASR initially described in tomato can be added to this list because of its relationship to a maize expressed sequence tag.
Six proteins are newly described in maize in this paper, but have been described previously in other plant species: soluble inorganic pyrophosphatase (potato, EC 3.6.1.1), glutamate-1-semialdehyde 2,1-aminotransferase (barley, EC 5.4.3.8), phosphoribulokinase (wheat, EC 2.7.1.19), chloroplastic Cys synthase (spinach, EC 4.2.99.8), chloroplastic Fru 1,6-bisphosphate aldolase (pea and rice, EC 4.1.2.13), and an ABA-induced protein (rice). Although potato, spinach, and pea belong to taxonomic groups very distant from maize, 81% to 100% identity in amino acid sequences was found between the peptide fragments and pyrophosphatase, Cys synthase, and aldolase. Very good probabilities were given by the BLAST program for these identifications. For the tomato ASR protein, the probability value was only 0.85, but three other amino acid sequences obtained from maize coleoptiles confirm the relationship with this protein (Touzet et al., 1996b). Finally, the internal amino acid sequences of three other proteins did not match any sequence in the databases.
DISCUSSION
We described protein changes occurring in maize leaves after progressive dehydration of the plants. Quantitative analysis revealed 78 proteins showing a significant alteration. Forty proteins detected in the controls and in stressed plants exhibited a 1.1- to 5.0-fold increase on average in the three genotypes upon water stress, whereas 10 others were reproducibly detected only in stressed plants. However, this should not be interpreted as specific expression in stressed plants; some protein spots were present at such a low intensity in controls that they were not detected by the computer in most two-dimensional gels. The relative quantity of 23 proteins was found to decrease to 50% to 80% of the control in stressed plants: this could be due to the repression of the synthesis of some proteins, but also to differential turnover.
Comparison between protein responses of the three genotypes studied revealed different kinds of genetic variations (Table I). Several induced proteins were specific to one of the two parental lines. In two instances (s494/s733 and s37/s53), two proteins were close to each other on two-dimensional patterns, were mutually exclusive in the two parental lines, and were both present in the hybrid; therefore, these are likely to be allelic products (for review, see de Vienne et al., 1996). For the remaining genotype-specific increased proteins (e.g. s714 and s171, identified as ASR protein and malate dehydrogenase, respectively), no alternate allelic form was detected in the genotype in which they were missing: the absence of the protein could be due to a null allele or the presence/absence variation could be controlled by another locus. Also, the effect of water stress was accompanied by a genotype effect for some other proteins: although their constitutive level was different, these proteins were similarly increased or decreased by drought in the three genotypes. Finally, the response of a few other proteins exhibited a genotype × treatment interaction, i.e. the protein quantity was differentially modified by stress according to the genotype.
Microsequencing was performed to tentatively identify 19 proteins reproducibly induced upon water stress in growth cabinet and/or in greenhouse conditions. Two of these proteins, detected only in stressed plants, were already known to be induced under various conditions of water stress and/or by exogenous ABA in different species, but have only putative functions.
RAB 17 (s1306), first described in maize embryos during the maturation phase (Vilardell et al., 1990), is induced in immature embryos and plantlet leaves by exogenous ABA and water stress (Close et al., 1989; Pla et al., 1989; Vilardell et al., 1990). This protein, located in the nucleus and in the cytosol, may play a role in nuclear protein transport through binding with nuclear-localization signal peptides (Goday et al., 1994).
ASR protein (s714), an ABA/water-stress/ripening-related protein exclusively induced in the Io genotype, was initially described in tomato (Iusem et al., 1993; Amitai-Zeigerson et al., 1994; Rossi and Iusem, 1994). It is also induced by water stress in Solanum chacoense (Silhavy et al., 1995) and loblolly pine (Chang et al., 1995). Subcellular fractionation (Iusem et al., 1993) and the presence of a nuclear-targeting sequence motif (Silhavy et al., 1995) led the latter authors to suggest that this basic protein (pH 7.9) may be involved in the protection of DNA structure during water loss or in gene regulation upon stress by changing DNA topology. However, s714 and the pine protein are more acidic (pH 6.1, Fig. 1). ASR protein accumulated in both growing conditions, whereas RAB17 was synthesized only in the greenhouse, i.e. in more drastic conditions of water stress.
The functions of two other induced proteins can be related to stress, although not exclusively to water stress. Ferritin (s137) is an iron-storage protein encoded in maize by at least two different genes, fm1 and fm2. The expression of these two genes is induced by iron in roots and leaves, but only fm2 was found to be induced by ABA or rapid dehydration of plantlets (Lobreaux et al., 1992, 1993; Fobis-Loisy et al., 1995). Progressive water stress increased the protein quantity by a factor of 2.1 in our conditions. The protein pair s53/s37, a supposed pair of allelic proteins, was not microsequenced but the amino acid composition is close to that of oryzain, a thiol-protease (Touzet et al., 1996a). Thiol proteases have been found to be induced by water stress (Guerrero et al., 1990; Williams et al., 1994).
In most cases the other identified proteins were enzymes involved in important metabolic pathways of higher plants. Triose phosphate isomerase (s234) and enolase (s754), which were 1.3- and 1.4 fold-induced, respectively, and NAD-malate dehydrogenase (s171) are enzymes involved in glycolysis, the Krebs cycle, and/or the oxydative pentose phosphate pathway. Several enzymes of these ATP-generating pathways, including triose phosphate isomerase, were shown to be induced upon saline and water stress in cultured cells of rice (Umeda et al., 1994). This coordinated induction is thought to be essential for activation of the entire energy-producing pathway to maintain homeostasis in stressed cells. It is notable that enolase is also involved in the response to other environmental stresses, such as anaerobic stress of maize roots (Lal et al., 1991), heat shock in yeast (Iida and Yahara, 1985), heat shock, salt stress, ABA treatment, and water stress in the common ice plant (Forsthoefel et al., 1995). Like the ASR protein, enolase seems to be induced during tomato fruit ripening (Van der Straeten et al., 1991).
Protein s383, which was detected only in stressed plants, is related to COMT, a key enzyme in the biosynthesis of lignin monomers catalyzing methylation of cinnamic acids. In normal growth conditions the gene has been shown to be expressed in elongating tissues, where active lignification of vascular system elements occurs (Collazo et al., 1992; Vignols et al., 1995). COMT genes are induced by pathogen attacks (Gowri et al., 1991; Jaeck et al., 1992; Pellegrini et al., 1993; Gregersen et al., 1994), i.e. in conditions also inducing active lignification (Lange et al., 1995). Other identified enzymes induced upon progressive dehydration are involved in general cell metabolism such as glycolysis and photosynthesis, but their function can be related to the phenylpropanoid pathway and lignin biosynthesis.
Soluble inorganic pyrophosphatase (s720), among other functions, participates in the assimilation of mineral nutrients, especially in sulfate activation (Schmidt and Jäger, 1992), and can then be connected to another induced protein, Cys synthase (s674, induced 1.7-fold). The latter is directly involved in sulfate assimilation through conversion of O-acetyl-Ser and sulfide into Cys and acetate. Cys is a precursor of numerous sulfur-containing compounds in the cell, especially Met and SAM. SAM is a widespread compound required in various methylation reactions, particularly for the methylation of several derivatives of the phenylpropanoid pathway, including the methylation catalyzed by COMT. Several authors have shown that the SAM-synthetase gene and/or activity are stimulated under different stress conditions: salt stress in tomato (Espartero et al., 1994), water stress in pine (Chang et al., 1995), and fungal elicitor application in parsley (Kawalleck et al., 1992). Therefore, because the increased quantity of COMT suggests the induction of a lignification process during progressive water stress, it can be hypothesized that the increase of Cys synthase (and maybe also of inorganic pyrophosphatase) contributes to this process by providing a greater quantity of SAM precursors.
Another important enzyme implicated in the general metabolism of the cell was strongly induced upon water stress (induction factor of 5.0): glutamate semialdehyde aminotransferase (s252) catalyzes 5-aminolevulinate formation via the C5 pathway. 5-Aminolevulinate is a precursor of tetrapyrrole compounds: chlorophylls, phytochrome, and porphyric proteins (cytochromes, catalases, and peroxidases). Peroxidases are known to be involved in the final step of lignin biosynthesis through the oxidative polymerization of monolignols and are induced by different stresses such as saline stress (Botella et al., 1994). Thus, induction of glutamate semialdehyde aminotransferase by water deprivation could also be linked to a stress-induced-lignification response of plants. It should be pointed out that this enzyme can also be induced by light (Grimm, 1990; Sangwan and O'Brian, 1993; Ilag et al., 1994). A few enzymes induced in our conditions are located in chloroplasts or are related to photosynthesis: phosphoribulokinase, Cys synthase, and aldolase. It is thus possible that for unknown reasons, a differentiation of chloroplasts occurred in stressed, etiolated tissues. Note that an Early Light Induced Protein (ELIP) has been shown to be induced by water stress in green leaves (Bartels et al., 1992; Ouvrard et al., 1996). It cannot be excluded, however, that some of the detected variations are not a direct response to water stress but, rather, are secondary effects of the reduction of leaf growth, cell division, and elongation.
The three β-glucosidase spots revealed by 2DE as increasing upon water stress are products of two different maize genes: s1305, exclusively present in stressed plants grown in the greenhouse, exhibited an amino acid sequence 100% identical to the glu1 gene product, and the partial sequence of s159 was identical to the newly described protein encoded by the so-called glu2 gene (Bandaranayake and Esen, 1996). The partial sequence of s92 is related to a region common to the two gene products s92 and s159, but according to their molecular masses (52 kD) and pI (respectively, 6.3 and 6.4) they could correspond to different products of the same glu2 gene. However, s159 is detected only in stressed plants, whereas s92 presented a 3.3-fold induction in stressed leaves.
The glu1 gene has been shown to be expressed in young maize tissues, where active cell division takes place (Brzobohaty et al., 1993), and it has been proposed that its activity is the release of active phytohormones (auxins or cytokinins) from glycosylated inactive storage forms (Campos et al., 1992; Brzobohaty et al., 1993; Falk and Rask, 1995). This expression pattern is close to the one observed for s1305, which is detectable in two-dimensional patterns from coleoptiles (Touzet et al., 1996b) and root tips but not in mature, green leaves (data not shown). However, it seems difficult to connect the hormone-regulating activity of β-glucosidase to its induction by water stress. Its activity has more recently been proposed to be the release of toxic hydroxamic acids as a defense response of plants against pests (Babcock and Esen, 1994). Another suggestion, which may be of particular interest in our context, is the implication of this enzyme in the lignification process. Freudenberg and Harkin (1963) hypothesized that the lignin-monomer precursors (cinnamic alcohols) are translocated toward the cell wall as β-glucoside esters, and are released in situ by a β-glucosidase to allow their polymerization into lignin. Recently, a β-glucosidase extracted from lodgepole pine xylem has been shown to hydrolyze coniferin, a β-glucoside of coniferyl alcohol and one of the major lignin monomers in pine, into the free alcohol (Dharmawardhana et al., 1995).
Considering the increased amounts of several enzymes observed upon dehydration (COMT, β-glucosidase, Cys synthase, and glutamate semialdehyde aminotransferase), activation of the lignin-biosynthesis pathway by water stress can be suggested. A cell wall reinforcement has already been reported in sorghum subjected to severe water stress: the lignosuberization occurring in roots during stress could reflect an increased resistance of the plant against a decrease in turgor and the necessity to restrict water loss from internal tissues (Cruz et al., 1992). In a study using loblolly pine treated with progressive water stress (as in the present study), Chang et al. (1995) also observed the induction of an ASR-like protein and of lignification-related proteins (SAM synthetase). COMT, Cys synthase, and SAM synthetase are related to the phenylpropanoid pathway but not specifically to lignin; their induction could also be related to the accumulation of ferulic acid, the product of the reaction catalyzed by the COMT. The quantity of ferulic acid bound to wall matrix polysaccharides is negatively correlated to cell wall extensibility (Wakabayashi et al., 1997). It has been proposed that cell wall hardening, a phenomenon occurring in maize leaves exposed to osmotic stress (Chazen and Neuman, 1994), is a response to water deficit and allows the reduction of growth rates (Neuman, 1995).
CONCLUSION
The computation of spot relative intensities and the use of two two-dimensional gels per genotype in the different conditions made it possible to use statistical tests for the detection of protein changes in response to water stress. This allowed us to quantify the variation in response, even when of low amplitude, and to compare the responses of the different genotypes. A set of proteins that increased from 1.3- to 5.0-fold upon water stress has been characterized. The main trend appears to be toward enzymes involved in basic metabolic pathways such as glycolysis and the Krebs cycle and a more specialized pathway, the phenylpropanoid pathway. Additional studies are currently in progress on COMT activity and its induction in other tissues to confirm the latter hypothesis.
Associations between protein variations and morphological and physiological traits affected by drought are being analyzed in a population of recombinant lines derived from the hybrid Io × Lc. Possible colocations of quantitative trait loci of induced proteins (Damerval et al., 1994) with those of responsive traits would be consistent with a causal relationship between the proteins and the phenotypic traits.
ACKNOWLEDGMENTS
We thank B. Piegu and V. Combes for invaluable technical assistance, M. Le Guilloux for illustrations, Dr. J. d'Alayer and M. Davi for peptide sequencing, and Agnès Leonardi and Catherine Damerval for their helpful comments on the manuscript.
Abbreviations:
- 2DE
two-dimensional electrophoresis
- ASR
ABA-water stress-ripening-induced protein
- COMT
caffeate O-methyltransferase
- SAM
S-adenosyl-l-Met
Footnotes
This work was funded by a grant from the European Communities BIOTECH Program, as part of the Project of Technological Priority 1993–1996.
LITERATURE CITED
- Almoguera C, Jordano J. Developmental and environmental concurrent expression of sunflower dry-seed-stored low-molecular-weight heat-shock protein and Lea mRNAs. Plant Mol Biol. 1992;19:781–792. doi: 10.1007/BF00027074. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amitai-Zeigerson H, Scolnik PA, Bar-Zvi D. Genomic nucleotide sequence of tomato Asr2, a second member of the stress/ripening-induced Asr1 gene family. Plant Physiol. 1994;106:1699–1700. doi: 10.1104/pp.106.4.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock GD, Esen A. Substrate specificity of maize β-glucosidase. Plant Sci. 1994;101:31–39. [Google Scholar]
- Bandaranayake H, Esen A (1996) Nucleotide sequence of a β-glucosidase (glu2) cDNA from maize (accession no. U44087). EMBL Data Library
- Bartels D, Hanke C, Schneider K, Michel D, Salamini F. A desiccation-related Elip-like gene from the resurrection plant Craterostigma plantagineum is regulated by light and ABA. EMBO J. 1992;11:2771–2778. doi: 10.1002/j.1460-2075.1992.tb05344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella MA, Quesada MA, Kononowicz AK, Bressan RA, Pliego F, Hasegawa PM, Valpuesta V. Characterization and in situ localization of a salt-induced tomato peroxidase mRNA. Plant Mol Biol. 1994;25:105–114. doi: 10.1007/BF00024202. [DOI] [PubMed] [Google Scholar]
- Brzobohaty B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K. Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science. 1993;262:1051–1054. doi: 10.1126/science.8235622. [DOI] [PubMed] [Google Scholar]
- Burstin J, Zivy M, de Vienne D, Damerval C. Analysis of scaling methods to minimize experimental variations in two-dimensional electrophoresis quantitative data: applications to the comparison of maize inbred lines. Electrophoresis. 1993;14:1067–1073. doi: 10.1002/elps.11501401170. [DOI] [PubMed] [Google Scholar]
- Campos N, Bako L, Feldwisch J, Schell J, Palme K. A protein from maize labeled with azido-IAA has novel β-glucosidase activity. Plant J. 1992;2:675–684. [Google Scholar]
- Chang S, Puryear JD, Dias MADL, Funkhouser EA, Newton RJ, Cairney J. Gene expression under water deficit in loblolly pine (Pinus taeda L.): isolation and characterization of cDNA clones. Physiol Plant. 1995;95:1–10. [Google Scholar]
- Chazen O, Neuman PM. Hydraulic signals from the roots and rapid cell-wall hardening in growing maize (Zea mays L.) leaves are primary responses to polyethylene glycol-induced water deficits. Plant Physiol. 1994;104:1385–1392. doi: 10.1104/pp.104.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Cheng H, Bjerknes M. One-step Coomassie brilliant blue R-250 staining of proteins in polyacrylamide gel. Anal Biochem. 1993;212:295–296. doi: 10.1006/abio.1993.1330. [DOI] [PubMed] [Google Scholar]
- Close TJ, Kortt AA, Chandler PM. A cDNA-based comparison of dehydration-induced proteins (dehydrins) in barley and corn. Plant Mol Biol. 1989;13:95–108. doi: 10.1007/BF00027338. [DOI] [PubMed] [Google Scholar]
- Collazo P, Montoliu L, Puigdomènech P, Rigau J. Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Mol Biol. 1992;20:857–867. doi: 10.1007/BF00027157. [DOI] [PubMed] [Google Scholar]
- Cruz RT, Jordan WR, Drew MC. Structural changes and associated reduction of hydraulic conductance in roots of Sorghum bicolor L. following exposure to water deficit. Plant Physiol. 1992;99:203–212. doi: 10.1104/pp.99.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C. Quantification of silver-stained proteins resolved by two-dimensional electrophoresis: genetic variability as related to abundance and solubility in two maize lines. Electrophoresis. 1994;15:1573–1579. doi: 10.1002/elps.11501501226. [DOI] [PubMed] [Google Scholar]
- Damerval C, de Vienne D, Zivy M, Thiellement H. Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis. 1986;7:52–54. [Google Scholar]
- Damerval C, le Guilloux M, Blaisonneau J, de Vienne D. A simplification of Heukeshoven and Dernick's silver staining of proteins. Electrophoresis. 1987;8:158–159. [Google Scholar]
- Damerval C, Maurice A, Josse JM, de Vienne D. Quantitative trait loci underlying gene product variation: a novel perspective for analyzing regulation of genome expression. Genetics. 1994;137:289–301. doi: 10.1093/genetics/137.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne D, Burstin J, Gerber S, Leonardi A, le Guilloux M, Murigneux A, Beckert M, Bahrman N, Damerval C, Zivy M. Two-dimensional electrophoresis of proteins as a source of monogenic and codominant markers for population genetics and mapping the expressed genome. Heredity. 1996;76:166–177. [Google Scholar]
- Dharmawardhana DP, Ellis BE, Carlson JE. A β-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol. 1995;107:331–339. doi: 10.1104/pp.107.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing WL, Mauxion F, Fauvarque MO, Reviron MP, de Vienne D, Vartanian N, Giraudat J. A Brassica napus transcript encoding a protein related to the Künitz protease inhibitor family accumulates upon water stress in leaves, not in seeds. Plant J. 1992;2:685–693. [PubMed] [Google Scholar]
- du Jardin P, Rojas-Beltran J, Gebhardt C, Brasseur R. Molecular cloning and characterization of a soluble inorganic pyrophosphatase in potato. Plant Physiol. 1995;109:853–860. doi: 10.1104/pp.109.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L III (1993) Structural motifs in Lea proteins. In TJ Close, EA Bray, eds, Plant Responses to Cellular Dehydration during Environmental Stress. Current Topics in Plant Physiology, Vol 10. American Society of Plant Physiologists, Rockville, MD, pp 91–103
- Dure L, III, Crouch M, Harada J, Ho THD, Mundy J, Quatrano R, Thomas T, Sung ZR. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol. 1989;12:475–486. doi: 10.1007/BF00036962. [DOI] [PubMed] [Google Scholar]
- Esen A. Purification and partial characterization of maize (Zea mays L.) β-glucosidase. Plant Physiol. 1992;98:174–182. doi: 10.1104/pp.98.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espartero J, Pintor-Toro JA, Pardo JM. Differential accumulation of S-adenosylmethionine synthetase transcripts in response to salt stress. Plant Mol Biol. 1994;25:217–227. doi: 10.1007/BF00023239. [DOI] [PubMed] [Google Scholar]
- Falk A, Rask L. Expression of a zeatin-O-glucoside-degrading β-glucosidase in Brassica napus. Plant Physiol. 1995;108:1369–1377. doi: 10.1104/pp.108.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobis-Loisy I, Loridon K, Lobreaux S, Lebrun M, Briat JF. Structure and differential expression of two maize ferritin genes in response to iron and abscisic acid. Eur J Biochem. 1995;231:609–619. doi: 10.1111/j.1432-1033.1995.tb20739.x. [DOI] [PubMed] [Google Scholar]
- Forsthoefel NR, Cushman MAF, Cushman JC. Posttranscriptional and posttranslational control of enolase expression in the facultative crassulacean acid metabolism plant Mesembryanthemum crystallinum L. Plant Physiol. 1995;108:1185–1195. doi: 10.1104/pp.108.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fray RG, Wallace A, Grierson D, Lycett GW. Nucleotide sequence and expression of a ripening and water stress-related cDNA from tomato with homology to the MIP class of membrane channel proteins. Plant Mol Biol. 1994;24:539–543. doi: 10.1007/BF00024122. [DOI] [PubMed] [Google Scholar]
- Freudenberg K, Harkin JM. The glucosides of cambial sap of spruce. Phytochemistry. 1963;2:189–193. [Google Scholar]
- Glaser HU, Thomas D, Gaxiola R, Montrichard F, Surdin-Kerjan Y, Serrano R. Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 1993;12:3105–3110. doi: 10.1002/j.1460-2075.1993.tb05979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goday A, Jensen AB, Culianez-Macia FA, Alba MM, Figueras M, Serratosa J, Torrent M, Pagès M. The maize abscisic acid-responsive protein rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell. 1994;6:351–360. doi: 10.1105/tpc.6.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowri G, Bugos RC, Campbell WH, Maxwell CA, Dixon RA. Stress responses in alfalfa (Medicago sativa L.). X. Molecular cloning and expression of S-adenosyl-l-methionine: caffeic acid 3-O-methyltransferase, a key enzyme of lignin biosynthesis. Plant Physiol. 1991;97:7–14. doi: 10.1104/pp.97.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen L, Christensen AB, Sommer-Knudsen J, Collinge DB. A putative O-methyltransferase from barley is induced by fungal pathogens and UV light. Plant Mol Biol. 1994;26:1797–1806. doi: 10.1007/BF00019493. [DOI] [PubMed] [Google Scholar]
- Grimm B. Primary structure of a key enzyme in plant tetrapyrrole synthesis: glutamate 1-semialdehyde aminotransferase. Proc Natl Acad Sci USA. 1990;87:4169–4173. doi: 10.1073/pnas.87.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero FD, Jones JT, Mullet JE. Turgor-responsive gene transcription and RNA levels increase rapidly when pea shoots are wilted: sequence and expression of three inducible genes. Plant Mol Biol. 1990;15:11–26. doi: 10.1007/BF00017720. [DOI] [PubMed] [Google Scholar]
- Heikkila JJ, Papp JET, Schultz GA, Bewley JD. Induction of heat shock protein messenger RNA in maize mesocotyls by water stress, abscisic acid, and wounding. Plant Physiol. 1984;76:270–274. doi: 10.1104/pp.76.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman WJ, Fornari CS, Tanaka CK. A comparison of the effect of salt on polypeptides and translatable mRNAs in roots of a salt-tolerant and a salt-sensitive cultivar of barley. Plant Physiol. 1989;90:1444–1456. doi: 10.1104/pp.90.4.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H, Yahara I. Yeast heat-shock protein of Mr 48,000 is an isoprotein of enolase. Nature. 1985;315:688–690. [Google Scholar]
- Ilag LL, Kumar AM, Söll D. Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell. 1994;6:265–275. doi: 10.1105/tpc.6.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Nakamura T, Han SY, Takabe T. Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic stress and abscisic acid. Plant Mol Biol. 1995;27:307–315. doi: 10.1007/BF00020185. [DOI] [PubMed] [Google Scholar]
- Iusem ND, Bartholomew DM, Hitz WD, Scolnik PA. Tomato (Lycopersicon esculentum) transcript induced by water deficit and ripening. Plant Physiol. 1993;102:1353–1354. doi: 10.1104/pp.102.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeck E, Dumas B, Geoffroy P, Favet N, Inze D, Van Montagu M, Fritig B, Legrand M. Regulation of enzymes involved in lignin biosynthesis: induction of O-methyltransferase mRNAs during the hypersensitive reaction of tobacco to tobacco mosaic virus. Mol Plant Microbe Interact. 1992;5:294–300. doi: 10.1094/mpmi-5-294. [DOI] [PubMed] [Google Scholar]
- Kawalleck P, Plesch G, Hahlbrock K, Somssich IE. Induction by fungal elicitor of S-adenosyl-l-methionine synthetase and S-adenosyl-l-homocysteine hydrolase mRNAs in cultured cells and leaves of Petroselinum crispum. Proc Natl Acad Sci USA. 1992;89:4713–4717. doi: 10.1073/pnas.89.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith CS, Hoang DO, Barrett BM, Feigelman B, Nelson MC, Thai H, Baysdorfer C. Partial sequence analysis of 130 randomly selected maize cDNA clones. Plant Physiol. 1993;101:329–332. doi: 10.1104/pp.101.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol Biol. 1994;25:791–798. doi: 10.1007/BF00028874. [DOI] [PubMed] [Google Scholar]
- Kononowicz AK, Raghothama KG, Casa AM, Reuveni M, Watad AA, Liu D, Bressan RA, Hasegawa PM (1993) Osmotin: regulation of gene expression and function. In TJ Close, EA Bray, eds, Plant Responses to Cellular Dehydration during Environmental Stress. Current Topics in Plant Physiology, Vol 10. American Society of Plant Physiologists, Rockville, MD, pp 144–157
- Lal SK, Johnson S, Conway T, Kelley PM. Characterization of a maize cDNA that complements an enolase-deficient mutant of Escherichia coli. Plant Mol Biol. 1991;16:787–795. doi: 10.1007/BF00015071. [DOI] [PubMed] [Google Scholar]
- Lange BM, Lapierre C, Sandermann H., Jr Elicitor-induced spruce stress lignin: structural similarity to early developmental lignins. Plant Physiol. 1995;108:1277–1287. doi: 10.1104/pp.108.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone A, Costa A, Tucci M, Grillo S. Comparative analysis of short- and long-term changes in gene expression caused by low water potential in potato (Solanum tuberosum) cell-suspension cultures. Plant Physiol. 1994;106:703–712. doi: 10.1104/pp.106.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisse T, Bartels D, Kalbitzer HR, Jaenicke R. The recombinant dehydrin-like desiccation stress protein from the resurrection plant Craterostigma plantagineum displays no defined three-dimensional structure in its native state. Biol Chem. 1996;377:555–561. doi: 10.1515/bchm3.1996.377.9.555. [DOI] [PubMed] [Google Scholar]
- Lobreaux S, Hardy T, Briat JF. Abscisic acid is involved in the iron-induced synthesis of maize ferritin. EMBO J. 1993;12:651–657. doi: 10.1002/j.1460-2075.1993.tb05698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobreaux S, Massenet O, Briat JF. Iron induces ferritin synthesis in maize plantlets. Plant Mol Biol. 1992;19:563–575. doi: 10.1007/BF00026783. [DOI] [PubMed] [Google Scholar]
- Marchionni M, Gilbert W. The triosephosphate isomerase gene from maize: introns antedate the plant-animal divergence. Cell. 1986;46:133–141. doi: 10.1016/0092-8674(86)90867-6. [DOI] [PubMed] [Google Scholar]
- Moons A, Bauw G, Prinsen E, Van Montagu M, Van Der Straeten D. Molecular and physiological responses to abscisic acid and salts in roots of salt-sensitive and salt-tolerant indica rice varieties. Plant Physiol. 1995;107:177–186. doi: 10.1104/pp.107.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons AM, Vandekerkhove J, Dekeyser R, Gheysen G, Van Montagu M (1996) Isolation and characterization of a novel abscisic acid- and salt-responsive gene family of Oryza sativa L. and analysis of the gene family (accession no. X95402). EMBL Data Library
- Neuman PM. The role of cell wall adjustment in plant resistance to water deficits. Crop Sci. 1995;35:1258–1266. [Google Scholar]
- Ouvrard O, Cellier F, Ferrare K, Tousch D, Lamaze T, Dupuis JM, Casse-Delbart F. Identification and expression of water stress- and abscisic acid-regulated genes in a drought-tolerant sunflower genotype. Plant Mol Biol. 1996;31:819–829. doi: 10.1007/BF00019469. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Geoffroy P, Fritig B, Legrand M. Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco (Nicotiana tabacum L.) leaves by infection or elicitor treatment. Plant Physiol. 1993;103:509–517. doi: 10.1104/pp.103.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla M, Goday A, Vilardell J, Gomez J, Pagès M. Differential regulation of ABA-induced 23–25 kDa proteins in embryo and vegetative tissues of the viviparous mutants of maize. Plant Mol Biol. 1989;13:385–394. doi: 10.1007/BF00015550. [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Whitford PN, Appleford NEJ, Wang TL, Cook SK, Henson IE, Loveys BR. A monoclonal antibody to (S)-abscisic acid: its characterisation and use in a radioimmunoassay for measuring abscisic acid in crude extracts of cereal and lupin leaves. Planta. 1988;173:330–339. doi: 10.1007/BF00401020. [DOI] [PubMed] [Google Scholar]
- Raines CA, Longstaff M, Lloyd JC, Dyer TA. Complete coding sequence of wheat phosphoribulokinase: developmental and light-dependent expression of the mRNA. Mol Gen Genet. 1989;220:43–48. [PubMed] [Google Scholar]
- Ramagopal S. Salinity stress induced tissue-specific proteins in barley seedlings. Plant Physiol. 1987;84:324–331. doi: 10.1104/pp.84.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razdan K, Heinrikson RL, Zurcher-Neely H, Morris PW, Anderson LE. Chloroplast and cytoplasmic enzymes: isolation and sequencing of cDNAs coding for two distinct pea chloroplast aldolases. Arch Biochem Biophys. 1992;298:192–197. doi: 10.1016/0003-9861(92)90112-a. [DOI] [PubMed] [Google Scholar]
- Reviron MP, Vartanian N, Sallantin M, Huet JC, Pernollet JC, de Vienne D. Characterization of a novel protein induced by rapid or progressive drought and salinity in Brassica napus leaves. Plant Physiol. 1992;100:1486–1493. doi: 10.1104/pp.100.3.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Iusem ND. Tomato (Lycopersicon esculentum) genomic clone homologous to a gene encoding an abscisic acid-induced protein. Plant Physiol. 1994;104:1073–1074. doi: 10.1104/pp.104.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiter RK, van Eldik GJ, van Herpen MM, Schrauwen JA, Wullems GJ. Expression in anthers of two genes encoding Brassica oleracea transmembrane channel proteins. Plant Mol Biol. 1997;34:163–168. doi: 10.1023/a:1005828325425. [DOI] [PubMed] [Google Scholar]
- Saito K, Tatsuguchi K, Murakoshi I, Hirano H. cDNA cloning and expression of cysteine synthase B localized in chloroplasts of Spinacia oleracea. FEBS Lett. 1993;324:247–252. doi: 10.1016/0014-5793(93)80127-g. [DOI] [PubMed] [Google Scholar]
- Sangwan I, O'Brian MR. Expression of the soybean (Glycine max) glutamate 1-semialdehyde aminotransferase gene in symbiotic root nodules. Plant Physiol. 1993;102:829–834. doi: 10.1104/pp.102.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Jäger K. Open questions about sulfur metabolism in plants. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:325–349. [Google Scholar]
- Shen B, Carneiro N, Torres-Jerez I, Stevenson R, Helentjaris T, Habben J, Larkins B, Almira E, Ferl R, Baysdorfer C (1994) Single-pass sequencing and mapping of clones from two maize cDNA libraries (accession no. T15295). EMBL Data Library [DOI] [PubMed]
- Silhavy D, Hutvagner G, Barta E, Banfalvi Z. Isolation and characterization of a water-stress-inducible cDNA clone from Solanum chacoense. Plant Mol Biol. 1995;27:587–595. doi: 10.1007/BF00019324. [DOI] [PubMed] [Google Scholar]
- Torres-Schumann S, Godoy JA, Pintor-Toro JA. A probable lipid transfer protein gene is induced by NaCl in stems of tomato plants. Plant Mol Biol. 1992;18:749–757. doi: 10.1007/BF00020016. [DOI] [PubMed] [Google Scholar]
- Touzet P, de Vienne D, Huet JC, Ouali C, Bouet F, Zivy M. Amino acid analysis of proteins separated by two-dimensional electrophoresis in maize: isoform detection and function identification. Electrophoresis. 1996a;17:1393–1401. doi: 10.1002/elps.1150170819. [DOI] [PubMed] [Google Scholar]
- Touzet P, Riccardi F, Morin C, Damerval C, Huet JC, Pernollet JC, Zivy M, de Vienne D. The maize two-dimensional gel protein database: towards an integrated genome analysis program. Theor Appl Genet. 1996b;93:997–1005. doi: 10.1007/BF00224104. [DOI] [PubMed] [Google Scholar]
- Tsutsumi K, Kagaya Y, Hidaka S, Suzuji J, Tokairin Y, Hu DL, Ishikawa K, Ejiri S. Structural analysis of the chloroplastic and cytoplasmic aldolase-encoding genes implicated the occurrence of multiple loci in rice. Gene. 1994;141:215–220. doi: 10.1016/0378-1119(94)90574-6. [DOI] [PubMed] [Google Scholar]
- Umeda M, Hara C, Matsubayashi Y, Li HH, Liu Q, Tadokoro F, Aotsuka S, Uchimiya H. Expressed sequence tags from cultured cells of rice (Oryza sativa L.) under stressed conditions: analysis of genes engaged in ATP-generating pathways. Plant Mol Biol. 1994;25:469–478. doi: 10.1007/BF00043875. [DOI] [PubMed] [Google Scholar]
- Van der Straeten D, Rodrigues-Pousada RA, Goodman HM, Van Montagu M. Plant enolase: gene structure expression, and evolution. Plant Cell. 1991;3:719–735. doi: 10.1105/tpc.3.7.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Salamini F, Bartels D. Dehydration and ABA increase mRNA levels and enzyme activity of cytosolic GAPDH in the resurrection plant Craterostigma plantagineum. Plant Mol Biol. 1994;26:541–546. doi: 10.1007/BF00039567. [DOI] [PubMed] [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capellades M, Puigdomènech P. The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell. 1995;7:407–416. doi: 10.1105/tpc.7.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell J, Goday A, Freire MA, Torrent M, Martinez MC, Torné JM, Pagès M. Gene sequence, developmental expression, and protein phosphorylation of RAB-17 in maize. Plant Mol Biol. 1990;14:423–432. doi: 10.1007/BF00028778. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Hoson T, Kamisaka S. Osmotic stress suppresses cell wall stiffening and the increase in cell wall-bound ferulic and diferulic acids in wheat coleoptiles. Plant Physiol. 1997;113:967–973. doi: 10.1104/pp.113.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weretilnyk EA, Hanson AD. Molecular cloning of a plant betaine-aldehyde dehydrogenase, an enzyme implicated in adaptation to salinity and drought. Proc Natl Acad Sci USA. 1990;87:2745–2749. doi: 10.1073/pnas.87.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, Bulman M, Huttly A, Phillips A, Neill S. Characterization of a cDNA from Arabidopsis thaliana encoding a potential thiol protease whose expression is induced independently by wilting and abscisic acid. Plant Mol Biol. 1994;25:259–270. doi: 10.1007/BF00023242. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K. Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol. 1992;33:217–224. [Google Scholar]
- Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:439–473. [Google Scholar]
- Zivy M, Thiellement H, de Vienne D, Hofmann JP. Study on nuclear and cytoplasmic genome expression in wheat by two-dimensional electrophoresis. 1. First results on 18 alloplasmic lines. Theor Appl Genet. 1983;66:1–7. doi: 10.1007/BF00281838. [DOI] [PubMed] [Google Scholar]