Abstract
We have investigated the transfer of Tn916 among strains of Enterococcus faecalis OG1 colonizing in the intestines of gnotobiotic rats. This animal model allows a low limit of detection and efficient colonization of the chosen bacteria. The animals continuously received tetracycline in drinking water. A tetracycline-sensitive recipient strain was allowed to colonize the animals before the resistant donor was introduced. The numbers of donors, recipients, and transconjugants in fecal samples and intestinal segments were estimated. The bioavailable amounts of tetracycline in fecal samples and intestinal segments were monitored by using bacterial biosensors carrying a transcriptional fusion of a tetracycline-regulated promoter and a lacZ reporter gene. Chromosomal locations of Tn916 in transconjugants isolated either from the same animal or from different animals were compared by Southern blot analysis. Our results indicated that selection for the resistant phenotype was the major factor causing higher numbers of transconjugants in the presence of tetracycline. Tetracycline-sensitive E. faecalis cells colonized the intestine even when the concentrations of tetracycline in feces and intestinal luminal contents exceeded growth-inhibitory concentrations. This suggests the existence of tetracycline-depleted microhabitats in the intestinal environment.
Horizontal transfer of antibiotic resistance genes between bacteria has attracted much attention, and numerous investigations of gene transfer in environmental settings have been reported (18, 26, 36, 39). One concern is the dissemination and accumulation of resistance genes in the environment, which might be enhanced by use of antimicrobial agents and constitutes a risk for spread of resistance to pathogenic bacteria (41). Since the first report of conjugative transposons in 1981 (10), increasing attention has been given to these self-transmissible discrete DNA elements and their role in spreading antibiotic resistance between bacteria (34). Tn916, which confers resistance to tetracycline, belongs to the Tn916-Tn1545 family of conjugative transposons and was first identified in Enterococcus faecalis (formerly Streptococcus faecalis) DS16 (10), but it is known to replicate in a wide range of clinically important gram-positive and gram-negative species (4).
Transposition of Tn916 and other conjugative transposons is a three-step process involving (i) excision from the donor chromosome and formation of a covalently closed circular intermediate (33), (ii) conjugation by way of single-stranded transfer to the recipient cell (32), and (iii) nonspecific integration into the recipient chromosome (32). Multiple copies of Tn916 can exist on the same chromosome (25). Tetracycline in subinhibitory concentrations has been shown to have a positive effect on the regulation of Tn916 family transposition (7, 35); however, the rate of transfer is low compared to that of conjugative transfer of plasmids (2). One investigation found 7.5 × 10−6 transconjugants per initial donor cell after overnight in vitro mating on membrane filters, increasing to 1.1 × 10−5 when the donor strain was pregrown in medium containing tetracycline (35). Other experiments involving transfer of Tn916 family transposons from E. faecalis hosts on agar plates showed a slightly lower transconjugant-to-donor ratio (7, 38). The low transfer rate necessitates a high colonization of the donor and recipient strains in animal models in order for transfer events to occur and subsequently be detected. This can be achieved by using gnotobiotic animals (24) that also have the advantage of a well-defined microbial flora, which eases selection of specific strains and thus lowers the detection limit. The interpretation of results obtained in studies of gene transfer in the intestinal tract is complicated by the fact that the gut represents a dynamic ecosystem, where nutrients and bacteria are continuously produced and continuously removed from the system with feces (11, 12, 18).
The use of bacterial whole-cell biosensors, carrying fusions of a selected promoter and a reporter gene, to estimate concentrations of various compounds, such as antibiotics (14) and heavy metals (29), has proved to be a useful alternative to conventional chromatographic methods. One of the major advantages is that biosensors detect only the bioavailable fraction of the compound. This is especially relevant in the complex intestinal system, where many factors can mask the presence of a specific compound and thereby reduce the ambient concentration, which affects the microbial community.
In the present study, transfer of Tn916 from a donor E. faecalis strain to an isogenic resident recipient strain (carrying different resistance markers) was examined in the gastrointestinal environment of gnotobiotic rats treated with tetracycline. The bioavailable tetracycline concentrations present in the intestinal contents as well as in feces were estimated by use of a bacterial biosensor strain. The results presented here shed new light on the kinetics of transfer and establishment of Tn916 in the intestinal environment.
MATERIALS AND METHODS
Bacterial strains and growth media.
The rifampin- and fusidic acid-resistant strain E. faecalis OG1RF (8) was used as the recipient strain. The donor strain (a gift from S. Donabedian) was the streptomycin- and spectinomycin-resistant E. faecalis OG1SS::Tn916 (37), which also carries a chromosomal insertion of the conjugative transposon Tn916, containing the tet(M) gene encoding resistance to tetracycline (9). A kanamycin-resistant Escherichia coli MC4100::tetlac strain, carrying a chromosomally inserted fusion between a tetracycline-inducible promoter and the β-galactosidase gene (16), was used as a whole-cell biosensor to estimate the bioavailable tetracycline concentrations in fecal samples and in contents extracted from the gastrointestinal tract.
Brain heart infusion broth (Oxoid, Hampshire, England) was used for all broth cultures of the strains, except in β-galactosidase assays, where Luria-Bertani (LB) broth was used (30). The following solid media were used for selective plating: Slanetz and Bartley medium (Oxoid) for selection of E. faecalis and MacConkey agar no. 3 (Oxoid) for selection of E. coli. Recipients plus transconjugants, donors, and transconjugant bacteria were selected on Slanetz and Bartley plates containing either (i) rifampin and fusidic acid, (ii) spectinomycin and streptomycin, or (iii) rifampin, fusidic acid, and tetracycline. All plates were incubated aerobically at 37°C for either 1 day (E. coli) or 2 days (E. faecalis). Antibiotics (Sigma) were used at the following concentrations: fusidic acid, 25 μg/ml; kanamycin, 50 μg/ml; rifampin, 25 μg/ml; spectinomycin, 500 μg/ml; streptomycin, 1,000 μg/ml; and tetracycline hydrochloride, (Sigma catalog no. T3383), 10 μg/ml.
Grouping of animals and dosing with tetracycline.
Twelve female germfree Sprague-Dawley rats, approximately 2 months old and bred at the Institute of Food Safety and Nutrition were originally obtained from IFFA Credo, L'Arbresle, France. Housing, feed, temperature, and light conditions were as previously described (17). The germfree state of the animals was verified by testing fecal samples for aerobic and anaerobic growth of bacteria and yeasts. The 12 rats were caged individually and placed in four groups with three animals in each group. Starting from day 6, each group received drinking water containing tetracycline at various concentrations as follows: group A, 0.0 μg/ml; group B, 5 μg/ml; group C, 10 μg/ml; and group D, 50 μg/ml. Fresh drinking water was prepared every 2 or 3 days and kept dark at all times. The tetracycline concentration in the drinking water was tested with a bacterial biosensor (15) and confirmed on days 11, 20, and 27 of the experiment (data not shown).
Colonization of animals.
An overnight culture of the desired strain was washed twice in autoclaved 0.9% (wt/vol) saline water containing peptone, and 1 ml was given to each rat by oral gavage. On day 0, all animals were dosed with 7.7 × 108 CFU of the recipient strain, E. faecalis OG1RF. On day 7, all animals were dosed with 1.8 × 109 CFU of the donor strain, E. faecalis OG1SS::Tn916. In addition, on day −1 and the day before euthanatization, all animal were dosed with approximately 109 CFU of E. coli MC4100/pTGFP2 (15). Results related to the use of this strain are not reported here.
Sampling and enumeration of bacteria in samples.
Fresh fecal samples (100 to 300 mg) were obtained directly from the rats every 2 to 3 days by gently pressing the abdomens of the animals. After euthanatization of the animals (six on day 40 and six on day 41), samples were immediately taken from the contents of the stomach, jejunum, ileum, cecum, and colon. All samples were initially diluted 10-fold (wt/vol) in saline water containing peptone, thoroughly homogenized, further diluted, and plated on appropriate selective agar plates for CFU counting.
Verification of transconjugants by PCR.
Eight colonies were isolated from fecal samples between days 13 and 22 on agar plates selective for the transconjugant resistance phenotype. Two putative transconjugant colonies from each group, all originating from different animals, were randomly picked. Extraction and purification of genomic DNA were carried out on these isolates, followed by PCRs with primers Tn916-2 and ReverseTet(M)-2, which were designed to verify the presence of tet(M) in connection with Tn916 as previously described (1). DNAs from the donor and recipient strains were included in the PCR as positive and negative controls, respectively.
Southern blot analysis.
Two separate Southern blots were prepared, using (i) the eight PCR-verified transconjugants originating from different rats as described above and (ii) a different set of eight transconjugants all isolated from the same animal (group B) on day 20. Chromosomal DNAs from all isolates and from the donor and recipient strains were prepared by using the Wizard genomic DNA purification kit (catalog no. A1120; Promega, Madison, Wis.), and then 2 μg of DNA was HindIII restricted and Southern blot analysis was performed with a digoxigenin-labeled tet(M) probe as described by the manufacturer (catalog no. 1093657; Roche Applied Science, Indianapolis, Ind.). The 700-bp DNA probe was the amplification product obtained from a PCR with primers: tet(M)-1 and tet(M)-2, which have been described previously (1).
Growth of E. faecalis OG1RF at various tetracycline concentrations.
In vitro growth rates of the tetracycline-sensitive E. faecalis OG1RF recipient strain at six different tetracycline concentrations were estimated by measuring the optical density at 600 nm at various times during balanced growth (approximately 15 generations) in brain heart infusion medium at 37°C. Three replicas were performed for each concentration.
Determination of bioavailable tetracycline.
Measurements of bioavailable tetracycline were performed with fecal samples from days 7, 15, 20, 25, and 29 and with samples obtained from five segments of the gastrointestinal tract as described above. The homogenized 10-fold dilutions of the samples were centrifuged (15,000 × g, 5 min), and the supernatant containing tetracycline was filtered through a 0.22-μm-pore-size filter (MILLEX-GV; Millipore, Carrigtwohill, Ireland). The tetracycline concentrations in the samples were estimated by using the E. coli MC4100::tetlac biosensor strain as previously described (16). Briefly, the samples were appropriately diluted and mixed with LB broth (1:10). The biosensor was then inoculated, and samples were incubated on a shaker (200 rpm) for 3 h at 37°C. After this period, the cells were disrupted by toluene treatment and β-galactosidase assays (23) were performed; the results were compared to a standard curve, based on measurements of known tetracycline concentrations.
Statistics.
In all graphs error bars indicate the standard errors of the means, calculated by dividing the standard deviation by the square root of the number of measurements. In order to be able to log transform the data shown in Fig. 1, measurements of no CFU on a plate were set to 1 CFU/g of feces (corresponding to detection of 0.04 colonies instead of zero). Transconjugant numbers observed within the four treatment groups were compared by using the Wilcoxon test.
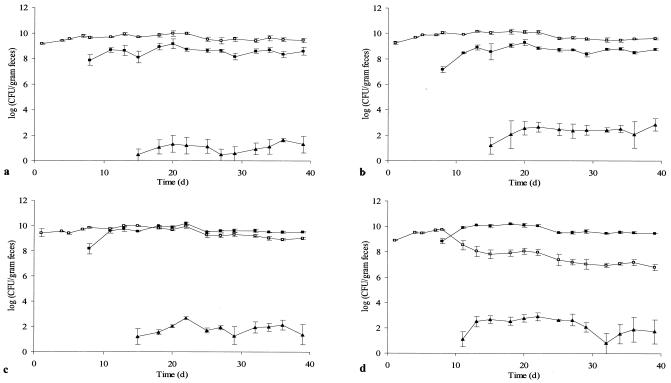
FIG. 1.
CFU of recipient E. faecalis OG1RF (open squares), donor E. faecalis OG1SS::Tn916 (closed squares), and transconjugant E. faecalis OG1RF::Tn916 (triangles) in fecal samples from rats receiving tetracycline at either 0.0 μg/ml (a), 5 μg/ml (b), 10 μg/ml (c), or 50 μg/ml (d) in drinking water. At days 0 and 7, respectively, the recipient and the donor strains were introduced. Each point represents the geometric average of values obtained from three animals. Error bars indicate standard errors of the means.
RESULTS
Colonization experiments.
The recipient strain as well as the donor strain readily colonized the guts of the germfree animals, and total E. faecalis CFU counts were between 109 and 1011 CFU/g (wet weight) of feces throughout the experiment (Fig. 1). In groups A (0 μg of tetracycline per ml) and B (5 μg of tetracycline per ml), the number of donor bacteria remained between 1 and 2 orders of magnitude lower than the number of recipients. In group C (10 μg of tetracycline per ml), the numbers of donor and recipient bacteria were almost identical. In group D (50 μg of tetracycline per ml), a significant decrease in the number of recipients was observed after introduction of the donor strain (Fig. 1). Transconjugant bacteria (E. faecalis OG1RF::Tn916) were first detected in group D (50 μg of tetracycline per ml), appearing 4 days after donor inoculation. In the other three groups, transconjugant bacteria were detected 8 days after introduction of the donor strain. The numbers of transconjugant cells in all four groups reached stable levels after a few days, and no major changes in CFU per gram occurred during the rest of the experiment. The lowest levels of transconjugants were observed in group A, where approximately 10 CFU/g of feces was detected. In the other three groups, the numbers of transconjugants remained significantly higher (P < 0.002) throughout the experiment, reaching 300 CFU/g of feces. However, transconjugant numbers were not higher in group D than in group B (P > 0.1). No colonies were detected on the donor or transconjugant selective plates prior to introduction of the donor (the limit of detection was 8 CFU/g of feces).
Enumeration of recipients, donors, and transconjugants in all examined segments of the gastrointestinal system after euthanatization showed that the donor/recipient ratio increased with increasing tetracycline concentration (data not shown), as observed in the fecal samples. Transconjugants were observed in the colon and the cecum in all four groups but were present at lower concentrations than in the fecal samples. In some instances transconjugant cells were found in the ileum, jejunum, and stomach segments.
Verification of transconjugants and Southern blot analysis.
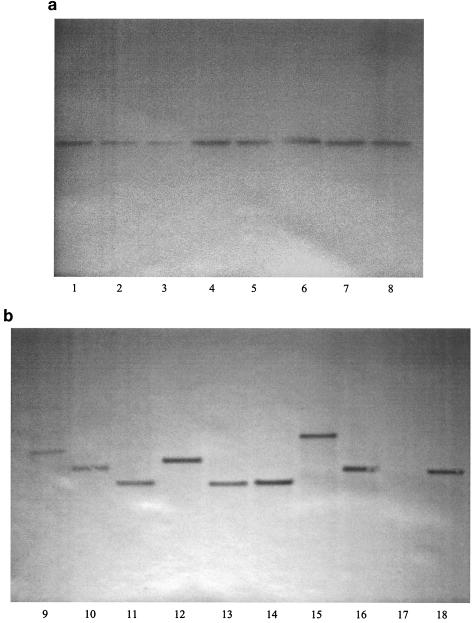
DNAs from eight transconjugants randomly isolated from two different animals in each group were shown by PCR analysis to contain Tn916. The same eight transconjugants were sensitive to a combination of streptomycin and spectinomycin. Southern blot analysis revealed that the tet(M) gene was located on HindIII-HindIII fragments of visibly different sizes for nearly all of the eight examined transconjugants originating from different animals (Fig. 2b). All fragments were different from the corresponding fragment from the donor strain. In all transconjugants as well as in the donor strain, only one band was visible, indicating a single chromosomal Tn916 insertion. The lengths of the bands were between 7 and 14 kb, consistent with the expected minimum size of 5.8 kb for a fragment containing the tet(M) gene. No band was detected by similar analysis of DNA from the recipient strain.
FIG. 2.
Southern blots of HindIII-digested transconjugant DNAs isolated from the same animal (a) and from eight different animals (b). The fragment lengths are between 7 and 14 kb in all lanes. Lanes 1 to 8, transconjugants isolated from the same animal in group B. Lanes 9 and 10, transconjugants from group D. Lanes 11 and 12, transconjugants from group C. Lanes 13 and 14, transconjugants from group B. Lanes 15 and 16, transconjugants from group A. Lane 17, DNA from recipient OG1RF. Lane 18, DNA from donor OG1SS::Tn916. The DNAs in lanes 7 and 14 originate from the same transconjugant.
Eight separate transconjugants isolated from the same animal (group B) on the same day revealed that the tet(M) gene was located on fragments of the same size, different from that of the donor strain, in the eight transconjugants (Fig. 2a), indicating that Tn916 was located at the same position on the chromosome for all of these isolates.
Growth of the recipient strain at various tetracycline concentrations.
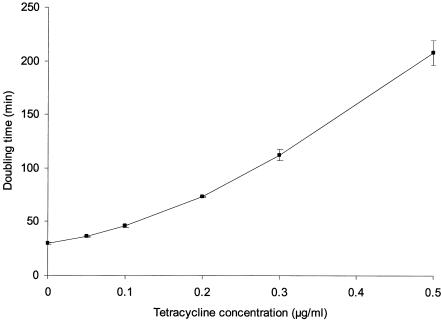
The doubling time of the tetracycline-sensitive recipient strain increased with increasing tetracycline concentration (Fig. 3). At 0.5 μg of tetracycline per ml the doubling time was nearly 3.5 h, compared to 30 min for growth in unsupplemented medium. The tetracycline concentration at which the doubling time was twice that of the strain grown without antibiotic was found to be approximately 0.16 μg of tetracycline per ml.
FIG. 3.
Doubling times (generation times) of the tetracycline-sensitive recipient E. faecalis OG1RF obtained with different tetracycline concentrations during balanced growth in LB broth. Each point represents the average of three values. Error bars indicate standard errors of the means.
Quantification of tetracycline concentrations in feces and in contents of the intestinal tract.
The tetracycline levels in the feces of rats in groups B, C, and D estimated by using the MC4100::tetlac biosensor followed by a β-galactosidase assay showed that the average levels of bioavailable tetracycline in feces were 0.65 μg/g, 2.85 μg/g, and 17.2 μg/g, respectively, for groups B (5 μg of tetracycline per ml), C (10 μg of tetracycline per ml), and D (50 μg of tetracycline per ml). Levels of tetracycline were nearly constant or slightly decreasing throughout the period. The background level of biosensor β-galactosidase expression measured in pure water was equal to that measured in fecal samples of animals belonging to group A, showing that the biosensor was not affected by the presence of fecal components. Concentrations of bioavailable tetracycline in various segments of the gastrointestinal tract were generally lower than those in fecal samples. A higher level of tetracycline than in other intestinal segments was observed in the jejunum segment for all animals examined (Table 1).
TABLE 1.
Bioavailable tetracycline concentrations measured in intestinal contents from various segments of the gastrointestinal tract and in fecal samples by use of biosensor E. coli MC4100::tetlac
| Groupa (tetracycline received, μg/ml) | Tetracycline concn (μg/g)b in:
|
|||||
|---|---|---|---|---|---|---|
| Stomach | Jejunum | Ileum | Cecum | Colon | Fecal samplec | |
| B (5) | 0.19 ± 0.04 | 1.78 ± 1.00 | 0.19 ± 0.10 | 0.35 ± 0.03 | 0.47 ± 0.17 | 0.65 ± 0.06 |
| C (10) | 0.19 ± 0.15 | 2.10 ± 0.17 | 0.73 ± 0.40 | 0.57 ± 0.07 | 0.83 ± 0.08 | 2.85 ± 0.38 |
| D (50) | 1.60 ± 0.77 | 22.9 ± 8.53 | 3.14 ± 2.52 | 4.86 ± 1.97 | 3.91 ± 0.40 | 17.19 ± 1.87 |
No measurable induction of the biosensor was observed in fecal samples from animals belonging to group A.
Values are means ± standard errors of the means from the three animals in each group.
The mean value ± standard error of the mean obtained from measurements for three animals on days 7, 15, 20, 25, 29 is given.
DISCUSSION
Significantly higher (P < 0.002) numbers of transconjugant cells per gram of feces were observed for animals dosed with tetracycline (groups B, C, and D) than for those not dosed with tetracycline (group A). However, higher tetracycline concentrations (above 5 μg/ml) did not result in higher numbers (P > 0.1) of transconjugant cells (Fig. 1). This finding is consistent with a study demonstrating that tetracycline treatment resulted in increased numbers of Listeria monocytogenes::Tn1545 transconjugants in the guts of gnotobiotic mice (7). Since the numbers of donor and recipient cells were the same in group A (0 μg of tetracycline per ml) and group B (5 μg of tetracycline per ml), our data indicate that the altered transconjugant numbers observed are not the result of an altered donor/recipient ratio. The observed higher transconjugant numbers caused by tetracycline can be a result of either (i) increased horizontal transfer, leading to more initial transfer events; (ii) increased vertical spread due to increased selective pressure; or (iii) a combination of these two factors. In many investigations of gene transfer in environmental settings it is difficult to determine the contribution of each of these factors to an observed increase in numbers of transconjugants. It is known that pregrowth of a Tn916 donor strain with tetracycline increases the transfer rate under laboratory conditions (35), and a proposed molecular mechanism for this has been published (3). Southern blot analysis of transconjugants originating from the same animal (group B, day 20) revealed that all of the examined isolates had the same Tn916 insert fragment (Fig. 2a). However, in the Southern blot analysis of eight transconjugants originating from eight different animals (Fig. 2b), nearly all fragments were of different lengths, in accordance with the partially random insertion properties of Tn916 (22). These observations indicate that all transconjugants isolated from one animal were formed by proliferation of one initial transconjugant cell. Multitransposition of Tn916, as previously described (25), was not seen in any of the 15 separately isolated transconjugants, also indicating a low transfer frequency of the conjugative transposon. Furthermore, the numbers of transconjugants in feces after the initial formation remained stable in all groups of animals (Fig. 1). Hence, we conclude that ongoing transfer from transconjugant cells to new recipients occurred at negligible levels.
Increasing concentrations of tetracycline in drinking water caused an increase in the donor/recipient ratio (Fig. 1), but the total number of E. faecalis organisms remained at the same constant level in all groups, reflecting the maximal bacterial number which can be maintained in the gastrointestinal tract. This finding demonstrates that the bacteriostatic antibiotic tetracycline provides a selective advantage to the tetracycline-resistant donors. It seemed that the selective advantage was most pronounced at the initial stage of colonization, as the donor/recipient ratio remained constant. A continued selective advantage would have caused complete replacement of the tetracycline-sensitive recipient strain over time (11, 12, 18).
In all animals the donor and the recipient strains colonized the intestine together throughout the experiment. Supported by a few previous publications from our laboratories (18, 20), this challenges the hypothesis that isogenic strains in the gut, like in a chemostat, are not able to coexist due to the highly competitive intestinal environment (11, 17, 19, 31). Chemostat predictions show that the cells must grow fast enough to compensate for the continuous washout of bacteria with feces and faster than other bacterial species competing for the same nutrients in order to persist in the dynamic system (12). Incomplete mixing of the bacterial population in the intestine might be the reason why this environment appears to be less competitive than a chemostat, and the kinetics observed in the intestinal environment might be more similar to those observed in a biofilm (18).
One feature of bacteria which would allow them to persist in the intestine even in the presence of a competing strain is attachment to the epithelial surface (11, 12). It is possible that Tn916 conveys an advantage to the donor strain due to expression of an adhesive factor in the donor strain facilitating attachment to recipients and perhaps also to the intestinal epithelium. Colonization of the two strains in different segments of the gastrointestinal tract would also explain their coexistence; however, CFU counts in five segments showed no sign of differences between the two strains. A final explanation, which is supported by older literature (13), could be that the E. faecalis strains do not remain isogenic in the gnotobiotic animals. In any case, one should bear in mind that germfree animal models, as used in the present study, offer more available niches for the introduced bacteria and hence a less competitive environment than is found in conventional animals. We must conclude that our present understanding of the principles decisive for the coexistence of isogenic strains in the gastrointestinal tract is not complete. It is clear that the kinetics in the gut are different and, not surprisingly, more complicated to predict than the kinetics in a chemostat. This emphasizes the importance of in situ and in vivo model systems to gain insight into transfer kinetics and establishment of mobile genetic elements in natural systems.
In this study it was possible to use a lacZ biosensor strain to estimate amounts of bioavailable tetracycline in fecal samples and in five segments of the gastrointestinal tract (Table 1). This is, to our knowledge, the first time that a whole-cell biosensor has been used to estimate intraintestinal concentrations, and we found that a change of tetracycline concentration in drinking water causes a proportional change in bioavailable amounts in the luminal contents of the intestine. The measured tetracycline concentrations in feces are comparable to those found in an earlier investigation using an agar diffusion method to estimate residual tetracycline in human-flora-associated mice (27). The peak in tetracycline concentration observed in the jejunums of all animals (Table 1) can be explained by the routes of secretion and absorption of this drug in the animal body. The contents of the jejunum hold more water than the contents of the large intestine, and it is likely that a large part of the water-soluble tetracycline is absorbed along with the water in the colonic section.
The bioavailable tetracycline concentrations measured in intestinal contents from rats drinking the highest concentration of tetracycline, group D (50 μg of tetracycline per ml), ranged between 1.6 and 22.9 μg/g (Table 1). No growth of the sensitive recipient strain is possible at these high tetracycline concentrations (Fig. 3), which thus would be expected to lead to complete loss of the recipient strain. Nevertheless, at least 107 CFU/g of feces was isolated from group D throughout the experiment. This is even more remarkable because the competing tetracycline-resistant donor strain was also present in the intestine. The ability of the recipient strain to proliferate in the intestine could be caused by the existence of tetracycline-depleted compartments in the gut. We propose that the mucus layer, constituting the interface between the lumen and the epithelial cells in the intestine, might represent such a compartment, since the physical and chemical properties of this layer are markedly different from those of the luminal contents. Supporting this hypothesis, previous studies of gram negative strains colonizing the animal intestine have suggested that the majority of bacterial growth occurs in the mucus layer and not in the luminal contents (21, 28, 40). However, it remains to be shown whether the structural matrix constituted by the intestinal mucus might prevent diffusion of tetracycline. A different possibility, which would allow colonization of the sensitive bacteria, is that the colonizing bacteria express a phenotype rendering them resistant to the drug, similar to what has been described for biofilm communities (5, 6). The fact that growth of tetracycline-sensitive bacteria occurs in the intestinal system when inhibitory concentrations of the drug are present is potentially of importance for antimicrobial therapy. Further work involving biosensor technology is needed to fully understand the interactions between antibiotics and bacteria in the gastrointestinal environment.
Acknowledgments
This work was financed by a grant from The Directorate for Food, Fisheries and Agro Business under the Danish Ministry of Food, Agriculture and Fisheries. M.I.B. was the recipient of a scholarship from Carlsberg's bequest in memory of brewer J. C. Jacobsen.
We thank Susan Donabedian for the gift of E. faecalis OG1SS::Tn916 and Mansour Badaki for technical assistance. We also thank Maja Danielsen and Sarah G. Nielsen for assistance with the animal experiments.
REFERENCES
- 1.Agersø, Y., L. B. Jensen, M. Givskov, and M. C. Roberts. 2002. The identification of a tetracycline resistance gene tet(M), on a Tn916-like transposon, in the Bacillus cereus group. FEMS Microbiol. Lett. 214:251-256. [DOI] [PubMed] [Google Scholar]
- 2.Andrup, L., and K. Andersen. 1999. A comparison of the kinetics of plasmid transfer in the conjugation systems encoded by the F plasmid from Escherichia coli and plasmid pCF10 from Enterococcus faecalis. Microbiology 145:2001-2009. [DOI] [PubMed] [Google Scholar]
- 3.Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. [DOI] [PubMed] [Google Scholar]
- 4.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 6.Davies, D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2:114-122. [DOI] [PubMed] [Google Scholar]
- 7.Doucet-Populaire, F., P. Trieu-Cuot, I. Dosbaa, A. Andremont, and P. Courvalin. 1991. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob. Agents Chemother. 35:185-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunny, G. M., and D. B. Clewell. 1975. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J. Bacteriol. 124:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flannagan, S. E., L. A. Zitzow, Y. A. Su, and D. B. Clewell. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32:350-354. [DOI] [PubMed] [Google Scholar]
- 10.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freter, R., H. Brickner, J. Fekete, M. M. Vickerman, and K. E. Carey. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 39:686-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freter, R., E. Stauffer, D. Cleven, L. V. Holdeman, and W. E. Moore. 1983. Continuous-flow cultures as in vitro models of the ecology of large intestinal flora. Infect. Immun. 39:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, R. J., and J. V. Qureshi. 1980. Virulence-related physiological changes and antigenic variation in populations of Streptococcus mutans colonizing gnotobiotic rats. Infect. Immun. 29:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen, L. H., F. Aarestrup, and S. J. Sørensen. 2002. Quantification of bioavailable chlortetracycline in pig feces using a bacterial whole-cell biosensor. Vet. Microbiol. 87:51-57. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, L. H., B. Ferrari, A. H. Sørensen, D. Veal, and S. J. Sørensen. 2001. Detection of oxytetracycline production by Streptomyces rimosus in soil microcosms by combining whole-cell biosensors and flow cytometry. Appl. Environ. Microbiol. 67:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, L. H., and S. J. Sørensen. 2000. Detection and quantification of tetracyclines by whole cell biosensors. FEMS Microbiol. Lett. 190:273-278. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsen, B. L., M. Skou, A. M. Hammerum, and L. B. Jensen. 1999. Horizontal transfer of the satA gene encoding streptogramin A resistance between isogenic Enterococcus faecium strains in the gastrointestinal tract of gnotobiotic rats. Microb. Ecol. Health Dis. 11:241-247. [Google Scholar]
- 18.Licht, T. R., B. B. Christensen, K. A. Krogfelt, and S. Molin. 1999. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145:2615-2622. [DOI] [PubMed] [Google Scholar]
- 19.Licht, T. R., D. Laugesen, L. B. Jensen, and B. L. Jacobsen. 2002. Transfer of the pheromone-inducible plasmid pCF10 among Enterococcus faecalis microorganisms colonizing the intestine of mini-pigs. Appl. Environ. Microbiol. 68:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Licht, T. R., C. Struve, B. B. Christensen, L. K. Poulsen, S. Molin, and K. A. Krogfelt. 2003. Evidence of increased spread and establishment of plasmid RP4 in the intestine under sub-inhibitory tetracycline concentrations. FEMS Microbiol. Ecol. 44:217-223. [DOI] [PubMed] [Google Scholar]
- 21.Licht, T. R., T. Tolker-Nielsen, K. Holmstrom, K. A. Krogfelt, and S. Molin. 1999. Inhibition of Escherichia coli precursor-16S rRNA processing by mouse intestinal contents. Environ. Microbiol. 1:23-32. [DOI] [PubMed] [Google Scholar]
- 22.Lu, F., and G. Churchward. 1995. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J. Bacteriol. 177:1938-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Moubareck, C., N. Bourgeois, P. Courvalin, and F. Doucet-Populaire. 2003. Multiple antibiotic resistance gene transfer from animal to human enterococci in the digestive tract of gnotobiotic mice. Antimicrob. Agents Chemother. 47:2993-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norgren, M., and J. R. Scott. 1991. The presence of conjugative transposon Tn916 in the recipient strain does not impede transfer of a second copy of the element. J. Bacteriol. 173:319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul, J. H. 1999. Microbial gene transfer: an ecological perspective. J. Mol. Microbiol. Biotechnol. 1:45-50. [PubMed] [Google Scholar]
- 27.Perrin-Guyomard, A., S. Cottin, D. E. Corpet, J. Boisseau, and J. M. Poul. 2001. Evaluation of residual and therapeutic doses of tetracycline in the human-flora-associated (HFA) mice model. Regul. Toxicol. Pharmacol. 34:125-136. [DOI] [PubMed] [Google Scholar]
- 28.Poulsen, L. K., T. R. Licht, C. Rang, K. A. Krogfelt, and S. Molin. 1995. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J. Bacteriol. 177:5840-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen, L. D., S. J. Sørensen, R. R. Turner, and T. Barkay. 2000. Application of a mer-lux biosensor for estimating bioavailable mercury in soil. Soil Biol. Biochem. 32:639-646. [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schlundt, J., P. Saadbye, B. Lohmann, B. L. Jacobsen, and E. M. Nielsen. 1994. Conjugal transfer of plasmid DNA between Lactococcus lactis strains and distribution of transconjugants in the digestive tract of gnotobiotic rats. Microb. Ecol. Health Dis. 7:59-69. [Google Scholar]
- 32.Scott, J. R., F. Bringel, D. Marra, G. Van Alstine, and C. K. Rudy. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11:1099-1108. [DOI] [PubMed] [Google Scholar]
- 33.Scott, J. R., P. A. Kirchman, and M. G. Caparon. 1988. An intermediate in transposition of the conjugative transposon Tn916. Proc. Natl. Acad. Sci. USA 85:4809-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott, K. P. 2002. The role of conjugative transposons in spreading antibiotic resistance between bacteria that inhabit the gastrointestinal tract. Cell Mol. Life Sci. 59:2071-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Showsh, S. A., and R. E. Andrews, Jr. 1992. Tetracycline enhances Tn916-mediated conjugal transfer. Plasmid 28:213-224. [DOI] [PubMed] [Google Scholar]
- 36.Søorensen, S. J., and L. E. Jensen. 1998. Transfer of plasmid RP4 in bulk soil and barley seedling microcosms. Antonie Leeuwenhoek J. Microbiol. 1:1-9. [DOI] [PubMed] [Google Scholar]
- 37.Thal, L. A., J. Silverman, S. Donabedian, and M. J. Zervos. 1997. The effect of Tn916 insertions on contour-clamped homogeneous electrophoresis patterns of Enterococcus faecalis. J. Clin. Microbiol. 35:969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres, O. R., R. Z. Korman, S. A. Zahler, and G. M. Dunny. 1991. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol. Gen. Genet. 225:395-400. [DOI] [PubMed] [Google Scholar]
- 39.van Elsas, J. D., and M. J. Bailey. 2002. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 42:187-197. [DOI] [PubMed] [Google Scholar]
- 40.Wadolkowski, E. A., D. C. Laux, and P. S. Cohen. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect. Immun. 56:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996-997. [DOI] [PubMed] [Google Scholar]