Abstract
The measurement of 1H transverse paramagnetic relaxation enhancement (PRE) has been used in biomolecular systems to determine long-range distance restraints and to visualize sparsely-populated transient states. The intrinsic flexibility of most nitroxide and metal-chelating paramagnetic spin-labels, however, complicates the quantitative interpretation of PREs due to delocalization of the paramagnetic center. Here, we present a novel, disulfide-linked nitroxide spin label, R1p, as an alternative to these flexible labels for PRE studies. When introduced at solvent-exposed α-helical positions in two model proteins, calmodulin (CaM) and T4 lysozyme (T4L), EPR measurements show that the R1p side chain exhibits dramatically reduced internal motion compared to the commonly used R1 spin label (generated by reacting cysteine with the spin labeling compound often referred to as MTSL). Further, only a single nitroxide position is necessary to account for the PREs arising from CaM S17R1p, while an ensemble comprising multiple conformations is necessary for those observed for CaM S17R1. Together, these observations suggest that the nitroxide adopts a single, fixed position when R1p is placed at solvent-exposed α-helical positions, greatly simplifying the interpretation of PRE data by removing the need to account for the intrinsic flexibility of the spin label.
Keywords: Paramagnetic relaxation enhancement, Site-directed spin-labeling, Electron paramagnetic resonance
Introduction
In recent years, a number of research groups have developed a variety of NMR methods to probe biomolecular structure and dynamics based on the site-specific introduction of paramagnetic species (Clore 2008, 2011; Clore and Iwahara 2009; Keizers and Ubbink 2011; Otting 2010). The most generally applicable method for introducing paramagnetic species is site-directed spin labeling (SDSL) (Hubbell et al. 2000; Todd et al. 1989), in which a cysteine is introduced at a site of interest by site-directed muta-genesis, and then modified with a sulfhydryl-specific compound to generate a spin label side chain. Due to the large dipolar interaction between the unpaired electrons of the paramagnetic species and the nuclear spins of the biomolecule, position-specific distance, angular information, or both can be extracted. Among the various paramagnetic NMR techniques, the measurement of transverse paramagnetic relaxation enhancements rates (PREs), particularly of 1HN nuclei, has shown broad applicability, allowing a number of important questions in structure and dynamics to be addressed (Fawzi et al. 2010; Iwahara and Clore 2006; Iwahara et al. 2004a, 2006; Takayama and Clore 2011; Tang et al. 2006, 2008, 2007; Volkov et al. 2010, 2006). PREs arising from site-directed introduction of nitroxide spin labels or paramagnetic metal ions are large enough to reliably measure distances up to 35 Å from the unpaired electron and to probe transient, sparsely-populated intra- or inter-molecular interactions that occasionally bring the spin label position in close proximity to 1H nuclei of interest (Iwahara et al. 2004b; 2007).
Up to this point, however, the motion of the spin label relative to the protein has complicated direct interpretation of both intra- and intermolecular PREs due to the conformational space that can be sampled by the paramagnetic center (Iwahara et al. 2004b). Among the myriad spin labels available, R1 (Fig. 1a) is the most widely used for EPR studies, and has also been used successfully in several PRE studies (Bermejo et al. 2009; Volkov et al. 2006). At solvent-exposed helical sites, the R1 side chain occupies a set of rotamers that are limited from the backbone through the disulfide linkage region due in part to interactions of the disulfide with the peptide backbone (Fleissner et al. 2009; Guo et al. 2008; 2007; Warshaviak et al. 2011). Transitions between rotamers in this region of the side chain change the spatial position of the nitroxide relative to the protein on the time scale of microseconds (Bridges et al. 2010), though additional rotamer transitions for the terminal rotatable bonds are site-dependent and on a much faster (likely in the ns range) time scale (Chou et al. 2003). NMR studies attempting to quantitatively interpret the PREs arising from R1 (Bermejo et al. 2009), like any other label with internal motions, must take into account the ensemble of nitroxide positions, their interconversion dynamics and the order parameters for the interspin (nitroxide-nucleus) vectors (Clore and Iwahara 2009; Iwahara et al. 2004b).
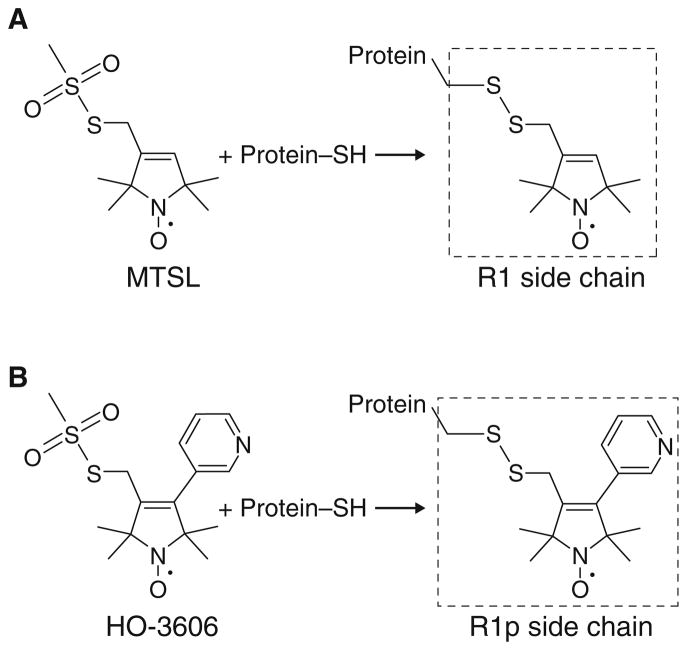
Fig. 1.
Reaction of the widely used spin-labeling compound (“MTSL”) and HO-3606 with a protein sulfhydryl group to generate the R1 and R1p side chains, respectively
To simplify the interpretation of NMR PRE data, an ideal spin label would not perturb the intrinsic structure and dynamics of the protein, and would have a single paramagnetic center that is rigidly attached to the protein. Indeed, several relatively rigid spin labels have been evaluated, but their general use for PRE studies appears to be limited. One such label is TOAC, an amino acid containing a stable nitroxide radical that can be introduced into synthetic peptides and has been used successfully for PRE studies (Lindfors et al. 2011). Although both the Cα and the nitroxide of TOAC are contained within a single piperidine ring, the radical can occupy at least two distinct positions because of ring isomerizations (Crisma et al. 2005; Flippen-Anderson et al. 1996). Moreover, the utility of TOAC for PRE studies is limited because its incorporation into large or isotope labeled proteins is not practical, and also risks perturbing local protein structure and dynamics. Two strategies, primarily used for the measurement of pseudo-contact chemical shifts (PCS), rely on multiple functional groups, either engineered or existing, in order to introduce paramagnetic ion chelating moieties. One is CLaNP-5 (Keizers et al. 2007), a large, organic molecule pre-chelated to a lanthanide ion (when cysteine bound, 614 Da plus a 139–175 Da lanthanide ion, compared to 185 Da for R1), that can be introduced via two proximally located, surface cysteine residues. The other is 4-mercaptomethyl-dipicolinic acid (Su et al. 2008), which requires (i) a carboxyl bearing side chain to be located near a single cysteine residue for incorporation and (ii) the addition of lanthanide ions to the protein sample. This labeling strategy is incompatible with metal-binding proteins, and may cause protein aggregation due to non-specific protein-lanthanide binding. Another recent strategy also designed for generating PCS is DOTA-M8, a stereospecifically methylated DOTA lanthanide chelating label (Haussinger et al. 2009). Although this group is relatively rigid and attachment requires only a single cysteine, it is relatively large (572 Da when cysteine bound, plus the lanthanide ion) and hydrophobic due to methylation to increase protein surface interactions, and the authors report that some motion of the paramagnetic center with respect to the backbone is evident by comparing the magnitude of the PCS and RDC susceptibility tensors. The requirement for multiple functional groups to incorporate the dipicolinic acid and CLaNP-5 labels, can limit those strategies to specific protein topologies while the large size of the CLaNP and DOTA based labels may perturb the structure, dynamics, and stability of the protein under investigation. Thus, a nitroxide label that can be incorporated via a single surface cysteine residue that combines the reliable, sequence-context independent rigidity of a lanthanide-based label with the ease of conjugation and small chemical structure of R1, would be of greater general use for PRE studies.
A suitable candidate may come from a series of R1 analogs, which show more restricted internal motions than R1 at solvent-exposed sites (Columbus et al. 2001) and presumably have fewer allowed conformations due to the presence of a rigid substituent at position 4 of the pyrroline ring. In particular, the 4-phenyl analog exhibits highly restricted motions and adopts a single conformer at a model solvent-exposed helical site (Fleissner 2007) (PDB code 1ZUR). However, spin labeled proteins bearing this particular derivative are prone to aggregation, probably due to an increase in surface hydrophobicity. To circumvent this problem, we synthesized a reagent (HO-3606) that generates a more hydrophilic, 4-pyridyl derivative of R1 (designated R1p, Fig. 1b) and evaluated its general use for PRE studies.
First, we examined the effect of the pyridyl group on the internal flexibility of the spin label side chain by EPR spectroscopy by introducing it into solvent-exposed helical sites in T4 lysozyme (T4L) and human calmodulin (CaM) as model systems. We demonstrate that the EPR spectra of R1p mutants of both proteins are consistent with a single, highly ordered component, and, most importantly, that the PREs arising from R1p can be accounted for by a single label conformation, greatly simplifying the procedure for using PREs as long range distance restraints or as probes of transient states.
Experimental methods
Synthesis of the methanethiosulfonate reagent HO-3606
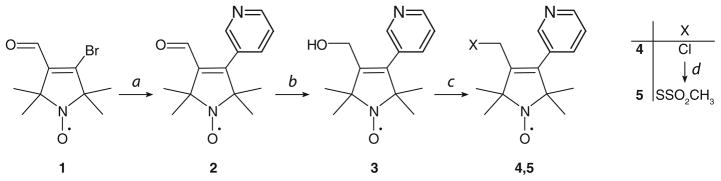
The reagent HO-3606 (radical 5) was prepared according to the general procedure of Scheme 1. Melting points were determined with a Boetius micro melting point apparatus and are uncorrected. Elemental analyses (C, H, N, S) were performed on Fisons EA 1110 CHNS elemental analyzer. Mass spectra were recorded on a Finnigan Automass-Multi instrument in the EI mode. The IR (Specord 85) spectra were in each case consistent with the assigned structure. Flash column chromatography was performed on Merck Kieselgel 60 (0.040–0.063 mm). Qualitative TLC was carried out on commercially prepared plates (20 × 20 × 0.02 cm) coated with Merck Kieselgel GF254. Compound 1 was prepared according to published procedures (Kalai et al. 1998); 3-pyridineboronic acid and other reagents were purchased from Aldrich.
Scheme 1.
Reagents and conditions: a 1 (1.0 equiv.), Pd(PPh3)4 (0.025 eq.), dioxane, under N2, r.t. 10 min., then 3-Pyridineboronic acid (1.025 equiv.), aq. 10% Na2CO3 (2.35 equiv.), reflux, 2h, 63%; b NaBH4 (1.25 equiv.), EtOH, r.t. 15 min. 84%; c MsCl (1.1 equiv.), Et3N (1.1 equiv.), CH2Cl2, 0°C to r.t. 30 min., acetone, LiCl (2.0 equiv.), reflux 30 min., 45–52%; d NaSSO2CH3 (2.0 equiv.), acetone, H2O, 40°C, 30–60 min., 36–24%
Synthesis of 3-Formyl-4-(pyridin-3-yl)-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxyl radical [2]
To a deoxygenated, stirred solution of aldehyde 1 (988 mg, 4.0 mmol) in dioxane (15 mL), Pd(PPh3)4 (100 mg, 0.1 mmol) was added in one portion and the mixture was stirred at room temperature (r.t.) for 10 min. Next, 3-pyridineboronic acid (504 mg, 4.1 mmol) was added followed by the addition of aqueous 10% Na2CO3 (10 mL, 9.4 mmol), and the mixture was stirred and refluxed for 2 h under N2. After cooling, the solvents were evaporated off in vacuo, and the residue was partitioned between CHCl3 (25 mL) and water (10 mL). The organic phase was separated, dried (MgSO4), filtered and evaporated to give aldehyde 2. Yield: 617 mg (63%) after flash chromatography purification (CHCl3/Et2O, 2:1), yellow solid, mp 98–99°C, Rf : 0.21 (CHCl3/Et2O, 2:1). MS EI m/z (%): 245 (M+, 100), 230(30), 200 (42), 172(88). IR (Nujol): 1670 (C=O), 1640, 1560, 1535 (C=C) cm−1. Composition calculated for C14H17N2O2: C, 68.55; H, 6.99; N, 11.42. Found: C, 68.50; H, 7.00; N, 11.36.
Synthesis of 3-Hydroxymethyl-4-(pyridin-3-yl)-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxyl radical [3]
To a solution of aldehyde 2 (980 mg, 4.0 mmol) in EtOH (15 mL), NaBH4 (190 mg, 5.0 mmol) was added in one portion, and the mixture was stirred at r.t. for 15 min. and then quenched with water (10 mL). The solution was extracted with CHCl3 (2 × 10 mL), the organic phase was dried (MgSO4), filtered and evaporated and the residue was purified by flash column chromatography (CHCl3/MeOH, 9:1) to offer alcohol 3. Yield: 830 mg (84%), off-white solid, mp 145–147°C, Rf : 0.14 (CHCl3/MeOH, 9:1). MS EI m/z (%): 247 (M+, 100), 232(61), 217 (16), 184(35). IR (Nujol): 3150 (OH), 1620, 1560, 1535 (C=C) cm−1. Composition calculated for C14H19N2O2: C, 67.99; H, 7.74; N, 11.33. Found: C, 67.92; H, 7.71; N, 11.25.
Synthesis of 3-Chloromethyl-4-(pyridin-3-yl)-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxyl radical [4]
To a solution of alcohol 3 (1.23 g, 5.0 mmol) in CH2Cl2 (15 mL) and Et3N (555 mg, 5.5 mmol), MeSO2Cl (630 mg, 5.5 mmol) was added at 0°C. The mixture was stirred at r.t. for 30 min., then washed with water (10 mL), and the organic phase was separated, dried (MgSO4), filtered and evaporated. The residue was dissolved in dry acetone (40 mL), then LiCl (420 mg, 10 mmol) was added and the mixture stirred and refluxed for 30 min. The solvent was evaporated off in vacuo, and the residue was partitioned between EtOAc (30 mL and water (10 mL). The organic phase was separated, dried (MgSO4), filtered, and evaporated, and the crude residue was further purified by flash column chromatography (CHCl3/Et2O, 2:1) to provide radical 4. Yield: 689 mg (52%), pale yellow solid, mp 87–88°C, Rf : 0.31 (CHCl3/Et2O, 2:1). MS EI m/z (%): 265/267 (M+, 82/28), 250/252(56/21), 235/237 (11/4), 215(100), 184(17). IR (Nujol): 1620, 1560, 1535 (C=C) cm−1. Composition calculated for C14H18ClN2O: C, 63.27; H, 6.83; N, 10.54. Found: C, 63.20; H, 6.85; N, 10.52.
Synthesis of 3-Methanesulfonilthiomethyl -4-(pyridin-3-yl)-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxyl Radical [5] (i.e., HO-3606)
To a solution of compound 4 (532 mg, 2.0 mmol) in acetone (10 mL), NaSSO2CH3 (536 mg, 4.0 mmol) in water (5 mL) was added and the mixture was kept at 40°C. The reaction mixture was monitored by TLC. After consumption of most of the starting material (~30 min), the acetone was evaporated off in vacuo, and the residue was partitioned between CHCl3 (30 mL) and brine (10 mL). The organic phase was separated, dried (MgSO4), filtered, and evaporated. The resulting residue was further purified by flash column chromatography (hexane/EtOAc and CHCl3/Et2O) to give the methanethiosulfonate [5]. Yield: 245 mg (36%), pale yellow solid, mp-125–127°C, Rf 0.12 (CHCl3/Et2O, 2:1). MS EI m/z (%): 341 (M+, 29), 326(10), 346 (39), 215(100), 79(42). IR (Nujol): 1615, 1560, 1535 (C=C) cm−1. Composition calculated for C15H21N2 O3S2: C, 52.76; H, 6.20; N, 8.20; S, 18.78. Found: C, 52.53; H, 6.13; N, 8.05; S, 18.55.
Sample preparation
The genetic construct used for expressing single cysteine mutants of T4 lysozyme (T4L) in the WT* background (C54T, C97A) has been described previously (Mchaourab et al. 1996). The mutants were expressed, purified, and spin labeled with either the common spin label (1-Oxyl-2,2,5,5-tetramethyl-2,5-dihydropyrrol-3-ylmethyl) methane thiosulfonate (sometimes referred to as “MTSL” or “MTSSL”) (Berliner et al. 1982) or HO-3606 according to the method described previously (Fleissner et al. 2009).
For experiments on calmodulin (CaM), a pET21a (Novagen) vector encoding the 148-residue human (CaM) protein was created by cloning the gene for CaM from Xenopus laevis (whose amino acid sequence is identical to the human protein) into the vector using PCR primers with BamH1 and NdeI restriction sites. A CaM variant for site-directed spin labeling, CaM S17C was created with the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
CaM S17C with uniform 2H/13C/15N labeling was expressed in BL21-CodonPlus (DE3) RIPL cells (Strata-gene) at 37°C using standard methods. Cultures of 1 L of minimal M9 medium using 2H2O, 15NH4, Cl and 2H7/13C-glucose as the sole sources of hydrogen, nitrogen and carbon, respectively, were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at A600nm ~0.6, and harvested by centrifugation 6–9 h later.
Cells were resuspended in ~25 mL of 40 mM Tris pH 7.4, 2 mM EDTA, 2 mM dithiothreitol (DTT), and 1× complete protease inhibitor (Roche), and lysed using a microfluidizer. The lysate was cleared by centrifugation and loaded onto a DEAE ion-exchange column (GE Life Sciences) pre-equilibrated with 20 mM Tris, pH 7.4, 80 mM (NH4)2SO4 and 1 mM MgCl2. Protein was eluted with a linear 0–100% gradient over 10 column volumes with a high-salt buffer containing 1.2 M (NH4)2SO4. Fractions containing CaM were pooled and loaded onto phenyl-Sepharose resin (GE Life Sciences) pre-equilibrated with 50 mM Tris, pH 7.4, 2 mM CaCl2 and 100 mM NaCl. CaM was eluted with the addition of 2 mM EGTA. Protein was stored in the elution buffer plus 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) and 1× complete protease inhibitor.
CaM S17C (~200 μM) was spin labeled by the addition of a tenfold molar excess of either “MTSL,” HO-3606, or (1-Acetoxy-2,2,5,5-tetramethyl-δ-3-pyrroline-3-methyl) methanethiosulfonate (Toronto Research Chemicals, Altenbach et al. 2001), to generate the spin label side chains R1, R1p, and a diamagnetic analog of R1, respectively. Labeling reactions were allowed to proceed at room temperature in the dark for ~2 h and tested for completion by liquid chromatography/mass spectrometry. Unreacted spin-label was removed with a HiPrep 26/10 Desalting Column (GE Life Sciences) and the labeled protein was exchanged into NMR buffer, comprising 5% 2H2O/95% 1H2O, 100 mM KCl, 0.1× EDTA-free complete protease inhibitor (Roche), 25 mM HEPES, pH 6.5, and 8 mM CaCl2, or into EPR buffer containing the same salts and buffering agents with 1H2O only. NMR samples were concentrated to 0.3 mM with Amicon Ultra Centrifugal Filter Units (3 kDa molecular weight cutoff). EPR experiments were performed at a concentration of 0.1 mM. Experiments were performed in the presence of 1.2-fold excess (added prior to centrifugation) of a tightly binding synthetic peptide (KRRWKKNFIAVSAANRFKKIS SSGAL; Anaspec) corresponding to the CaM-binding domain (CBD) of human skeletal muscle myosin-light-chain kinase (skMLCK).
EPR spectroscopy
For EPR experiments, ~5 μL samples of T4L (~0.5 mM protein in 50 mM MOPS pH 6.8, 25 mM NaCl) or CaM (~0.1 mM protein in NMR buffer without 2H2O) were loaded into glass capillaries (0.6 mm × 0.84 mm) sealed at one end. Where indicated, the sample also included 30% w/v sucrose to increase the viscosity of the medium, a 1.2 molar excess of the previously described CBD synthetic peptide from human skMLCK, or both. Spectra were collected at X-band (9 GHz) over 100 G on a Bruker Elexys 580 spectrometer fitted with a high-sensitivity resonator using 20 mW incident microwave power and 1 G field modulation amplitude at 100 kHz. Sample temperature was maintained within a single degree of the reported value using a Bruker ER4131VT temperature controller. EPR spectra were simulated using the MOMD model (Budil et al. 1996), using spatially averaged A and g tensor principal values corresponding to the nitroxide motion relative to the protein (the time-independent, effective Hamiltonian) (Hubbell and McConnell 1971).
NMR spectroscopy
Backbone resonance assignments for peptide bound CaM were transferred from known assignments and verified by three-dimensional triple resonance heteronuclear correlation spectroscopy (Clore and Gronenborn 1994) using standard HNCO, HNCACB, and CBCA(CO)NH pulse sequences. Transverse 1HN-Γ2 PRE rates were obtained from the differences in the 1HN-R2 transverse relaxation rates between the paramagnetic and diamagnetic samples (Clore and Iwahara 2009) using a 3D HNCO-based pulse scheme similar to a previously described scheme (Hu et al. 2009) except without TROSY. Compared to most PRE experiments, this experiment adds a 13C′ dimension to resolve overlapping resonances present in the 2D 1H-15N correlation spectrum of CaM. Data were recorded at 27°C on a Bruker 600 MHz spectrometer equipped with a triple resonance z-gradient cryoprobe. Two time points (separated by 20 ms) were used for the 1HN-R2 measurements (Iwahara et al. 2007). In this case, no significant differences in 1HN-R2 were observed for CaM incorporating the diamagnetic control label and without a label (free-cysteine) in NMR buffer containing 5 mM DTT.
PRE data analysis
The position, or ensemble of positions, of the nitroxide spin label that best fits the observed 1HN-Γ2 PRE rates was computed by comparing predicted and observed 1HN-Γ2 PREs using the coordinates of the structure of Ca2+-loaded, peptide-bound CaM in complex with smooth muscle MLCK (PDB 1CDL) (Meador et al. 1992). 1HN-Γ2 PREs were back-calculated in Xplor-NIH (Schwieters et al. 2006; Schwieters et al. 2003) with either a single conformer or a five-conformer ensemble for the spin label together with the Solomon-Bloembergen Model Free (SBMF) representation (Iwahara et al. 2004b). The label and immediately adjacent side chain coordinate positions were optimized in torsion angle space by simulated annealing to minimize the difference between observed and calculated 1HN-Γ2 PREs, as previously described (Iwahara et al. 2004b) with the addition of a weighting factor for the PRE force constant equal to the square of the inverse of the expected error in the observed PRE at each residue (Xplor-NIH PRE force constant option sigma). The error in the observed PRE was calculated from the peak intensities at the two time points in the paramagnetic and diamagnetic samples (Iwahara et al. 2004b) and a minimum error of ±1.5 s−1 was used to account for small sample-to-sample variations. In addition to the PRE pseudo-potential, the target function includes stereochemical restraints, a quartic van der Waals repulsion term to prevent atomic overlap between the spin-label and the protein, and a multidimensional torsion angle database potential of mean force (Clore and Kuszewski 2002). (Note atomic overlap between spin labels in the five-conformer ensemble is allowed since the ensemble represents a distribution of states.) To evaluate the goodness-of-fit, the appropriate reduced χ2:
| (1) |
was computed. The number of degrees of freedom M is given by:
| (2) |
where N is the number of observed PREs, and C, the number of spin label conformers in the fitting ensemble, one or five. C is multiplied by three, for the three translational degrees of freedom involved in fitting each nitroxide position. Because the effective electron relaxation time τs, which is equal to the electron spin–lattice relaxation time T1e in the case of 1H-Γ2 PREs for macromolecules, is much longer than the protein rotational correlation time τr (Iwahara et al. 2007), the PRE correlation time for the calculation of PRE rates was assumed to be approximately the same as τr (9.9 ns) for CaM (Chang et al. 2003).
Complete Xplor-NIH scripts and documentation to introduce the spin label into a protein of known structure, find the label position(s) that best-fits the intramolecular PRE data, and compute χ2 are available online at http://nmr.cit.nih.gov/xplor-nih/doc/current.
Results
EPR demonstrates restricted internal motion of R1p relative to R1
All “free” nitroxides have similar isotropic three-line spectra independent of the substituent at the 3 or 4 positions of the ring. The spin density at these positions is quite low, and the presence of the pyridyl ring in R1p has little effect on the EPR spectrum.
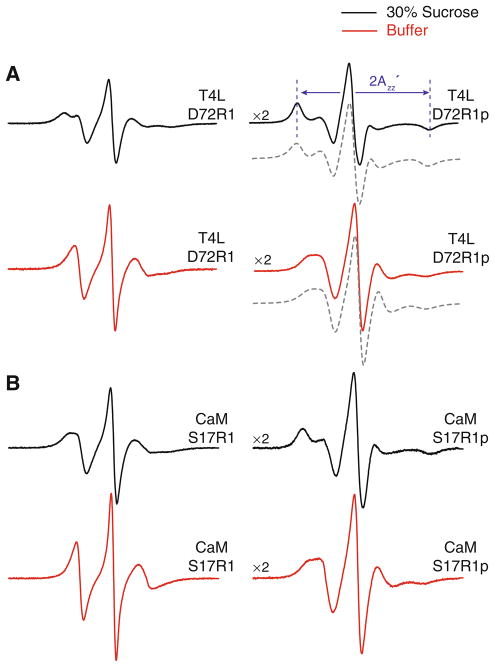
To compare the internal flexibility of the R1p side chain to that of R1, the solvent-exposed site D72 of T4L was selected for introduction of the spin labels. This site has previously been used as a reference since contributions of backbone fluctuations to the motion of the nitroxide are minimal (Columbus et al. 2001). The room temperature EPR spectra of the T4L D72R1p mutant and the previously published D72R1 mutant (Mchaourab et al. 1996) are presented in Fig. 2a. Both mutants were studied in buffer (Fig. 2a, red traces), but were also studied in a 30% sucrose solution (Fig. 2a, black traces), which reduces the effect of protein rotational diffusion on the spectrum and allows the motion of the nitroxide relative to the protein to be examined (Mchaourab et al. 1996). Assuming a spherical geometry for T4L, an estimate of its rotational correlation time (τR) at room temperature according to the Stokes–Einstein relation is 6–8 ns in buffer and 18–24 ns in 30% sucrose. Fits to multifrequency EPR spectra of spin labeled T4L yield τR ≈ 10 ns in buffer and approximately 3 times longer in a viscosity corresponding to 30% sucrose, consistent with previously reported values (Zhang et al. 2010). Regardless of the exact values, rotational diffusion within this time regime will strongly influence the EPR spectrum as shown in Fig. 2.
Fig. 2.
EPR spectra of R1 and R1p labeled proteins. a EPR spectra of T4L D72R1 (left) and T4L D72R1p (right) at 298 K. b EPR spectra of CaM S17R1 (left) and CaM S17R1p (right) at 298 K. Spectra shown in red are of the mutants in buffer, and those in black are in buffer containing 30% (w/v) sucrose. Shown below the T4L D72R1p spectra (gray, dashed trace) are simulated spectra according to a modified effective Hamiltonian model using values for the magnetic tensor derived assuming an order parameter of 0.76 and no diffusion axis tilt (Axx = Ayy = 7.07 G, Azz = 34.9 G, gxx = gyy = 2.00667, gzz = 2.00267). Best-fit values for τr were 5.4 ns (buffer) and 17 ns (30% sucrose). All spectra are normalized to the same concentration of nitroxide and have a magnetic field scan width of 100 G. Vertical scaling factors (×2, left side of spectrum) were applied to select spectra for display purposes
In solutions of 30% sucrose, the T4L mutants D72R1 and D72R1p have spectra characterized by well-resolved outer hyperfine extrema with splitting of (Fig. 2) that correspond to nitroxides with the 2p orbital on nitrogen oriented along the laboratory magnetic field. For T4L D72R1, the overall lineshape arises from a fast anisotropic internal motion of the nitroxide that can be accurately described by an order parameter (S) and correlation time (τi) on the ns time scale (Columbus 2001). In the fast motion limit, the time-dependent Hamiltonian describing the motion can be replaced by an effective, time-independent, Hamiltonian with the A and g magnetic interactions tensors averaged over motion within a cone. In this limit, which has been shown to be a reasonable approximation for T4L D72R1, the value of is a quantitative measure of the order parameter, increasing with increased order (Columbus et al. 2001).
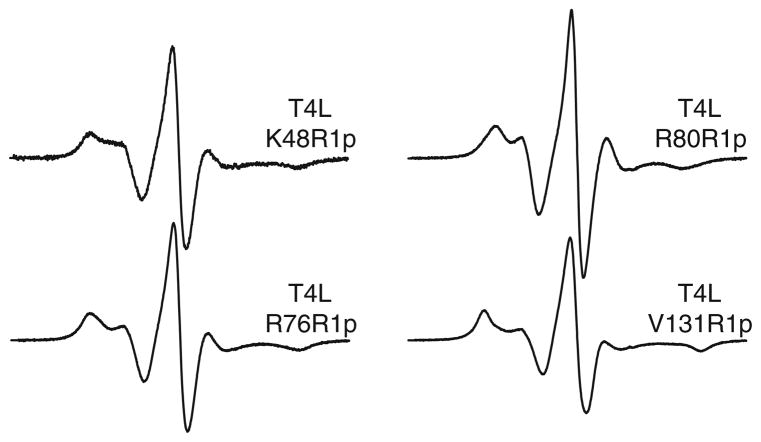
As is evident in Fig. 2a, the spectra of T4L D72R1p and T4L D72R1 in sucrose are similar except that is considerably larger for that of T4L D72R1p (64 G for D72R1p and 53 G for D72R1), indicating a higher order for the internal motion of R1p. This result is consistent with expectations from the crystal structure of the 4-phenyl analog of R1 in which the entire side chain was resolved (see Discussion). Due to this increase in order, the rotational diffusion of the protein, even in 30% sucrose, will contribute to the EPR spectrum of T4L D72R1p by reducing . To deduce the value corresponding to purely internal motion of the side chain, spectra of T4L D72R1p were recorded as a function of sucrose concentration from 15 to 35% wt/wt, and then analyzed (Timofeev and Tsetlin 1983) to give less than that in the rigid limit (Kusnetzow et al. 2006). Within the context of the effective Hamiltonian model for motion in a cone (Griffith and Jost 1976), this result gives an order parameter for the internal motion of R1p of S = 0.76, compared to 0.43 for R1 (Columbus et al. 2001). If this simple model has merit, then it should be possible to reproduce the experimental spectra of T4L D72R1p in both buffer and 30% sucrose using the effective Hamiltonian parameters corresponding to S = 0.76 and computing the effect of protein rotational diffusion according to the stochastic Louisville method that is widely used for simulating the effect of motion on EPR spectra (Budil et al. 1996). The dashed traces shown in Fig. 2 are fits to the experimental spectra using the effective Hamiltonian parameters given in the legend and allowing the isotropic τR of the protein to vary. The best-fit τR values are 5 and 17 ns for buffer and 30% sucrose, respectively, and both are within the expected range, considering the simple nature of the model and the assumed isotropic diffusion of the protein. Thus, a first-order working model for the overall motion of the R1p sidechain is a fast, highly-ordered internal motion of small amplitude modulated by slow rotational diffusion of the protein. To explore the generality of the ordered internal motion of R1p, it was placed at several other solvent-exposed helical sites in T4L (Fig. 3). For R1p at helical sites 48, 76 and 131 the spectra are very similar to that at 72, suggesting that the above results will be general for R1p in rigid helical structures. The spectrum of R80R1p has a small , indicating higher nitroxide mobility compared to the others. This is likely due to contributions from backbone flexibility at this C-terminal site where the crystallographic thermal factors are relatively high. The spectrum of CaM S17R1 is similar to other non-interacting, solvent exposed helical sites that exhibit fast (~2 ns), weakly ordered (S < 0.5) motion. As for T4L, the spectra of CaM S17R1p exhibit reduced mobility compared to those of the analogous R1 mutant (Fig. 2b). Remarkably, the spectra of CaM S17R1p are similar to those of T4L D72R1p. Because T4L and CaM are of similar size and the sites in each case are located at the solvent exposed surface of a helix, the internal motions relative to the protein must be comparable.
Fig. 3.
EPR spectra of T4L R1p mutants in a buffered solution containing 30% sucrose at 295 K. Solvent-exposed helical sites in T4L were chosen for incorporation of the R1p spin label. The magnetic field scan width is 100 G
A single label conformation accounts for the experimental PREs
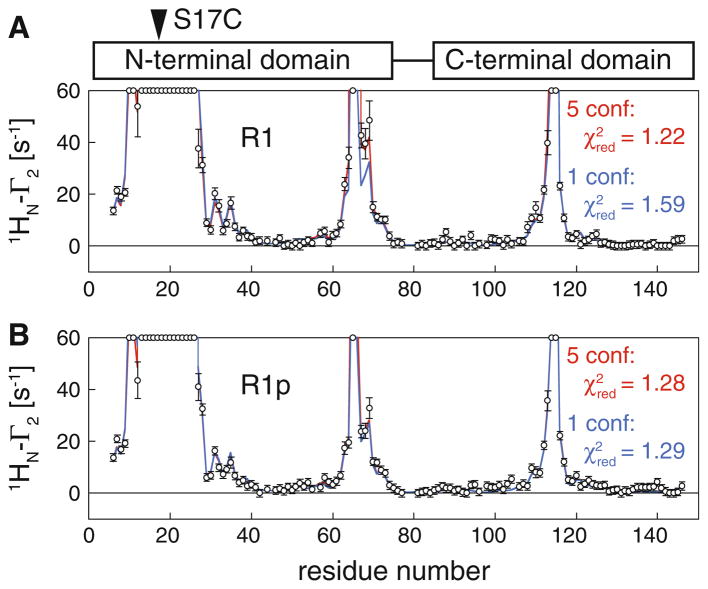
The 1HN-Γ2 PRE rates for CaM S17C reacted with the R1 side chain label, CaM S17R1, and with the pyridyl label, CaM S17R1p, are presented in Fig. 4. In the Ca2+-loaded and peptide-bound state, CaM adopts a rigid structure in which the hydrophobic clefts of the N- and C-terminal domains of CaM close around the target peptide (Ikura et al. 1992; Meador et al. 1992). Since the labels are attached in the same S17C position, the pattern of PREs arising from S17R1 and S17R1p are similar. Quantitatively, however, the effect of the larger conformational dynamics of R1 compared to R1p on the observed PREs can be easily distinguished.
Fig. 4.
Comparison of observed and calculated PRE data for Ca2+-loaded, peptide-bound CaM with a spin label conjugated to S17C: a S17R1; b S17R1p. Experimental PRE profiles are shown as circles (error bar, 1 SD). PREs too large (>60 s−1) to be accurately measured are plotted at the top of the charts. PRE profiles back-calculated from the structures of CaM (Ca2+-loaded, peptide-bound) are displayed as solid lines using either a single conformer (blue) or 5 conformer ensemble (red) to represent the spin label. Reduced χ2 values are given for each of the fits. Note that use of a multiple-conformer ensemble representation significantly improves the fit to the experimental data in the case of R1 but not R1p
Assuming a single, static conformation of the side chain, CaM S17R1 shows a reasonable fit with a reduced χ2 of 1.59 (Fig. 4, top, blue line) (a value of 1.0 suggesting a perfect fit within the error of the measurement). Though the fit is good overall, the observed PREs at positions 67, 68, and 69 are clearly not well fit, suggesting that the nitroxide position may sample many different states with distinct PREs that cannot be captured by a single structure. By introducing an ensemble of five (equally weighted) conformations, the fit to the data for S17R1 can be improved such that the predicted PREs are within the experimental error in this region (Fig. 4a, red line). The reduced χ2 for the five-conformer ensemble drops to 1.22, indicating that the fit is quantitatively superior to the fit using a single conformation, and hence a multiple conformer ensemble is necessary to quantitatively treat the PREs for S17R1.
In contrast, the PREs for CaM S17R1p can be quantitatively accounted for by a single conformation of the label, resulting in a χ2 of 1.29 (Fig. 4b, blue line). This value is considerably smaller than the χ2 for a single conformer representation of S17R1 and comparable to the χ2 for the five-conformer ensemble. Unlike in the case of S17R1, fitting an ensemble of five conformations of the label to the observed PREs for S17R1p leaves the fit essentially unchanged (red and blue lines overlap) and does not improve the reduced χ2, indicating that a single conformation of the R1p spin-label side chain is sufficient to account for the observed PREs.
Discussion
Recent studies have demonstrated that PREs can not only provide sensitive, long-range structural restraints (Iwahara et al. 2004b), but can also allow transient, sparsely-populated structures to be observed (Clore 2011; Clore and Iwahara 2009). However, a quantitative description of the PREs arising from most spin labels (e.g., R1) typically requires the use of an ensemble of spin label conformers to represent the positions of the nitroxides due to the inherent conformational flexibility of the spin label side chain. Here we introduce and characterize a new, rigid spin label for proteins that, when placed at solvent-exposed helical positions, can simplify PRE data analysis. In contrast to other rigidly attached, lanthanide labels developed for PCS, only a single cysteine is required to incorporate this new, disulfide-linked spin label, designated R1p; the thiol-specific reagent for generating R1p (HO-3606, Scheme 1) is a 4-pyridyl analog of the commonly used MTSL reagent that generates the R1 spin label. Although R1 and R1p differ only by the 4-subsituent, the EPR spectra of R1p at solvent-exposed sites in T4L and CaM are dramatically different from those for the analogous R1 mutants, exhibiting highly restricted motion relative to the protein (Fig. 2). This result is consistent with the crystal structure of the 4-phenyl analog at T4L V131, in which the entire side chain is resolved in a single rotamer (Fleissner 2007) (PDB accession code 1ZUR). In contrast, two rotamers were observed in the crystal structure of R1 at the same site though the nitroxide rings of neither rotamer was resolved due to disorder (Fleissner et al. 2009). Interestingly, the EPR spectra of the rigidly attached R1p mutants are strikingly similar to mutants bearing the 4-phenyl analog of R1 (Columbus et al. 2001), suggesting that the conformational space of the R1p side chain is also restricted. Indeed, the intramolecular PREs observed for CaM S17R1p are fully accounted for within experimental error by a single nitroxide position, and the fit cannot be improved by introducing an ensemble representation. This is in sharp contrast to the PREs arising from S17R1 where a multiple conformer ensemble is required and yields a significantly better fit than a single conformer representation.
At any surface helical site, the assumption that R1p can be treated as a occupying a single position for PRE studies can be quickly determined by combining a pair of 1HN-R2 experiments to compute intramolecular (or intradomain) PREs with the Xplor-NIH scripts we have provided (see Methods). Since quantitative calculation of PREs for a label with a single nitroxide position does not require the representation of a conformational ensemble of labels nor a model free analysis incorporating order-parameters for the nitroxide-proton vectors, we propose that site directed spin-labeling with HO-3606 to form the nitroxide bearing side chain R1p will dramatically simplify the quantitative procedures needed for analyzing intra- and intermolecular PRE data.
Acknowledgments
We thank Vincenzo Venditti and Mengli Cai for helpful suggestions, Dusty Baber and Jinfa Ying for assistance with NMR spectrometers, and Evan Brooks and Mária Balog for expert technical assistance. This work was in part supported by funds from the Intramural Program of the NIH, NIDDK and the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the NIH (to G.M.C.), NIH grants R01EY05216 (to W.L.H.), and the Jules Stein Professorship Endowment (to W.L.H.). The synthesis of new spin label reagents was supported by Hungarian National Research Funds (OTKA K81123).
Contributor Information
Nicolas L. Fawzi, Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MA 20892-0520, USA
Mark R. Fleissner, Jules Stein Eye Institute and Department of Chemistry and Biochemistry, University of California, Los Angeles, CA 90095, USA
Nicholas J. Anthis, Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MA 20892-0520, USA
Tamás Kálai, Institute of Organic and Medicinal Chemistry, University of Pécs, Szigeti str. 12, 7624 Pécs, Hungary.
Kálmán Hideg, Institute of Organic and Medicinal Chemistry, University of Pécs, Szigeti str. 12, 7624 Pécs, Hungary.
Wayne L. Hubbell, Email: hubbellw@jsei.ucla.edu, Jules Stein Eye Institute and Department of Chemistry and Biochemistry, University of California, Los Angeles, CA 90095, USA
G. Marius Clore, Email: mariusc@mail.nih.gov, Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MA 20892-0520, USA.
References
- Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- Berliner LJ, Grünwald J, Hankovszky HO, Hideg K. A novel reversible thiol-specific spin label: papain active site labeling and inhibition. Anal Biochem. 1982;119:450–455. doi: 10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- Bermejo GA, Strub MP, Ho C, Tjandra N. Determination of the solution-bound conformation of an amino acid binding protein by NMR paramagnetic relaxation enhancement: use of a single flexible paramagnetic probe with improved estimation of its sampling space. J Am Chem Soc. 2009;131:9532–9537. doi: 10.1021/ja902436g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges MD, Hideg K, Hubbell WL. Resolving conformational and rotameric exchange in spin-labeled proteins using saturation recovery EPR. Appl Magn Reson. 2010;37:363–390. doi: 10.1007/s00723-009-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J Magn Reson Ser A. 1996;120:155–189. [Google Scholar]
- Chang SL, Szabo A, Tjandra N. Temperature dependence of domain motions of calmodulin probed by NMR relaxation at multiple fields. J Am Chem Soc. 2003;125:11379–11384. doi: 10.1021/ja034064w. [DOI] [PubMed] [Google Scholar]
- Chou JJ, Case DA, Bax A. Insights into the mobility of methyl-bearing side chains in proteins from 3JCC and 3JCN couplings. J Am Chem Soc. 2003;125:8959–8966. doi: 10.1021/ja029972s. [DOI] [PubMed] [Google Scholar]
- Clore GM. Visualizing lowly-populated regions of the free energy landscape of macromolecular complexes by paramagnetic relaxation enhancement. Mol Biosyst. 2008;4:1058–1069. doi: 10.1039/b810232e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM. Exploring sparsely populated states of macromolecules by diamagnetic and paramagnetic NMR relaxation. Protein Sci. 2011;20:229–246. doi: 10.1002/pro.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Kuszewski J. χ1 rotamer populations and angles of mobile surface side chains are accurately predicted by a torsion angle database potential of mean force. J Am Chem Soc. 2002;124:2866–2867. doi: 10.1021/ja017712p. [DOI] [PubMed] [Google Scholar]
- Columbus L. PhD Dissertation. UCLA; 2001. Investigating backbone and side chain dynamics of α-helices in the nanosecond regime with site-directed spin labeling; pp. 51–58. [Google Scholar]
- Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in α-helices: analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- Crisma M, Deschamps JR, George C, Flippen-Anderson JL, Kaptein B, Broxterman QB, Moretto A, Oancea S, Jost M, Formaggio F, et al. A topographically and conformationally constrained, spin-labeled, α-amino acid: crystallographic characterization in peptides. J Pept Res. 2005;65:564–579. doi: 10.1111/j.1399-3011.2005.00258.x. [DOI] [PubMed] [Google Scholar]
- Fawzi NL, Doucleff M, Suh JY, Clore GM. Mechanistic details of a protein–protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc Natl Acad Sci USA. 2010;107:1379–1384. doi: 10.1073/pnas.0909370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner MR. PhD Thesis. UCLA; 2007. X-ray structures of nitroxide side chains in proteins: a basis for interpreting distance measurements and dynamic studies by electron paramagnetic resonance. [Google Scholar]
- Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flippen-Anderson JL, George C, Valle G, Valente E, Bianco A, Formaggio F, Crisma M, Toniolo C. Crystallographic characterization of geometry and conformation of TOAC, a nitroxide spin-labelled Cα,α-disubstituted glycine, in simple derivatives and model peptides. Int J Pept Protein Res. 1996;47:231–238. doi: 10.1111/j.1399-3011.1996.tb01350.x. [DOI] [PubMed] [Google Scholar]
- Griffith OH, Jost PC. Lipid spin labels in biological membranes. In: Berliner LJ, editor. Spin labeling: theory and applications. Academic Press; New York: 1976. pp. 454–523. [Google Scholar]
- Guo ZF, Cascio D, Hideg K, Kalai T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16:1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZF, Cascio D, Hideg K, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17:228–239. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussinger D, Huang JR, Grzesiek S. DOTA-M8: an extremely rigid, high-affinity lanthanide chelating tag for PCS NMR spectroscopy. J Am Chem Soc. 2009;131:14761–14767. doi: 10.1021/ja903233w. [DOI] [PubMed] [Google Scholar]
- Hu K, Doucleff M, Clore GM. Using multiple quantum coherence to increase the 15N resolution in a three-dimensional TROSY HNCO experiment for accurate PRE and RDC measurements. J Magn Reson. 2009;200:173–177. doi: 10.1016/j.jmr.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell WL, McConnell HM. Molecular motion in spin-labeled phospholipids and membranes. J Am Chem Soc. 1971;93:314–326. doi: 10.1021/ja00731a005. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Schwieters CD, Clore GM. Characterization of nonspecific protein-DNA interactions by 1H paramagnetic relaxation enhancement. J Am Chem Soc. 2004a;126:12800–12808. doi: 10.1021/ja046246b. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J Am Chem Soc. 2004b;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara J, Tang C, Clore GM. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalai T, Balog M, Jeko J, Hideg K. 3-substituted 4-bromo-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxyl radicals as versatile synthons for synthesis of new paramagnetic heterocycles. Synthesis-Stuttgart. 1998:1476–1482. [Google Scholar]
- Keizers PHJ, Ubbink M. Paramagnetic tagging for protein structure and dynamics analysis. Prog Nucl Magn Reson Spectrosc. 2011;58:88–96. doi: 10.1016/j.pnmrs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Keizers PHJ, Desreux JF, Overhand M, Ubbink M. Increased paramagnetic effect of a lanthanide protein probe by two-point attachment. J Am Chem Soc. 2007;129:9292–9293. doi: 10.1021/ja0725201. [DOI] [PubMed] [Google Scholar]
- Kusnetzow AK, Altenbach C, Hubbell WL. Conformational states and dynamics of rhodopsin in micelles and bilayers. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindfors HE, Venkata BS, Drijfhout JW, Ubbink M. Linker length dependent binding of a focal adhesion kinase derived peptide to the src SH3-SH2 domains. FEBS Lett. 2011;585:601–605. doi: 10.1016/j.febslet.2011.01.026. [DOI] [PubMed] [Google Scholar]
- Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme, correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin—2.4Å structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- Otting G. Protein NMR using paramagnetic ions. Ann Rev Biophys. 2010;39:387–405. doi: 10.1146/annurev.biophys.093008.131321. [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
- Su XC, Man B, Beeren S, Liang H, Simonsen S, Schmitz C, Huber T, Messerle BA, Otting G. A dipicolinic acid tag for rigid lanthanide tagging of proteins and paramagnetic NMR spectroscopy. J Am Chem Soc. 2008;130:10486–10847. doi: 10.1021/ja803741f. [DOI] [PubMed] [Google Scholar]
- Takayama Y, Clore GM. Intra- and intermolecular translocation of the bi-domain transcription factor Oct1 characterized by liquid crystal and paramagnetic NMR. Proc Natl Acad Sci USA. 2011;108:E169–E176. doi: 10.1073/pnas.1100050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein–protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- Tang C, Louis JM, Aniana A, Suh JY, Clore GM. Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease. Nature. 2008;455:693–696. doi: 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeev VP, Tsetlin VI. Analysis of mobility of protein side-chains by spin label technique. Biophys Struct Mech. 1983;10:93–108. [Google Scholar]
- Todd AP, Cong J, Levinthal F, Levinthal C, Hubbell WL. Site-directed mutagenesis of colicin E1 provides specific attachment sites for spin labels whose spectra are sensitive to local conformation. Proteins. 1989;6:294–305. doi: 10.1002/prot.340060312. [DOI] [PubMed] [Google Scholar]
- Volkov AN, Worrall JAR, Holtzmann E, Ubbink M. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc Natl Acad Sci USA. 2006;103:18945–18950. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov AN, Ubbink M, van Nuland NAJ. Mapping the encounter state of a transient protein complex by PRE NMR spectroscopy. J Biomol NMR. 2010;48:225–236. doi: 10.1007/s10858-010-9452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshaviak DT, Serbulea L, Houk KN, Hubbell WL. Conformational analysis of a nitroxide side chain in an a-helix with density functional theory. J Phys Chem B. 2011;115:397–405. doi: 10.1021/jp108871m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZW, Fleissner MR, Tipikin DS, Liang ZC, Moscicki JK, Earle KA, Hubbell WL, Freed JH. Multifrequency electron spin resonance study of the dynamics of spin labeled T4 lysozyme. J Phys Chem B. 2010;114:5503–5521. doi: 10.1021/jp910606h. [DOI] [PMC free article] [PubMed] [Google Scholar]