Abstract
High-performance affinity chromatography (HPAC) was used to examine the changes in binding that occur for the sulfonylurea drug glibenclamide with human serum albumin (HSA) at various stages of glycation for HSA. Frontal analysis on columns containing normal HSA or glycated HSA indicated glibenclamide was interacting through both high affinity sites (association equilibrium constant, Ka, 1.4–1.9 × 106 M−1 at pH 7.4 and 37°C) and lower affinity sites (Ka, 4.4–7.2 × 104 M−1). Competition studies were used to examine the effect of glycation at specific binding sites of HSA. An increase in affinity of 1.7- to 1.9-fold was seen at Sudlow site I with moderate to high levels of glycation. An even larger increase of 4.3- to 6.0-fold in affinity was noted at Sudlow site II for all of the tested samples of glycated HSA. A slight decrease in affinity may have occurred at the digitoxin site, but this change was not significant for any individual glycated HSA sample. These results illustrate how HPAC can be used as tool for examining the interactions of relatively non-polar drugs like glibenclamide with modified proteins and should lead to a more complete understanding of how glycation can alter the binding of drugs in blood.
Keywords: High-performance affinity chromatography, Drug-protein binding, Glibenclamide, Human serum albumin, Glycation
1 Introduction
The International Diabetes Federation reported in 2011 that 366.2 million people in the world are affected by diabetes [1]. A total of 25.8 million are affected by diabetes in the United States alone [2]. Diabetes is a condition that is caused by high glucose levels in blood and has two major forms [3]. Type I diabetes (i.e., juvenile onset or insulin-dependent diabetes) affects 5–10% of diabetic patients and is caused when pancreatic beta cells (i.e., insulin-producing cells) are attacked by the immune system [2]. Most of the remaining 90–95% of diabetic patients suffer from type II diabetes (i.e., non-insulin dependent or adult onset diabetes), which is caused by insulin resistance [1,2].
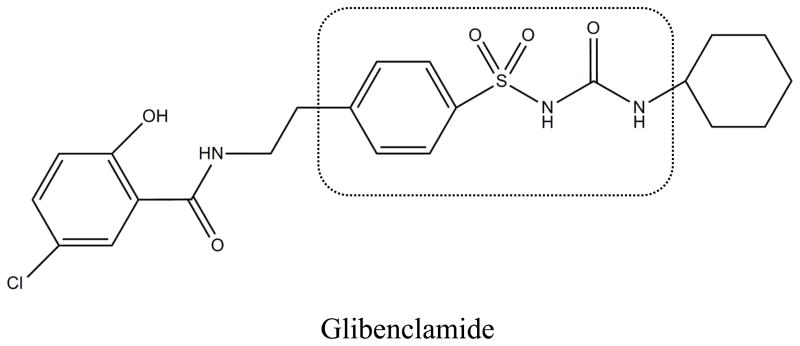
Sulfonylurea drugs are oral medications that are commonly used to treat type II diabetes. These drugs lower blood glucose levels by increasing the amount of insulin that is produced by pancreatic beta cells [4]. According to the biopharmaceutical classification system, sulfonylurea drugs are listed as class II drugs with high permeability and low solubility [5]. Figure 1 shows the core structure of a sulfonylurea drug, which is composed of phenylsulfonyl and urea groups, both which are hydrophilic [6,7]. Various non-polar functional groups can be found on either side of this core structure and contribute to both the effectiveness and metabolism of these drugs [6–8]. Glibenclamide, or glyburide (see Figure 1), is a popular second-generation sulfonylurea drug. Second-generation sulfonylurea drugs like glibenclamide are more easily excreted and can be prescribed in lower doses than first-generation sulfonylureas (e.g., tolbutamide and acetohexamide) [8]. For instance, glibenclamide has a therapeutic level in serum of 0.08–0.4 μM versus 60–215 or 185–370 μM for acetohexamide and tolbutamide, respectively [9].
Figure 1.
Structure of glibenclamide. The region within the dashed box shows the core structure of a sulfonylurea drug.
Although sulfonylurea drugs are used to lower blood glucose levels, hypoglycemia is a relatively common side effect if the apparent dose of these drugs is too high. This situation occurs in 2%–20% of patients, depending on the type of sulfonylurea being used [10]. One factor that will affect the free fractions of these drugs in the circulation, and their apparent dose, is the level of binding by these drugs with serum proteins, especially with human serum albumin (HSA) [6,9–12]. HSA (molar mass, 66.5 kDa) is the most abundant protein in plasma and is responsible for transporting various fatty acids, low mass hormones, and drugs in the circulation [11–19]. There are two main drug binding sites on HSA: Sudlow sites I and II. Sudlow site I is found in subdomain IIA and binds to bulky heterocyclic anionic drugs such as warfarin, azapropazone, phenylbutazone and salicylate [12,13]. Sudlow site II is located in subdomain IIIA and can bind to ibuprofen, fenoprofen, ketoprofen, benzodiazepine, and L-tryptophan, among other solutes [12,13]. Recent studies have shown that both of these sites have interactions with first-generation sulfonylurea drugs, tolbutamide and acetohexamide, and the second-generation drug gliclazide [14–16]. Other studies have found that some drugs can bind to a separate region on HSA known as the digitoxin site [17–22], but no previous reports have examined the interactions of sulfonylureas at this site.
The elevated levels of glucose in blood during diabetes can lead to a protein modification known as glycation [23–28]. Glycation is a non-enzymatic process in which amine groups can undergo the formation of a reversible Schiff base with the open chain form of a reducing sugar. The Schiff base can later rearrange to form a more stable Amadori product. Patients with diabetes are estimated to have 20–30% or more of HSA in a glycated form, while individuals without diabetes may have 6–13% of HSA glycated [23,24,26]. Previous work has suggested that glycation can alter the interactions between HSA and some solutes. For instance, it has been shown in chromatographic studies that glycation can alter the ability of Sudlow sites I and II on HSA to bind with sulfonylurea drugs [14–16]. Experiments based on fluorescence spectroscopy, circular dichroism, and theoretical calculations have also found that glycation and related modifications can affect intermolecular interactions between drugs and HSA, including possible changes in the binding of drugs at Sudlow site I of this protein [29,30].
The purpose of this study is to use high-performance affinity chromatography (HPAC) to examine the binding of glibenclamide to HSA at various stages of glycation. HPAC is a chromatographic technique that uses an immobilized biological molecule as the stationary phase [31–33]. Previous studies have shown that columns containing normal HSA or glycated HSA can provide good precision and fast analysis times for studies of drug-protein interactions [14–16], while also giving good correlation with solution-based methods (e.g., equilibrium dialysis, ultrafiltration, or spectroscopic methods) [32,33]. In addition, HPAC is easy to automate and can be used with various detection schemes, including absorbance, fluorescence or mass spectrometry [31–38]. HPAC has been used previously in detailed studies that have examined the effects of glycation on the binding of HSA to other sulfonylurea drugs [14–16], as well as the effects of some specific modifications on drug-protein interactions (e.g., the selective modification of Tyr-411, Trp-214, or Cys-34 on HSA) [39–41]. However, this approach has only been used in screening studies with glibenclamide and required a solubilizing agent for work with this relatively low solubility drug [42]. These conditions and the lack of more complete binding information have, in turn, made it difficult in prior work to directly compare the overall interactions of HSA with this drug and with other sulfonylureas.
This report will seek to overcome these prior limitations by examining how HPAC can be adapted for providing more complete information on the protein binding of relatively low solubility drugs such as glibenclamide. For instance, this approach will be modified and explored for use with such a drug in the methods of frontal analysis and zonal elution to look at both the global and site-specific changes in binding that may occur for glibenclamide with normal HSA versus in vitro glycated HSA. These experiments should help indicate how HPAC can then be modified for use with other non-polar drugs. In addition, this work should provide useful data on how glycation can alter the binding of glibenclamide or related drugs to HSA and lead to a more complete understanding of how glycation can alter the binding and transport of such drugs in the circulation.
2 Experimental
2.1 Reagents
The glibenclamide (≥ 99.9% pure), R-warfarin (≥ 97%), L-tryptophan (≥ 97%), digitoxin (97% pure), β-cyclodextrin (> 98% pure), D-(+)-glucose (≥ 99.5%), sodium azide (95%), HSA (essentially fatty acid free, ≥ 96%), and commercial sample of in vitro glycated HSA (Lot 058K6087) were obtained from Sigma-Aldrich (St. Louis, MO, USA). The Nucleosil Si-300 (7 μm particle diameter, 300 Å pore size) was purchased from Macherey-Nagel (Düren, Germany). Reagents for the bicinchoninic acid (BCA) protein assay were from Pierce (Rockford, IL, USA). For the measurement of glycation levels, a fructosamine assay kit was obtained from Diazyme Laboratories (San Diego, CA, USA). All aqueous solutions were prepared with water from a Nanopure system (Barnstead, Dubuque, IA, USA) and filtered through a 0.2 μm GNWP nylon membrane from Millipore (Billerica, MA, USA).
2.2 Apparatus
The chromatographic system was comprised of a DC-2080 degasser, two PU-2080 pumps, an AS-2057 autosampler, a CO-2060 column oven, and a UV-2075 absorbance detector from Jasco (Tokyo, Japan), which included a Rheodyne Advantage PF six-port valve (Cotati, CA, USA). EZ Chrom Elite software v3.21 (Scientific Software, Pleasanton, CA, USA) and Jasco LC Net were used to control the system. Chromatograms were analyzed through the use of Peak-Fit 4.12 (Jandel Scientific Software, San Rafael, CA, USA). Data Fit 8.1.69 (Oakdale, PA, USA) was utilized to perform non-linear regression.
2.3 Methods
For the sake of comparison, the in vitro glycated HSA samples used in this work were the same as employed in previous HPAC studies with other sulfonylurea drugs [14–16,43]. Although many prior studies have used in vitro glycated HSA for binding studies like those described in this report [14–16,30,44], it has also been suggested in recent work with related modifications that in vivo glycated HSA may provide a better model for representing drug-protein interactions that occur at physiological conditions [44]. To minimize differences due to the use of in vitro glycated HSA in this study, conditions for preparation of the glycated HSA samples were selected to closely mimic the glucose and protein concentrations and reaction conditions that are present in blood. The gHSA1 was purchased from Sigma and was glycated under proprietary conditions; this support had a measured glycation level of 1.31 (± 0.05) mol hexose/mol HSA, as might be found in a patient with prediabetes, and represented mildly glycated HSA [14–16,43]. The gHSA2 and gHSA3 samples were prepared in vitro as described previously [14,15,43] and represented glycation levels of patients with controlled or advanced diabetes [45,46]. These two preparations contained 2.34 (± 0.13) and 3.35 (± 0.14) mol hexose/mol HSA, respectively [14,15]. It has been found in separate, ongoing studies based on mass spectrometry and ultrafiltration and HPAC that these protein preparations have similar modification patterns and binding properties to samples of in vivo HSA with comparable levels of glycation [47–51], thus making these preparations reasonable models for the types of binding studies that are described in this report.
Diol-bonded silica was produced from Nucleosil Si-300 silica according to the literature [52]. The Schiff base method was used to immobilize HSA or glycated HSA onto the diol-bonded silica, also as reported previously [53–55]. Using the same procedure, control supports were prepared with no protein being added during the immobilization step. Although both the Schiff base immobilization method and glycation involve free amine groups on proteins, these processes tend to involve different residues on HSA [49–51]. A BCA assay was carried out in triplicate to determine the protein content of each support, using soluble HSA as the standard and samples containing the control support as the blanks. The support containing normal HSA was found previously with this assay to have a protein content of 38 (± 3) mg HSA/g silica. The three types of glycated HSA supports that were prepared (referred to later in this report as gHSA1, gHSA2 and gHSA3) were found to have protein contents of 29 (± 4), 47 (± 8), or 40 (± 3) mg HSA/g silica [14,43].
All of the supports were downward slurry packed into separate 2.0 cm × 2.1 mm I.D. columns at 3500 psi (24 MPa) using pH 7.4, 0.067 M potassium phosphate buffer. These columns were then stored at 4°C in the same pH 7.4 phosphate buffer. Each column was used in fewer than 500 sample applications and was routinely washed with pH 7.4, 0.067 M phosphate buffer. Throughout the course of this study, no significant changes in binding properties were noted for any of these columns, as reported previously for similar systems [53].
The R-warfarin, L-tryptophan, glibenclamide and digitoxin samples were prepared in pH 7.4, 0.067 M potassium phosphate buffer. The limited solubility of digitoxin (i.e., roughly 4 mg/L) [56] required the addition of 0.88 mM β-cyclodextrin to increase the solubility of this drug, as described in prior work [20–22]. The R-warfarin and digitoxin solutions were used within two weeks of preparation, and the L-tryptophan solutions were used within one day of preparation, respectively [43,57]. Due to the low solubility of glibenclamide in water, the procedure for preparing solutions containing this drug were altered from those used in previous studies with other, more soluble sulfonylurea drugs. Previous HPAC studies with acetohexamide, tolbutamide and gliclazide could be carried out by preparing overnight, with stirring at room temperature, solutions that contained up to 200–1000 μM of these drugs in a pH 7.4, 0.067 M phosphate buffer [14–16]. Glibenclamide was much less soluble under the same conditions and additional steps had to be taken to provide a suitably broad range of concentrations for use in methods such as frontal analysis. β-Cyclodextrin has previously been employed as a solubilizing agent for glibenclamide in screening studies [42]; however, this approach alters the apparent retention of the applied drug onto the column and requires relatively complex procedures to correct for this effect and carry out more detailed binding studies [31–38]. In this current study, solutions of glibenclamide were instead prepared without the use of any solubilizing agent by utilizing both stirring and sonication in a covered container that was kept for 5–7 days at 35–50°C. It was found that a stable stock solution containing up to 50 μM glibenclamide could be prepared under these conditions, as confirmed by absorbance measurements and dilution studies. This stock solution was then used directly or diluted to make working solutions for frontal analysis and zonal elution experiments involving HPAC.
The mobile phases used in the chromatographic studies were based on pH 7.4, 0.067 M potassium phosphate buffer, which was used to apply the samples and to elute retained analytes under isocratic conditions. The solutions used in the chromatographic system were filtered through a 0.2 μm nylon filter and were degassed for 10–15 min before use. All experiments were carried out at a physiological temperature of 37°C and using a flow rate of 0.5 mL/min. Prior work on similar columns has shown that frontal analysis and zonal elution studies performed under these flow rate conditions allow for the reproducible measurements of retention factors, binding capacities, and association equilibrium constants [14–16,43,55,57].
In the frontal analysis experiments, the columns were first equilibrated with pH 7.4, 0.067 M potassium phosphate buffer. Using a six-port valve, a switch was made from this pH 7.4 buffer to a solution containing a known concentration of glibenclamide in the same buffer. After a breakthrough curve was formed and a stable plateau had been reached, a switch was made back to the pH 7.4, 0.067 M potassium phosphate buffer to elute the retained drug. These frontal analysis experiments were carried out using sixteen sample solutions that contained 1–50 μM glibenclamide, with the elution of glibenclamide being monitored at 250 nm. All experiments were performed in quadruplicate and the central point of each breakthrough curve was determined by using the equal area function of Peak Fit 4.12. A correction for non-specific binding (e.g., 41% of the total binding seen on the HSA column for 50 μM glibenclamide) was made by subtracting the results for the control column from the data for each column containing normal HSA or glycated HSA, according to methods described in Refs. [14,16].
The zonal elution competition studies were carried out by utilizing R-warfarin as a site-specific probe for Sudlow site I of HSA, L-tryptophan as a probe for Sudlow site II, and digitoxin as a probe for the digitoxin site [12,13,20–22]. Mobile phases containing 1–20 μM glibenclamide were used in these experiments. These concentrations were used to dilute the site-specific probes to a concentration of 5 μM. During these studies, a 20 μL sample of 5 μM R-warfarin, L-tryptophan and digitoxin was injected onto each column and monitored at 308 nm, 280 nm, or 205 nm, respectively. Sodium nitrate, which was monitored at 205 nm, was injected at a concentration of 20 μM and used as a non-retained solute [32,38,42,43]. All of these injections were performed in quadruplicate on each protein column or control column. The central moments for the resulting peaks were determined by using PeakFit v4.12 and an exponentially modified Gaussian curve model.
3. Results and Discussion
3.1 Frontal analysis studies
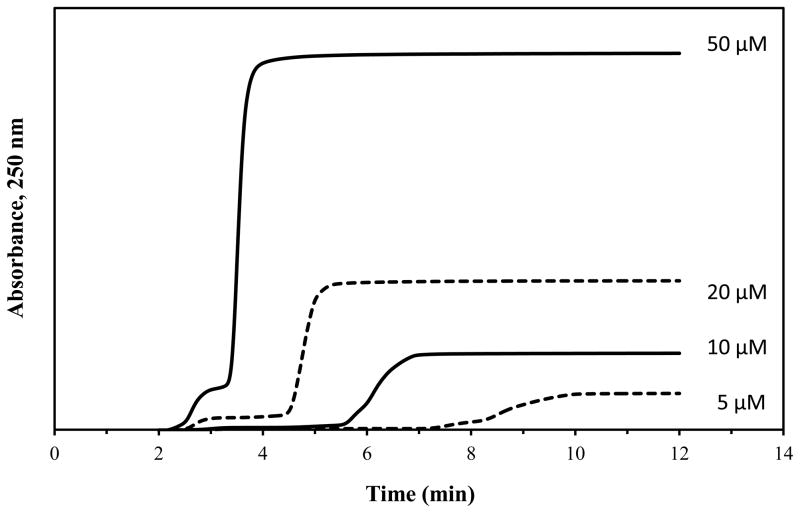
Frontal analysis was used to first examine the overall binding of glibenclamide to the normal HSA and glycated HSA columns. These experiments used HPAC to examine the global binding of glibenclamide with these proteins by providing information on the number of binding sites and association equilibrium constants for these sites. Figure 2 shows some typical breakthrough curves that were produced in these studies, which typically required 5–20 min to obtain. The moles of drug that were required to reach the central point of each breakthrough curve were measured as a function of the concentration of the applied drug. The results were then fit to various binding models.
Figure 2.
Example of frontal analysis studies for glibenclamide on a normal HSA column at pH 7.4 and 37°C. These results were obtained at 0.5 mL/min and using glibenclamide concentrations of 50, 20, 10, and 5 μM (top-to-bottom). The small initial step changes shown to the left occurred at or near the column void time and probably represent only a small difference in composition and background absorbance of each drug solution versus the application buffer; these small step changes were not included in the data analysis and integration of the much larger frontal analysis curves that are shown to the right.
Eqs. (1) or (2) were used to see how such data agreed with a model that involved the interactions of glibenclamide at a single type of binding region on HSA or glycated HSA [32,38].
One-site model:
| (1) |
| (2) |
In these equations, the term mLapp represents the measured moles of applied drug that was required to reach the central point of the breakthrough curve at a given molar concentration of the applied drug, [A] [32,38]. The association equilibrium constant and total moles of binding sites for this interaction are represented by terms Ka1 and mL1, respectively.
A two-site binding model was also used to examine the data, as described by Eqs. (3) and (4) [32,38].
Two-site model:
| (3) |
| (4) |
In these equations, Ka1 and Ka2 represent the association equilibrium constants for the two classes of sites, while the moles of these two groups of sites are described by the terms mL1 and mL2. Eq. (4) also includes the term α1, which represents the fraction of all binding sites for the drug that are made up of its highest affinity regions (e.g., α1 = mL1/mLtot if Ka1 and mL1 are used to refer to the highest affinity sites). In a similar manner, the ratio of the association equilibrium constants for the low versus high affinity sites is represented by β2, where β2 = Ka2/Ka1 in the case where Ka1 refers to the highest affinity sites and Ka2 refers to the lower affinity sites [32,38].
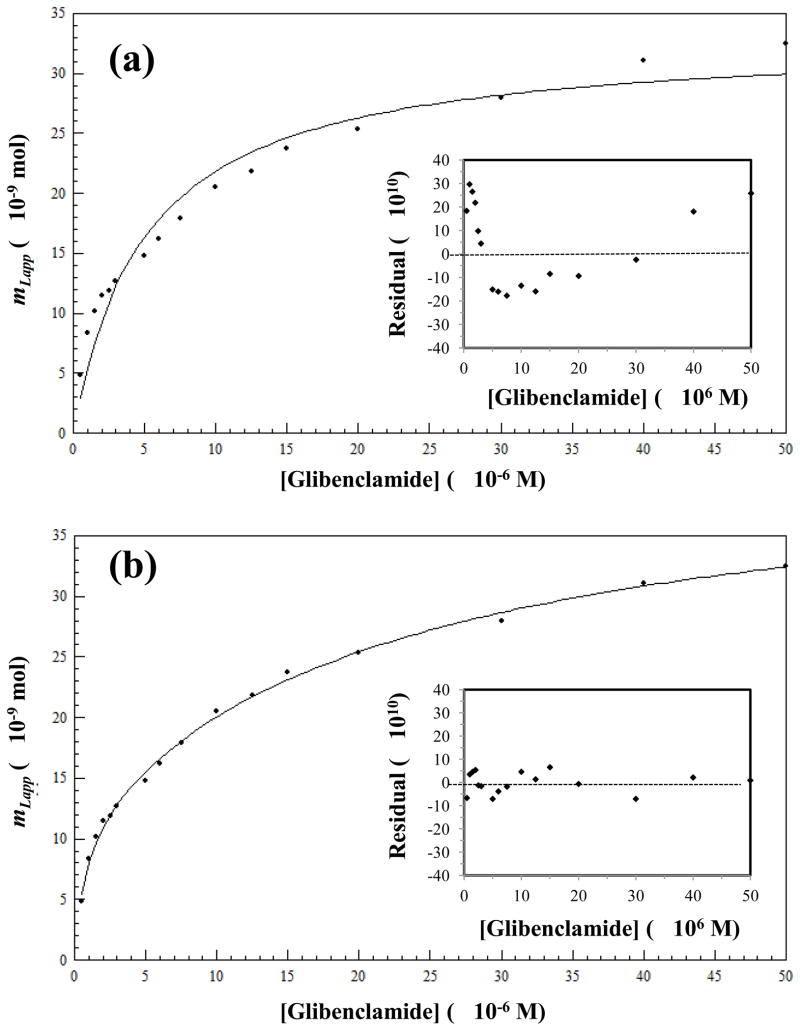
Previous results with other sulfonylurea drugs have indicated that these solutes tend to follow a two-site binding model during their interactions with both normal HSA and glycated HSA [14–16,58]. Figure 3 shows the results that were obtained when using non-linear regression and the non-transformed data from a frontal analysis experiment with glibenclamide and the normal HSA column; similar trends were seen for glycated HSA. In each case, lower concentrations were used in this work with glibenclamide than in previous experiments with other sulfonylurea drugs [14–16] due to the lower solubility of glibenclamide in an aqueous solution. To help compensate for this lower range and provide good estimates of the binding parameters for the system, a greater number of concentrations were tested in the given range and more replicates were used at each concentration (e.g., sixteen tested concentrations versus nine to fifteen and four replicates versus three for most of the previous experiments in Refs. [14–16]). Furthermore, several samples were at the lower end of this concentration range to better examine the stronger binding that was observed for glibenclamide with HSA when compared with previously-examined sulfonylureas [14–16]. Under these conditions, a two-site binding model was again found to give a better fit than a one-site binding model for the glibenclamide data in Figure 3, with a correlation value of 0.997 (n = 16) versus 0.952, respectively. As shown by the insets to Figure 3, residual plots for the two-site model gave a more random distribution of the residuals about the best-fit line when compared to the results for the one-site model and a smaller sum of the squares of the residuals (i.e., 3.0 × 10−18 vs. 4.9 × 10−17).
Figure 3.
Data obtained for glibenclamide on a normal HSA column as fit to (a) a one-site binding model generated by using Eq. (1) or (b) a two-site binding model generated by using Eq. (3). The insets in (a) and (b) show the corresponding residual plots. Each point represents the average of four runs, with typical relative standard deviations that ranged from ±0.02% to ±7.9% (average, ±1.9%).
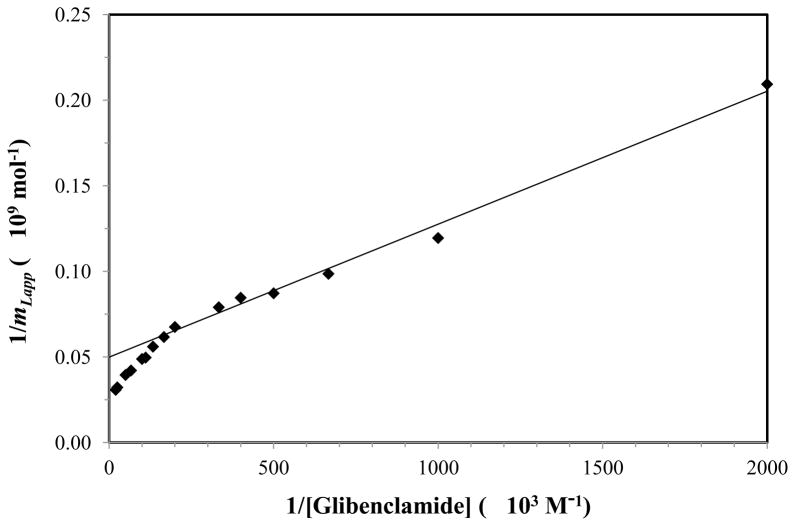
The presence of more than one group of binding sites for glibenclamide on normal HSA was confirmed when using Eqs. (2) and (4) and a double reciprocal plot of 1/mLapp vs. 1/[A] as an alternative route to examine the data. When this type of plot was made (see Figure 4), a linear relationship was seen as the value of 1/[A] increased, with clear deviations being noted at smaller values of 1/[A]. If one-site binding were present, a linear response at all values of 1/[A] would have been expected in such a plot, as predicted by Eq. (2). The fact that deviations from linearity were seen at small values for 1/[A] meant that at least two groups of sites were involved in this interaction, as indicated by Eq. (4) (note: similar deviations from linearity are expected for higher-order binding models) [32,38]. The linear response that was noted at higher values of 1/[A] in Figure 4 is predicted even for models involving multiple binding sites, as shown by Eq. (5) [59,60].
Figure 4.
A double-reciprocal plot for frontal analysis experiments that examined the binding of glibenclamide with normal HSA. The data in this plot were the same as utilized in Figure 3. The best-fit line was generated by using the data for 0.5–5 μM glibenclamide and gave a correlation coefficient of 0.996 (n = 7).
| (5) |
In addition, it is known from this linear region that an estimate can be made for the association equilibrium constant of the highest affinity sites in the column [59,60]. Using the linear region of Figure 4, the value of Ka1 was found to be approximately 6.4 (± 0.5) × 105 M−1 for the normal HSA column, where the value in parentheses represents a range of ± 1 S.D. This value for Ka1 is comparable to a binding constant of 7.6 × 105 M−1 at 37°C and pH 7.4 that has been previously reported for glibenclamide with normal HSA when using a one-site binding model [61].
Use of the two-site model in Eq. (3) and the non-transformed frontal analysis data provided association equilibrium constants of 1.4 (± 0.5) × 106 M−1 and 4.4 (± 1.0) × 104 M−1 for the binding of normal HSA with glibenclamide at pH 7.4 and 37°C (see Table 1). The mL values for these sites were 1.1 (± 0.2) × 10−8 mol and 3.1 (± 0.1) × 10−8 mol, respectively, in the column that was used for this experiment. It was noted in this case that the result for Ka1 was roughly 10-fold higher than the corresponding values for the high affinity sites that have been measured on an identical column and under the same mobile phase conditions for other sulfonylurea drugs (i.e., tolbutamide, acetohexamide and gliclazide) [16,58] (Note: a lower retention for glibenclamide on an HSA column was noted versus these other drugs in the screening studies described in Ref. [42], but in this earlier case the apparent retention for glibenclamide was lowered by the presence of β-cyclodextrin as a solubilizing agent). An increase in affinity for glibenclamide to HSA when compared to first-generation sulfonylurea drugs has been noted previously and was expected due to the larger aromatic groups that are present in glibenclamide versus these other agents [61].
Table 1.
Association equilibrium constants and moles of binding sites estimated when using a two-site model to describe the binding of glibenclamide with normal HSA and gHSA3 at pH 7.4 and 37 °Ca
| Type of HSAb | Ka1 (× 106 M−1) | mL1 (× 10−8 mol) | Ka2 (× 104 M−1) | mL2 (× 10−8 mol) |
|---|---|---|---|---|
| Normal HSA | 1.4 (± 0.5) | 1.1 (± 0.2) | 4.4 (± 1.0) | 3.1 (± 0.1) |
| gHSA3 | 1.9 (± 1.5) | 0.9 (± 0.3) | 7.2 (± 2.8) | 2.4 (± 0.2) |
The values in parentheses represent a range of ±1 S.D., as based on error propagation and the precisions of the best-fit slopes and intercepts obtained when using Eq. (3) for n = 16.
The level of glycation for the gHSA3 sample is given in Section 2.3.
The specific activity based on the protein content for this column at the high and lower affinity sites were 0.63 (± 0.09) and 1.73 (± 0.07) mol/mol normal HSA, respectively. This result suggested that one high affinity region existed on HSA with a binding constant in the range of 106 M−1, along with several lower affinity regions with binding constants between 104 and 105 M−1. For comparison, analysis of the same data by using a one-site model resulted in an intermediate value for Ka1 of 2.0 (± 0.3) × 105 M−1, a value for mL1 of 3.3 (± 0.1) × 10−8 mol, and a relative activity of 1.8 (± 0.1) mol/mol normal HSA.
Frontal analysis experiments were also performed with glibenclamide and a highly glycated HSA sample (gHSA3). Plots prepared according to Eqs. (1–4) gave trends similar to those shown for normal HSA in Figures 3 and 4. For each type of plot, the glycated HSA column again gave the best-fit to a two-site model (see summary in Table 1). The one-site model gave a correlation coefficient of 0.976 (n = 16) in comparison to a correlation coefficient of 0.994 for a two-site model for this column. Residual plot analysis for the two different models showed a random distribution for the two-site model and a smaller sum of the squares of the residuals when compared to the one-site model (i.e., 8.5 × 10−18 vs. 3.7 × 10−17). The association equilibrium constants obtained for this column when using Eq. (3) and a two-site model were 1.9 (± 1.5) × 106 M−1 and 7.2 (± 2.8) × 104 M−1. Based on the measured levels of these sites and the column’s known protein content, the relative activities for these sites were found to be 0.45 (± 0.16) and 1.20 (± 0.11) mol/mol gHSA3, which were consistent with the results observed for normal HSA. Analysis of the same data according to a double reciprocal plot gave deviations from linearity that confirmed at least two groups of binding sites were present. The linear region of this latter plot was again used with Eq. (5) to estimate Ka1 for the high affinity sites, giving a value of 6.8 (± 0.4) × 105 M−1. This result was comparable to the binding constant that was estimated by the same approach for the high affinity sites of glibenclamide with normal HSA.
3.2 Binding of glibenclamide at Sudlow site I
Competition studies and zonal elution were next used to test for any changes that may have occurred in the binding of glibenclamide at specific binding sites on normal HSA or glycated HSA. These experiments used site-specific probes to examine the competition between the injected probe and a competing agent, such as a drug that was placed at a known concentration in the mobile phase. R-Warfarin was employed as a site-specific probe for Sudlow site I in these studies, while the mobile phase contained glibenclamide as a competing agent. Sudlow site I was of interest in this work because previous studies with other sulfonylurea drugs have shown that this region has a relatively high affinity for these solutes on both normal HSA and glycated HSA [14–16,58]. Examples of some results that were obtained in this report with glibenclamide are given in Figure 5(a). In both these zonal elution studies and those described in the following sections, the same range in concentrations for glibenclamide as a competing agent was used as in prior work with other sulfonylurea drugs [14–16,58] because this range was well within the solubility limit of glibenclamide. However, the number of concentrations that were tested in this range was increased (from five-to-six up to seven-to-eight) and the number of replicates was increased (from three to four) to allow for more precise estimates to be obtained for the binding parameters of this system.
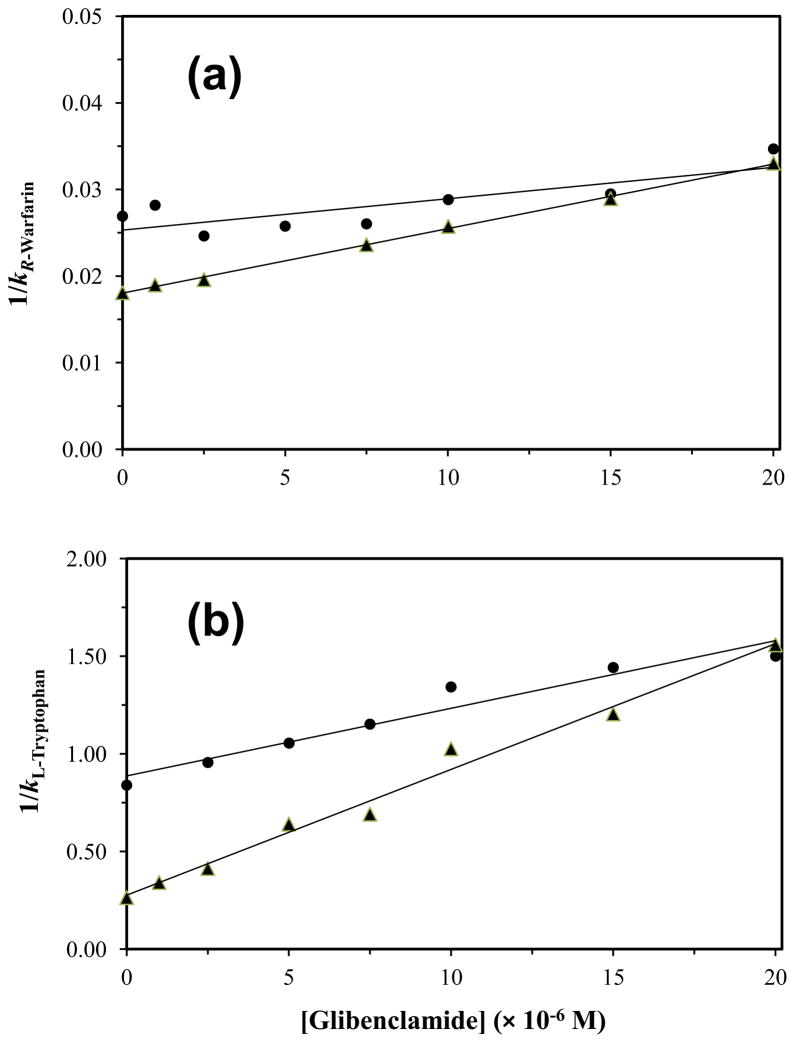
Figure 5.
Plots prepared according to Eq. (6) that show how the reciprocal of the retention factor for (a) R-warfarin or (b) L-tryptophan changed on normal HSA or glycated HSA columns as the concentration of glibenclamide was varied in the mobile phase. These results are for normal HSA (●) and gHSA2 (▲). Each point in these plots is the mean of four runs, with relative standard deviations in (a) that ranged from ±0.3% to ±2.6 (average, ±2.1%) and in (b) that ranged from ±2.2% to ±15.4 (average, ±7.1%). The correlation coefficients for the normal HSA and gHSA2 plots in (a) were 0.966 (n = 6) and 0.975 (n = 8), respectively, while the correlation coefficients in (b) were 0.970 (n = 7) and 0.993 (n = 8).
When analyzing data like that given in Figure 5, direct competition between an injected site-specific probe and a competing agent for a common binding site on an immobilized protein should result in a decrease in the retention factor (k) for the probe as the molar concentration of the competing agent ([I]) is increased [15,32]. The change in k as a function of [I] for a system with competition at a single type of site is described by Eq. (6). This equation predicts that a linear relationship will be formed for such a system when a plot is made of 1/k versus [I].
| (6) |
In this equation, KaA and KaI are the association equilibrium constants for the injected probe and competing agent, respectively, at their site of competition. The term VM is the column void volume, and mL represents the moles of common binding sites in the column. According to this relationship, the ratio of the slope to the intercept for the best-fit line can be used to find KaI, which provides information on the association equilibrium constant for the agent in the mobile phase at its specific site of competition with the injected probe.
Examination of the specific changes in binding at Sudlow site I for glibenclamide by this approach gave linear relationships according to Eq. (6) for the columns that contained either normal HSA or glycated HSA and under the drug concentrations that could be employed in these studies. These lines had correlation coefficients that ranged from 0.966 to 0.992 (n = 6–8). Residual plots gave random distributions for the data about the best-fit lines and sums for the squares of the residuals that ranged from 2.9 × 10−7 to 2.3 × 10−5. The results for all the tested columns were found to fit with a direct competition model for glibenclamide and R-warfarin at Sudlow site I. Similar previous experiments have noted that related drugs such as tolbutamide, acetohexamide and gliclazide also bind to this site on HSA [14–16].
In the next stage of this work, the association equilibrium constants that were obtained from Eq. (6) were used to see how the binding of glibenclamide at Sudlow site I was affected as the level of glycation for HSA was varied. The results are summarized in Table 2. The site-specific association equilibrium constant that was measured by this approach for glibenclamide at Sudlow site I of normal HSA was 2.4 (± 0.3) × 104 M−1 at pH 7.4 and 37°C. This value was slightly smaller than association equilibrium constants of 4.2–5.5 × 104 M−1 that have been reported for normal HSA with the first-generation sulfonylurea drugs tolbutamide and acetohexamide but was similar to an association equilibrium constant of 1.9 × 104 M−1 that has been measured at this site for gliclazide, another second-generation sulfonylurea drug [14–16,58] This binding constant is lower than the values of 0.64–1.4 × 106 M−1 which were estimated in Section 3.1 for the high affinity sites of glibenclamide with normal HSA, indicating that other regions probably made up the high affinity sites for this drug. It was further noted that this affinity was in the same general range as the value of 4.4 (± 1.0) × 104 M−1 that was obtained in Section 3.1 for the lower affinity sites of glibenclamide with normal HSA.
Table 2.
Local association equilibrium constants obtained for glibenclamide at specific binding regions of normal HSA and glycated HSA at 37°C and pH 7.4a
| Type of HSAb | Association equilibrium constant, KaI | ||
|---|---|---|---|
| Sudlow site I, (× 104 M−1) | Sudlow site II (× 104 M−1) | Digitoxin site (× 106 M−1) | |
| Normal HSA | 2.4 (± 0.3) | 3.9 (± 0.2) | 2.1 ± (0.8) |
| gHSA1 | 2.5 (± 0.1) | 16.7 (± 0.4) | 1.7 ± (0.8) |
| gHSA2 | 4.1 (± 0.7) | 23.3 (± 0.8) | 1.1 ± (0.4) |
| gHSA3 | 4.5 (± 0.3) | 17.8 (± 0.4) | 1.2 ± (0.2) |
The values in parentheses represent a range of ±1 S.D., as based on error propagation and the precisions of the best-fit slopes and intercepts obtained when using Eq. (6) for n = 5–8.
gHSA1, gHSA2, and gHSA3 refer to glycated HSA samples with different levels of glycation, as described in more detail in Section 2.3.
The association equilibrium constant for glibenclamide at Sudlow site I for normal HSA was next compared to values measured at the same site for each glycated HSA sample. There was no significant increase in the association equilibrium constant for glibenclamide at Sudlow site I in going from normal HSA to gHSA1. However, a 1.7- to 1.9-fold increase in the association equilibrium constant was noted between normal HSA and gHSA2 or gHSA3, which was significant at the 95% confidence level. A change in affinity with the level of glycation has also been observed at Sudlow site I for tolbutamide, acetohexamide, and gliclazide [14–16], as well as for other drugs (e.g., meloxicam and warfarin) [30]. These changes have been suggested to be due to variations in the extent and types of modifications that occur at Sudlow site I during the glycation process [14–16,30,49–51].
3.3 Binding of glibenclamide at Sudlow siteII
Competition experiments were also performed for glibenclamide using L-tryptophan as a probe for Sudlow site II of HSA. Previous studies have shown that this site is another region that has moderately strong binding to other sulfonylurea drugs [14–16,58]. Figure 5(b) shows some examples of the data that were obtained for normal HSA and glycated HSA in these experiments. All of the results gave a linear responses when fit to Eq. (6), with correlation coefficients that ranged from 0.968 to 0.993 (n = 7–8). The corresponding residual plots gave a random distribution of the points about the best-fit lines. It was determined from these results that glibenclamide was competing with L-tryptophan and was binding directly to Sudlow site II on both normal HSA and glycated HSA.
The site-specific association equilibrium constants that were determined for glibenclamide at Sudlow site II through these competition studies are summarized in Table 2. The affinity for glibenclamide at Sudlow site II of normal HSA was 3.9 (± 0.2) × 104 M−1, which is similar to values of 5.3–13 × 104 M−1 that have been reported at this site for acetohexamide, tolbutamide and gliclazide [14–16,58]. However, this result is still much lower than the high affinity constant of 0.64–1.4 × 106 M−1 that was estimated for glibenclamide with HSA in Section 3.1. Thus, this indicated that another region on this protein besides Sudlow sites I or II also had strong interactions with glibenclamide. The binding constant for glibenclamide to Sudlow site II of normal HSA was instead a better fit with the value of 4.4 (± 1.0) × 104 M−1 that was measured in the frontal analysis studies for the lower affinity regions of HSA. This similarity, when combined with the results in Section 3.2 and the moles of low affinity regions that were determined in Section 3.1, suggested that both Sudlow sites I and II made up the lower affinity sites that were detected during the frontal analysis experiments. This model also fits with the observation that the value for Ka2 in Table 1 increased significantly in going from normal HSA to gHSA3, because a large increase was also seen in Table 2 for the association equilibrium constant of glibenclamide at Sudlow site II between normal HSA and gHSA3.
Table 2 shows how the affinity for glibenclamide with HSA at Sudlow site II, as determined from the best-fit lines obtained with Eq. (6), changed as the level of glycation for HSA was increased. The columns containing gHSA1, gHSA2 and gHSA3 had a 4.3-, 6.0- or 4.6-fold increase in affinity for glibenclamide at Sudlow site II versus normal HSA. All of these differences were significant at the 95% confidence level. This trend is similar to the large increases in affinity that have been observed for L-tryptophan with the same samples of glycated HSA [62]. Tolbutamide has also been noted to have a modest increase in affinity at this site during the glycation of HSA [15], while acetohexamide and gliclazide have been found to have a moderate decrease in affinity or a mixed change in binding strength as the levels of glycation for HSA are varied [14,16].
3.4 Binding of glibenclamide at the digitoxin site
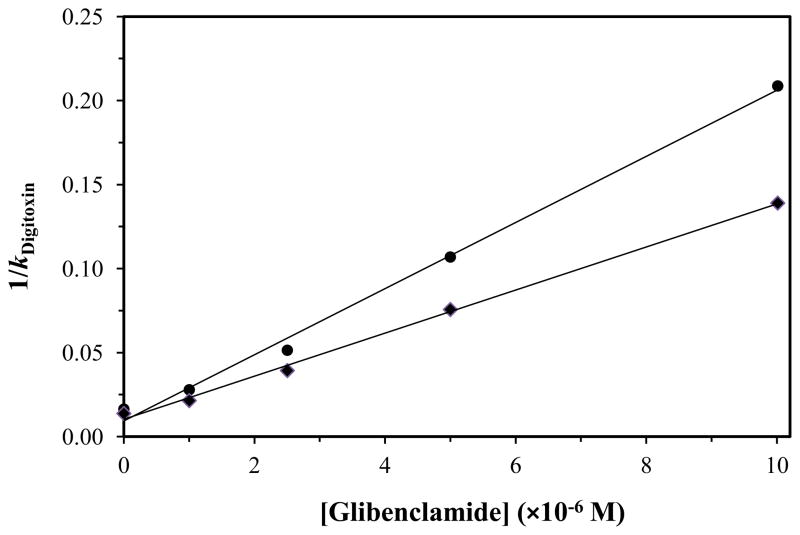
To help locate the high affinity region for glibenclamide on HSA, digitoxin was also used as a site-specific probe for HSA [17–22]. Figure 6 provides some typical results that were obtained for normal HSA and glycated HSA. When Eq. (6) was used to analyze these results, all of the normal and glycated HSA columns gave a linear response. The best-fit lines had correlation coefficients that ranged from 0.994 to 0.999 (n = 4–5), and the residual plots gave a random distribution for the data about the best-fit lines. These results indicated that glibenclamide and digitoxin had direct competition on the tested columns, confirming that glibenclamide was binding to the digitoxin site of HSA. This result is supported by a previous observation that glibenclamide and first-generation drugs such as tolbutamide appear to bind to different numbers of sites and through different mechanisms with HSA, with non-polar interactions being important for glibenclamide and ionic forces playing a greater role for tolbutamide [61]. This model and the observed binding of glibenclamide to the digitoxin site is also consistent with the fact that this site is known to bind other large, relatively hydrophobic drugs such as digitoxin and acetyldigitoxin [17–22].
Figure 6.
Plots prepared according to Eq. (6) that show how the reciprocal of the retention factor for digitoxin on HSA or glycated HSA columns changed as the concentration of glibenclamide was varied in the mobile phase. These results are for normal HSA (●) and gHSA3 (◆). The correlation coefficients for these plots were 0.998 (n = 5) and 0.999 (n = 5), respectively. Each point in these plots is the mean of four runs, with relative standard deviations that ranged from ±0.6% to ±4.6 (average, ±1.8%).
The association equilibrium constants for glibenclamide at the digitoxin site were next calculated from the best-fit lines of plots like those in Figure 6. The results are summarized in Table 2. In this case, the association equilibrium constant measured for glibenclamide at the digitoxin site of normal HSA was 2.1 (± 0.8) × 106 M−1. This value was now statistically identical to the association equilibrium constant of 1.4 (± 0.5) × 106 M−1 that had been previously estimated by frontal analysis for the high affinity sites of glibenclamide on normal HSA. The same group of experiments suggested that there may have been a decrease of 1.2- to 1.9-fold in affinity at the digitoxin site for glibenclamide when going from normal HSA to gHSA1, gHSA2 or gHSA3. None of these individual differences were significant at the 90% or 95% confidence level; however, the overall trend of a decrease in affinity with an increase in the level of HSA glycation was significant at the 90% confidence level when the complete set of samples was considered.
4 Conclusion
This report illustrated how HPAC could be modified for use with relatively low solubility drugs such as glibenclamide and used as a tool to examine variations in drug interactions with modified proteins, as demonstrated by using this approach to investigate the changes that occur in the binding of glibenclamide to HSA at various stages of glycation. Frontal analysis studies were used to estimate the affinity and moles of binding sites for glibenclamide with HSA. The results showed that binding with normal HSA and glycated HSA followed a two-site model in which interactions occurred at both high and lower affinity sites. The association equilibrium constants for the high affinity regions were in the range of 1.4–1.9 × 106 M−1 at pH 7.4 and 37°C for columns containing normal HSA or glycated HSA. The lower affinity regions had association equilibrium constants that increased from 4.4 × 104 M−1 to 7.2 × 104 M−1 for the same columns.
The binding of glibenclamide to normal HSA and glycated HSA at Sudlow sites I and II and at the digitoxin site was confirmed through the use of zonal elution competition studies. The affinities for glibenclamide at Sudlow sites I and II of normal HSA were 2.4 × 104 M−1 and 3.9 × 104 M−1, respectively. These values were consistent with values that were estimated for the lower affinity sites in the frontal analysis experiments. As the level of glycation was increased, a 1.7- to 1.9-fold increase in affinity was seen for glibenclamide at Sudlow site I for HSA with moderate to high levels of glycation. An even larger change was noted at Sudlow site II, in which an increase in affinity of 4.3- to 6.0-fold was observed versus normal HSA for all of the tested glycated HSA samples. The association equilibrium constant for glibenclamide at the digitoxin site of normal HSA was 2.1 × 106 M−1, which fit with the value that was measured for the high affinity sites by frontal analysis. Further studies indicated that glycation may have lead to a slight decrease in affinity for glibenclamide at the digitoxin site, but this change was not significant at the 95% confidence level for any individual sample of glycated HSA.
The results of this report are of clinical interest for several reasons. First, the large changes in binding seen for glibenclamide with glycated HSA, especially at Sudlow site II, are of clinical interest because they would be expected to alter the affective dose of the drug by changing the drug’s free fraction in the circulation. This effect is of potential importance for sulfonylurea drugs like glibenclamide because of the high level of plasma protein binding of this drug, the relatively narrow therapeutic range of this drug in serum, and the undesirable effects that occur if such the apparent activity of this drug levels fall below or above this range (i.e., hyperglycemia or hypoglycemia) [9–11]. The fact that the major drug binding regions on HSA are affected to different extents by glycation is also of interest in that it implies that drug-drug interactions at these sites will also vary for agents like glibenclamide. Finally, this work confirms that many sulfonylurea drugs can bind to Sudlow sites I and II but also demonstrates for the first time that the digitoxin site can play a major role in these interactions in the case of glibenclamide.
These results also illustrate how HPAC can be modified and used for examining the interactions of relatively non-polar drugs like glibenclamide with modified proteins and to provide a quantitative analysis of the changes in binding for such drugs that may occur both globally and at specific interaction sites. These efforts were aided by many of the potential advantages of HPAC for drug-binding studies. For instance, the ability of HPAC to be used with detection methods such as UV/visible absorbance spectroscopy [31–33,35,39–42] made it possible to look at the low-to-moderate concentrations of glibenclamide that were required for the frontal analysis and zonal elution experiments in this report. The good precision and fast analysis times of HPAC [31–33], along with the ability to reuse normal HSA or glycated HSA columns over hundreds of experiments [14–16], made it convenient to use more replicates and sample concentrations with these columns without the need for additional protein. Furthermore, this last feature made it possible to use the same protein preparations as in work with previous sulfonylurea drugs [14–16,58], allowing a direct comparison to now be made in the binding properties of glibenclamide versus these other drugs. The ability to easily combine this method with new probes (e.g., digitoxin) for examining newly-identified drug-protein interaction sites was further illustrated in this report. The approaches used here for such experiments are not limited to glibenclamide and HSA but could be adapted for use with systems that involve other drugs or modified proteins. Based on these features, it is expected that HPAC will continue to grow in applications and as a powerful technique for examining these and additional types of biological interactions.
Highlights.
HPLC affinity columns were used to study drug binding with modified proteins.
Binding of glibenclamide with glycated human serum albumin (HSA) was studied.
Frontal analysis was used to detect global changes in binding for glycated HSA.
Competition studies examined site-specific changes in binding by glibenclamide.
Local changes in binding upon HSA glycation were seen at Sudlow sites I and II.
Acknowledgments
This work was supported by the National Institutes of Health under grant R01 DK069629 and was conducted in facilities that were renovated under grant RR015468.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.International Diabetes Federation. IDF Diabetes Atlas. 5. Chap 2 Brussels, Belgium: International Diabetes Federation; 2011. [Google Scholar]
- 2.National Diabetes Fact Sheet: General Information and National Estimates on Diabetes in the United States, 2011. US Centers for Disease Control and Prevention; Atlanta, GA: 2011. pp. 1–12. [Google Scholar]
- 3.Nelson DL, Cox M. Lehninger Principles of Biochemistry. Chap 23 Freeman; New York: 2005. [Google Scholar]
- 4.Skillman TG, Feldman JM. Am J Med. 1981;70:36. doi: 10.1016/0002-9343(81)90773-7. [DOI] [PubMed] [Google Scholar]
- 5.Amidon GL, Lennernas H, Shah VP, Crison JR. Pharm Res. 1995;12:413. doi: 10.1023/a:1016212804288. [DOI] [PubMed] [Google Scholar]
- 6.Zavod RM, Krstenansky JL, Currie BL. In: Foye’s of Medicinal Chemistry. Lemke TL, Williams DA, editors. Chap 32 Lippincott Williams and Wilkins; Philadelphia: 2008. [Google Scholar]
- 7.Foster DW. In: Harrison’s Principles of Internal Medicine. Isselbacher KJ, Braunwald E, Wilson JD, Martin JB, Fauci AS, Kasper DL, editors. Chap 29 McGraw-Hill; New York: 1998. [Google Scholar]
- 8.Jakoby MG, Covey DF, Cistola DP. Biochemistry. 1995;34:8780. doi: 10.1021/bi00027a029. [DOI] [PubMed] [Google Scholar]
- 9.Regenthal R, Krueger M, Koeppel C, Preiss R. J Clin Monitor Computing. 1999;15:529. doi: 10.1023/a:1009935116877. [DOI] [PubMed] [Google Scholar]
- 10.Davis SN. In: Goodman and Gilman’s The Pharmacological Basis Of Therapeutics. 11. Brunton LL, Lazo JS, Parker KL, editors. Chap 60 McGraw-Hill; New York: 2006. [Google Scholar]
- 11.Tietz NW, editor. Clinical Guide to Laboratory Tests. 2. Saunders; Philadelphia: 1990. [Google Scholar]
- 12.Sudlow G, Birkett DJ, Wade DN. Mol Pharmacol. 1976;12:1052. [PubMed] [Google Scholar]
- 13.Ascoli GA, Domenici E, Bertucci C. Chirality. 2006;18:667. doi: 10.1002/chir.20301. [DOI] [PubMed] [Google Scholar]
- 14.Joseph KS, Anguizola J, Jackson AJ, Hage DS. J Chromatogr B. 2010;878:2775. doi: 10.1016/j.jchromb.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph KS, Anguizola J, Hage DS. J Pharm Biomed Anal. 2011;54:426. doi: 10.1016/j.jpba.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda R, Anguizola J, Joseph KS, Hage DS. Anal Bioanal Chem. 2011;401:2811. doi: 10.1007/s00216-011-5382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bros O, Fremstad D, Poulsson C. Pharmacol Toxicol. 1993;72:310. doi: 10.1111/j.1600-0773.1993.tb01656.x. [DOI] [PubMed] [Google Scholar]
- 18.Sjoholm I. In: Drug-Protein Binding. Reidenberg M, Erill S, editors. Chap 4 Praeger; New York: 1986. [Google Scholar]
- 19.Brok A. Acta Pharmacol Toxicol. 1975;36:13. doi: 10.1111/j.1600-0773.1975.tb00767.x. [DOI] [PubMed] [Google Scholar]
- 20.Hage DS, Sengupta A. J Chromatogr B. 1999;724:91. doi: 10.1016/s0378-4347(98)00589-1. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Ohnmacht C, Hage DS. J Chromatogr B. 2004;809:137. doi: 10.1016/j.jchromb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Ohnmacht CM, Chen S, Tong Z, Hage DS. J Chromatogr B. 2006;836:83. doi: 10.1016/j.jchromb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 23.Mendez DL, Jensen RA, McElroy LA, Pena JM, Esquerra RM. Arch Biochem Biophys. 2005;444:92. doi: 10.1016/j.abb.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 24.Colmenarejo G. Med Res Rev. 2003;23:275. doi: 10.1002/med.10039. [DOI] [PubMed] [Google Scholar]
- 25.Koyama H, Sugioka N, Uno A, Mori S, Nakajima K. Biopharm Drug Dispos. 1997;18:791. doi: 10.1002/(sici)1099-081x(199712)18:9<791::aid-bdd66>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Garlick RL, Mazer JS. J Biol Chem. 1983;258:6142. [PubMed] [Google Scholar]
- 27.Iberg N, Fluckiger R. J Biol Chem. 1986;261:13542. [PubMed] [Google Scholar]
- 28.Nakajou K, Watanabe H, Kragh-Hansen U, Maruyama T, Otagiri M. Biochim Biophys Acta. 2003;1623:88. doi: 10.1016/j.bbagen.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Khodarahmi R, Karimi SA, Kooshk MRA, Ghadami SA, Ghobadi S, Amani M. Spectrochimica Acta Part A. 2012;89:177. doi: 10.1016/j.saa.2011.12.058. [DOI] [PubMed] [Google Scholar]
- 30.Trynda-Lemiesz L, Wiglusz K. J Mol Structure. 2011;995:35. [Google Scholar]
- 31.Schiel JE, Joseph KS, Hage DS. In: Adv Chromatogr. Grinsberg N, Grushka E, editors. Chap 4 Taylor & Francis; New York: 2010. [Google Scholar]
- 32.Hage DS. J Chromatogr B. 2002;768:3. doi: 10.1016/s0378-4347(01)00482-0. [DOI] [PubMed] [Google Scholar]
- 33.Patel S, Wainer IW, Lough WJ. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. Chap 24 Taylor & Francis; New York: 2006. [Google Scholar]
- 34.Winzor DJ. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. Chap 23 Taylor & Francis; New York: 2006. [Google Scholar]
- 35.Domenici E, Bertucci C, Salvadori P, Wainer IW. J Pharm Sci. 1991;80:164. doi: 10.1002/jps.2600800216. [DOI] [PubMed] [Google Scholar]
- 36.Kovarik P, Hodgson RJ, Covey T, Brook MA, Brennan JD. Anal Chem. 2005;77:3340. doi: 10.1021/ac048263p. [DOI] [PubMed] [Google Scholar]
- 37.Schriemer DC. Anal Chem. 2004;76:440A. doi: 10.1021/ac041684m. [DOI] [PubMed] [Google Scholar]
- 38.Hage DS, Anguizola J, Barnaby O, Jackson A, Yoo MJ, Papastavros E, Pfaunmiller E, Sobansky M, Tong Z. Curr Drug Metab. 2011;12:313. doi: 10.2174/138920011795202938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noctor TAG, Wainer IW. Pharmaceut Res. 1992;9:480. doi: 10.1023/a:1015884112039. [DOI] [PubMed] [Google Scholar]
- 40.Chattopadhyay A, Tian T, Kortum L, Hage DS. J Chromatogr B. 1998;715:183. doi: 10.1016/s0378-4347(98)00140-6. [DOI] [PubMed] [Google Scholar]
- 41.Haginaka J, Kanasugi N. J Chromatogr A. 1997;769:215. doi: 10.1016/s0021-9673(97)00025-3. [DOI] [PubMed] [Google Scholar]
- 42.Basiaga SBG, Hage DS. J Chromatogr B. 2010;878:3193. doi: 10.1016/j.jchromb.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joseph KS, Hage DS. J Pharm Biomed Anal. 2010;53:811. doi: 10.1016/j.jpba.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baraka-Vidot J, Guerin-Dubourg A, Bourdon E, Rondeau P. Biochimie. 2012;94:1960. doi: 10.1016/j.biochi.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 45.Powers AC. In: Harrison’s Principles of Internal Medicine. Kasper DL, Fauci AS, Longo DL, Braunwald E, Hauser SL, Jameson JL, editors. Chap 323 McGraw-Hill; New York: 2005. [Google Scholar]
- 46.Lapolla A, Fedele D, Reitano R, Arico NC, Seraglia R, Traldi P, Marotta E, Tonani R. J Am Soc Mass Spectrom. 2004;15:496. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Barnaby O. PhD Dissertation. University of Nebraska; Lincoln, NE: 2010. Characterization of Glycation Sites on Human Serum Albumin using Mass Spectrometry. [Google Scholar]
- 48.Joseph KS. PhD Dissertation. University of Nebraska; Lincoln, NE: 2010. Chromatographic Studies of Drug-Protein Binding in Diabetes. [Google Scholar]
- 49.Barnaby OS, Cerny RL, Clarke W, Hage DS. Clin Chim Acta. 2011;412:277. doi: 10.1016/j.cca.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnaby OS, Cerny RL, Clarke W, Hage DS. Clin Chim Acta. 2011;412:1606. doi: 10.1016/j.cca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wa C, Cerny RL, Hage DS. Anal Chem. 2006;78:7967. doi: 10.1021/ac0609935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruhn PF, Garver S, Hage DS. J Chromatogr A. 1994;669:9. doi: 10.1016/0021-9673(94)80332-3. [DOI] [PubMed] [Google Scholar]
- 53.Yang J, Hage DS. J Chromatogr A. 1997;766:15. doi: 10.1016/s0021-9673(96)01040-0. [DOI] [PubMed] [Google Scholar]
- 54.Loun B, Hage DS. Anal Chem. 1994;66:3814. doi: 10.1021/ac00093a043. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Hage DS. J Chromatogr. 1993;645:241. doi: 10.1016/0021-9673(93)83383-4. [DOI] [PubMed] [Google Scholar]
- 56.Yalkowsky SH, Dannenfelser RM. Aquasol Database of Aqueous Solubility, Ver. 5. Univ. Ariz; Tucson: 1992. [Google Scholar]
- 57.Conrad ML, Moser AC, Hage DS. J Sep Sci. 2009;32:1145. doi: 10.1002/jssc.200800567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joseph KS, Hage DS. J Chromatogr B. 2010;878:1590. doi: 10.1016/j.jchromb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tweed SA, Loun B, Hage DS. Anal Chem. 1997;69:4790. doi: 10.1021/ac970565m. [DOI] [PubMed] [Google Scholar]
- 60.Tong Z, Schiel JE, Papastavros E, Ohnmacht CM, Smith QR, Hage DS. J Chromatogr A. 2011;1218:2065. doi: 10.1016/j.chroma.2010.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crooks MJ, Brown KF. J Pharm Pharmac. 1974;26:304. doi: 10.1111/j.2042-7158.1974.tb09280.x. [DOI] [PubMed] [Google Scholar]
- 62.Joseph KS, Hage DS. J Pharm Biomed Anal. 2010;53:811. doi: 10.1016/j.jpba.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]