Abstract
In organisms other than higher plants, family 19 chitinase was first discovered in Streptomyces griseus HUT6037, and later, the general occurrence of this enzyme in Streptomyces species was demonstrated. In the present study, the distribution of family 19 chitinases in the class Actinobacteria and the phylogenetic relationship of Actinobacteria family 19 chitinases with family 19 chitinases of other organisms were investigated. Forty-nine strains were chosen to cover almost all the suborders of the class Actinobacteria, and chitinase production was examined. Of the 49 strains, 22 formed cleared zones on agar plates containing colloidal chitin and thus appeared to produce chitinases. These 22 chitinase-positive strains were subjected to Southern hybridization analysis by using a labeled DNA fragment corresponding to the catalytic domain of ChiC, and the presence of genes similar to chiC of S. griseus HUT6037 in at least 13 strains was suggested by the results. PCR amplification and sequencing of the DNA fragments corresponding to the major part of the catalytic domains of the family 19 chitinase genes confirmed the presence of family 19 chitinase genes in these 13 strains. The strains possessing family 19 chitinase genes belong to 6 of the 10 suborders in the order Actinomycetales, which account for the greatest part of the Actinobacteria. Phylogenetic analysis suggested that there is a close evolutionary relationship between family 19 chitinases found in Actinobacteria and plant class IV chitinases. The general occurrence of family 19 chitinase genes in Streptomycineae and the high sequence similarity among the genes found in Actinobacteria suggest that the family 19 chitinase gene was first acquired by an ancestor of the Streptomycineae and spread among the Actinobacteria through horizontal gene transfer.
Chitinase (EC 3.2.1.14) is a glycosyl hydrolase which catalyzes the degradation of chitin, an insoluble linear β-1,4-linked polymer of N-acetylglucosamine. This enzyme is present in a wide range of organisms, including organisms that do not contain chitin, and it plays important physiological and ecological roles. Based on amino acid sequence similarity, chitinases are classified into families 18 and 19 of glycosyl hydrolases (13, 14). The members of the two different families differ in their amino acid sequences, three-dimensional (3D) structures (9, 10, 27, 36), and molecular mechanisms of catalytic reactions and are thus considered to have different evolutionary origins.
Family 18 chitinases are widely distributed in a variety of organisms, such as bacteria, fungi, viruses, animals, and higher plants (classes III and V). On the other hand, family 19 chitinases were found only in higher plants until recently. However, since chitinase C (ChiC) of Streptomyces griseus HUT6037 was identified as the first family 19 chitinase in an organism other than higher plants (25), the number of family 19 chitinases found in other organisms has increased. For example, the general occurrence of family 19 chitinases in Streptomyces species has been demonstrated (28, 40). In addition, the recent progress of genome-sequencing projects for various organisms has revealed the presence of family 19 chitinases in some other bacteria (5, 7, 8, 11, 21, 26, 32, 35, 39, 44) and a few other organisms, including a nematode (22, 43).
Plant family 19 chitinases are thought to be part of a mechanism of defense against fungal pathogens. This role has been deduced from the following observations: (i) chitin is the major component of the cell wall of plant pathogens, (ii) chitinase is one of the pathogenesis-related proteins (12, 18, 19), and (iii) some plant chitinases exhibit antifungal activity in vitro (4, 16, 20, 31). This hypothesis has been supported by the observation that transgenic plants constructed by introducing the plant chitinase gene expressed enhanced resistance against fungal diseases (3). Since S. griseus ChiC exhibited significant sequence similarity to plant family 19 chitinases in the catalytic domain, the antifungal activity of ChiC was examined, and a remarkable ability of ChiC to inhibit hyphal extension of Trichoderma reesei was demonstrated (40). Therefore, antifungal activity may be a common characteristic of family 19 chitinases.
Although the number of organisms that have family 19 chitinases is increasing, family 19 chitinases of Streptomyces are of special interest because of their high levels of similarity to plant class IV chitinases and because of their antifungal activity. To clarify why the distribution of family 19 chitinases in organisms is more restricted than that of family 18 chitinases and how these types of chitinases evolved, a detailed study of Streptomyces family 19 chitinases is critical. In this study, to see whether organisms closely related to Streptomyces possess family 19 chitinases, we searched for family 19 chitinase genes in Actinobacteria and studied the phylogenetic relationship of the genes with those of other organisms.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Strains belonging to the class Actinobacteria used in this study are listed in Table 1. These strains were obtained from the Japan Collection of Microorganisms (http://www.jcm.riken.go.jp/), the NITE Biological Resource Center (http://www.nbrc.nite.go.jp/e-home/index-e.html/), and the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (http://www.dsmz.de/); the only exception was Cellulomonas cellulans YCWD3. All of the strains except YCWD3 were grown under the conditions described at the websites. C. cellulans YCWD3 (6) was grown at 30°C in a medium containing 1.0% mannitol, 0.2% peptone, 0.1% meat extract, and 0.1% yeast extract (pH 7.0). Bacillus circulans WL-12 (42) and Serratia marcescens 2170 (41) were grown at 30°C in Luria-Bertani medium for chromosomal DNA extraction. Escherichia coli JM109 was used as a general host for gene cloning, and the pT7Blue T vector (Novagen, Madison, Wis.) was used for nucleotide sequencing of amplified DNA fragments obtained by PCR. E. coli JM109 cells carrying a recombinant plasmid were selected on Luria-Bertani medium containing 100 μg of ampicillin per ml. The DNA fragment used as a probe for Southern hybridization was prepared from recombinant plasmid pGC01A (40), which carries a 1-kb DNA fragment of the chiC gene of S. griseus HUT6037. pSK+-RCC2, used in the control experiment to test amplification of a plant chitinase gene by PCR, contains a gene encoding Oryza sativa RCCII (24).
TABLE 1.
Strains belonging to the class Actinobacteria used in this study
| Strain no. | Organism | Order | Suborder | Cleared zone | Signal in Southern hybridization | No. of detectable family 19 chitinase genes |
|---|---|---|---|---|---|---|
| 1 | Actinomyces naeslundii JCM 8349 | Actinomycetales | Actinomycineae | − | − | 0 |
| 2 | Arcanobacterium haemolyticum NBRC15585 | Actinomycetales | Actinomycineae | − | − | 0 |
| 3 | Corynebacterium glutamicum JCM 1318 | Actinomycetales | Corynebacterineae | − | − | 0 |
| 4 | Dietzia maris JCM 6166 | Actinomycetales | Corynebacterineae | − | − | 0 |
| 5 | Nocardia asteroides JCM 3384 | Actinomycetales | Corynebacterineae | − | NTa | NT |
| 6 | Rhodococcus rhodochrous JCM 3202 | Actinomycetales | Corynebacterineae | − | NT | NT |
| 7 | Geodermatophilus obscurus subsp. dictyosporus JCM 3154 | Actinomycetales | Frankineae | − | NT | NT |
| 8 | Microsphaera multipartita JCM 9543 | Actinomycetales | Frankineae | − | NT | NT |
| 9 | Glycomyces harbinensis JCM 7347 | Actinomycetales | Glycomycineae | + | + | 1 |
| 10 | Agromyces ramosus JCM 3108 | Actinomycetales | Micrococcineae | − | NT | NT |
| 11 | Arthrobacter crystallopoietes JCM 2522 | Actinomycetales | Micrococcineae | − | NT | NT |
| 12 | Arthrobacter ramosus JCM 1334 | Actinomycetales | Micrococcineae | − | − | 0 |
| 13 | Cellulomonas cellulans YCWD3 | Actinomycetales | Micrococcineae | + | + | 1 |
| 14 | Cellulomonas fimi JCM 1341 | Actinomycetales | Micrococcineae | + | − | 0 |
| 15 | Curtobacterium luteum JCM 1480 | Actinomycetales | Micrococcineae | + | − | 0 |
| 16 | Microbacterium imperiale JCM 1378 | Actinomycetales | Micrococcineae | − | − | 0 |
| 17 | Dermabacter hominis JCM 7448 | Actinomycetales | Micrococcineae | − | NT | NT |
| 18 | Kocuria kristinae JCM 7237 | Actinomycetales | Micrococcineae | − | − | 0 |
| 19 | Kocuria varians JCM 7238 | Actinomycetales | Micrococcineae | − | NT | NT |
| 20 | Promicromonospora sukumoe JCM 6845 | Actinomycetales | Micrococcineae | + | + | 1 |
| 21 | Terrabacter tumescens JCM 1365 | Actinomycetales | Micrococcineae | − | NT | NT |
| 22 | Actinoplanes brasiliensis JCM 3196 | Actinomycetales | Micromonosporineae | + | + | 2 |
| 23 | Catenuloplanes japonicus JCM 9106 | Actinomycetales | Micromonosporineae | + | + | 1 |
| 24 | Couchioplanes caeruleus subsp. caeruleus JCM 3195 | Actinomycetales | Micromonosporineae | + | − | 0 |
| 25 | Micromonospora chalcea JCM 3031 | Actinomycetales | Micromonosporineae | + | − | 0 |
| 26 | Pilimelia terevasa JCM 3091 | Actinomycetales | Micromonosporineae | − | NT | NT |
| 27 | Luteococcus japonicus JCM 9415 | Actinomycetales | Propionibacterineae | − | − | 0 |
| 28 | Microlunatus phosphovorus JCM 9379 | Actinomycetales | Propionibacterineae | − | NT | NT |
| 29 | Actinobispora yunnanensis JCM 9330 | Actinomycetales | Pseudonocardineae | − | NT | NT |
| 30 | Actinokineospora riparia JCM 7471 | Actinomycetales | Pseudonocardineae | + | + | 1 |
| 31 | Actinopolyspora mortivallis JCM 7550 | Actinomycetales | Pseudonocardineae | − | NT | NT |
| 32 | Amycolatopsis orientalis subsp. orientalis JCM 4600 | Actinomycetales | Pseudonocardineae | + | + | 1 |
| 33 | Kibdelosporangium aridum subsp. aridum JCM 7912 | Actinomycetales | Pseudonocardineae | + | + | 2 |
| 34 | Saccharopolyspora erythraea JCM 4026 | Actinomycetales | Pseudonocardineae | + | − | 0 |
| 35 | Streptoalloteichus hindustanus JCM 3268 | Actinomycetales | Pseudonocardineae | + | + | 1 |
| 36 | Kitasatospora setae JCM 3304 | Actinomycetales | Streptomycineae | + | + | 2 |
| 37 | Actinomadura kijaniata JCM 3306 | Actinomycetales | Streptosporangineae | + | − | 0 |
| 38 | Microbispora rosea subsp. aerata JCM 3076 | Actinomycetales | Streptosporangineae | + | − | 0 |
| 39 | Nocardiopsis lucentensis JCM 9420 | Actinomycetales | Streptosporangineae | + | − | 0 |
| 40 | Nonomuraea spiralis JCM 3286 | Actinomycetales | Streptosporangineae | + | + | 1 |
| 41 | Planobispora rosea JCM 3166 | Actinomycetales | Streptosporangineae | + | + | 2 |
| 42 | Planomonospora parontospora subsp. antibiotica JCM 3094 | Actinomycetales | Streptosporangineae | + | + | 2 |
| 43 | Streptosporangium nondiastaticum JCM 3114 | Actinomycetales | Streptosporangineae | + | − | 0 |
| 44 | Actinocorallia herbida JCM 9647 | Bifidobacteriales | − | NT | NT | |
| 45 | Atopobium minutum JCM 1118 | Coriobacteriales | − | − | 0 | |
| 46 | Collinsella aerofaciens JCM 10188 | Coriobacteriales | − | − | 0 | |
| 47 | Eggerthella lenta JCM 9979 | Coriobacteriales | − | − | 0 | |
| 48 | Rubrobacter radiotolerans JCM 2153 | Rubrobacterales | − | − | 0 | |
| 49 | Sphaerobacter thermophilus DSM 20745 | Sphaerobacterales | − | − | 0 |
NT, not tested.
Detection of chitinase activity.
Actinobacteria strains were streaked on agar plate medium containing 0.2% colloidal chitin, 0.05% KCl, 0.1% K2HPO4, 0.05% MgSO4 · 7H2O, 0.001% FeSO4, 0.05% yeast extract, and 2.0% agar (pH 7.0) and were incubated at 30°C for 3 to 14 days. Chitinase production was assessed by visual inspection of cleared zones that formed around colonies.
Gene manipulation.
Chromosomal DNAs of various Actinobacteria strains were extracted from the mycelia by the method described by Hopwood et al. (15), with minor modifications. Chromosomal DNAs of B. circulans WL-12 and S. marcescens 2170 were extracted from the cells as described by Silhavy et al. (33). Other gene manipulations were performed as described by Sambrook and Russell (30).
Southern hybridization.
Chromosomal DNAs (3 μg) of Actinobacteria strains were digested with restriction enzyme PstI or SalI, electrophoresed on a 1.0% agarose gel, and transferred onto a nylon membrane (MAGNA; OSMONICS). Probe DNA was prepared from pGC01A by digesting the plasmid with restriction enzymes ApaI and BamHI and was labeled and detected by using an AlkPhos direct labeling detection system with CDP-Star (Amersham Biosciences, Uppsala, Sweden) according to the supplier's instructions.
PCR amplification and determination of the nucleotide sequence of a portion of the family 19 chitinase genes of Actinobacteria strains.
A portion of the genes encoding family 19 chitinases of Actinobacteria strains was amplified by PCR by using LA Taq DNA polymerase (TaKaRa, Kyoto, Japan). The forward and reverse primers used for PCR were 5′-AAGCTCGCSGCSTTCCTSGC-3′ and 5′-GCACTCGAGSGCGCCGTTGAT-3′), respectively. Thirty amplification cycles of denaturation for 30 s at 98°C, primer annealing for 30 s at 50°C, and DNA synthesis for 1.0 min at 72°C were used. After the last cycle, DNA synthesis was performed for 10 min at 72°C (40). The amplified fragments were ligated with the T vector pT7Blue and were maintained in E. coli JM109. Nucleotide sequences of amplified fragments in the T vector were determined with an automated laser fluorescence sequencer (model 4200; LI-COR). Sequencing reactions were performed with a Thermo Sequenase primer cycle sequencing kit with 7-deaza-dGTP (Amersham Biosciences) used according to the supplier's instructions with double-stranded templates. Nucleotide sequence data were analyzed by using the GENETYX system (Software Kaihatsu Co.).
Phylogenetic analysis.
Multiple alignments were obtained with the Clustal X (37) program. The GeneDoc (23) program was used as an editing tool for multiple alignments. Phylogenetic trees were calculated by the neighbor-joining method (29) implemented in Clustal X and were drawn by using the program TreeView. Nucleotide and amino acid sequences of family 19 chitinases were obtained from the CAZy database (http://afmb.cnrs-mrs.fr/CAZY/).
Chemicals.
Colloidal chitin was prepared from powdered chitin purchased from Funakoshi Chemical Co. (Tokyo, Japan) by using the methods described by Jeuniaux (17). Other chemicals used in this study were obtained from Wako Pure Chemical Industries, Ltd. (Osaka, Japan).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been deposited in the DBBJ database under accession numbers AB125373 to AB125390.
RESULTS AND DISCUSSION
Chitinase productivity of strains belonging to the class Actinobacteria.
The class Actinobacteria has been classified into the following five subclasses based on 16S rRNA sequences: Actinobacteridae, Acidimicrobidae, Coriobacteridae, Sphaerobacteridae, and Rubrobacteridae (34). Subclasses Acidimicrobidae, Coriobacteridae, Sphaerobacteridae, and Rubrobacteridae each contain only one order, and each of the orders contains only one family. On the other hand, subclass Actinobacteridae consists of two orders, the Actinomycetales and Bifidobacteriales. The order Actinomycetales comprises 10 suborders, and most of them contain many families, while the order Bifidobacteriales contains only one family. For example, the Micrococcineae, one of the suborders in the Actinomycetales, contains nine families. Therefore, the order Actinomycetales accounts for the greatest part of the Actinobacteria.
To initiate a study to reveal the distribution of family 19 chitinases in the Actinobacteria, 49 strains of various species were chosen from five orders and all suborders in the class Actinobacteria, as shown in Table 1. The strain in the subclass Acidimicrobidae was not included in this study because of the difficulty of establishing and maintaining a culture of Acidimicrobium ferrooxidans DSM 10331, which is the only one strain in this subclass that is available. The chitinase production of the strains was tested by using agar plate medium containing colloidal chitin. Chitinase production was assessed by visual inspection of cleared zones that formed around colonies. As shown in Table 1, 22 of 49 strains formed cleared zones and thus appeared to produce chitinases. These 22 strains and 13 chitinase-negative strains, randomly chosen as controls, were used for further experiments.
Detection of family 19 chitinase genes by Southern hybridization.
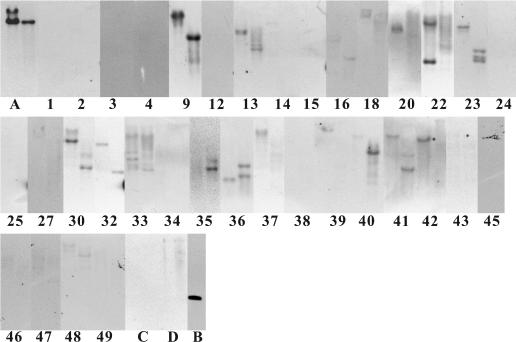
Southern hybridization was used to detect family 19 chitinase genes; part of the chiC gene of S. griseus HUT6037 was used as the probe. The probe used in these experiments contained the DNA region corresponding to the entire catalytic domain of ChiC and the 41-bp downstream region from the termination codon. Chromosomal DNA was extracted from the 35 strains and digested with either PstI or SalI. As shown in Fig. 1, clear signals were detected for 13 of the 22 strains that showed chitinase activity, suggesting that family 19 chitinase genes were present. Some strains showed only one signal, and the others showed two or three signals, indicating that multiple genes for family 19 chitinases were present. On the other hand, none of the chitinase-negative strains examined as controls showed clear signals, although very faint signals were observed with Actinomadura kijaniata, Eggerthella lenta, Kocuria kristinae, Microbacterium imperiale, Nocardiopsis lucentensis, and Rubrobacter radiotolerans.
FIG. 1.
Detection of family 19 chitinase genes by Southern hybridization. Chromosomal DNAs were digested with either PstI (left lane of each pair) or SalI (right lane of each pair). The numbers are the strain numbers in Table 1. (A) S. griseus HUT6037; (B) ApaI-BamHI-digested fragment of the chiC gene in plasmid pGC01A; (C) B. circulans WL-12; (D) S. marcescens 2170.
Detection of family 19 chitinase genes by PCR.
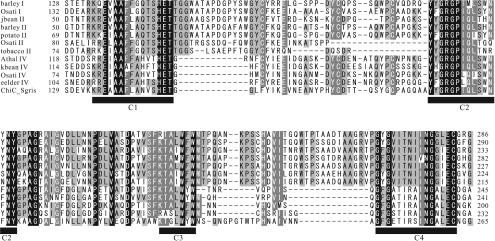
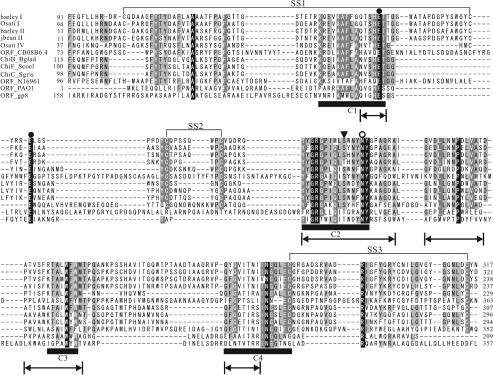
To confirm that the strains exhibiting hybridization signals really possess the genes for family 19 chitinases, PCR amplification of part of the family 19 chitinase genes was attempted. In order to design PCR primers, the amino acid sequences of S. griseus ChiC and plant class I, II, and IV chitinases were compared, and the four conserved regions were deduced from the alignment shown in Fig. 2. Forward and reverse primers were designed by using the regions corresponding to conserved regions C1 and C4, respectively, taking into account the high G+C content of Actinobacteria. By using this primer set, the corresponding region in the cDNA encoding class I chitinase of O. sativa (24) was successfully amplified in the control experiment. This fact indicates that the genes amplified with this primer set included not only genes that were very similar to the chiC gene of S. griseus HUT6037.
FIG. 2.
Alignment of the conserved regions of family 19 chitinases used to design PCR primers. Residues conserved in all sequences are indicated by white type on a black background, while residues conserved in >80 and >60% of the proteins examined are indicated by white type on a dark gray background and by black type on a light gray background, respectively. Conserved regions of family 19 chitinases are indicated by the solid bars (C1, C2, C3, and C4). Conserved regions 1 and 2 were used to design PCR primers. barley I, barley class I chitinase (amino acid sequence accession no. Q42839); Osati I, O. sativa class I chitinase (Q42992); jbean II, jack bean class II chitinase (O81934); barley II, barley class II chitinase (P11955); potato II, potato class II chitinase (Q43184); Osati II, O. sativa class II chitinase (O80423); tobacco II, tobacco class II chitinase (P17514); Athal IV, Arabidopsis thaliana class IV chitinase (O23248); kbean IV, kidney bean class IV chitinase (P27054); Osati IV, O. sativa class IV chitinase (O04138); eelder IV, European elder class IV chitinase (Q43150); ChiC_Sgris, S. griseus HUT6037 ChiC (O50152).
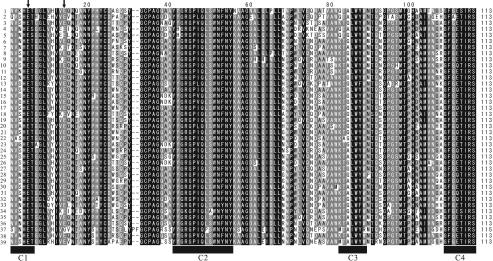
Chromosomal DNA was extracted from 22 chitinase-positive strains and 13 chitinase-negative control strains and then subjected to PCR amplification with the primers that we designed. All of the amplified fragments that were approximately 400 bp long were cloned and sequenced to exclude fragments of non-family 19 chitinase genes. As a result, 18 fragments of family 19 chitinase genes were obtained from 13 strains. These strains perfectly matched the strains that exhibited clear signals in the Southern hybridization experiments described above. In addition, no fragment of a family 19 chitinase was obtained from the 13 chitinase-negative strains. From Actinoplanes brasiliensis, Kibdelosporangium aridum subsp. aridum, Kitasatospora setae, Planobispora rosea, and Planomonospora parontospora subsp. antibiotica, two fragments with slightly different nucleotide sequences were amplified, indicating that two distinct family 19 genes were present. Amino acid sequences deduced from the nucleotide sequences of the amplified fragments indicated that the fragments are truly parts of the family 19 chitinase genes. Most of the amplified sequences without primer regions were 340 bp long and encoded 113 amino acid residues; the only exception was one of the two fragments amplified from P. rosea, fragment 2, which was 346 bp long and encoded 115 amino acids. The levels of identity of the nucleotide and deduced amino acid sequences of these amplified fragments with the sequences of the corresponding regions of S. griseus HUT6037 chiC and ChiC were 78 to 87% and 70 to 83%, respectively. Figure 3 shows an alignment of the amino acid sequences deduced from the amplified fragments and the corresponding regions of reported family 19 chitinases of Streptomyces species (40) and Nocardiopsis prasina OPC-131 (38). Four conserved regions characteristic of family 19 chitinases were found in all the deduced sequences. Except for fragment 2 from P. rosea, Glu residues at positions 5 and 14, which were two catalytic amino acid residues (1), were found in all sequences. In fragment 2 of P. rosea, a Glu residue at position 14 was replaced by Gly, and in addition, Pro and Phe were inserted at positions 31 and 32.
FIG. 3.
Alignment of the amino acid sequences deduced from the amplified fragments of family 19 chitinases from Actinobacteria strains determined in this study and those reported previously. Residues conserved in all sequences are indicated by white type on a black background, while residues conserved in >80 and >60% of the proteins examined are indicated by white type on a dark gray background and by black type on a light gray background, respectively. Conserved regions of family 19 chitinases are indicated by the solid bars (C1, C2, C3, and C4). Catalytic amino acid residues of family 19 chitinases are indicated by arrows. 1, Glycomyces harbinensis; 2, Cellulomonas cellulans YCWD3; 3, Promicromonospora sukumoe; 4, Actinoplanes brasiliensis fragment 1; 5, Actinoplanes brasiliensis fragment 2; 6, Catenuloplanes japonicus; 7, Actinokineospora riparia; 8, Amycolatopsis orientalis subsp. orientalis; 9, Kibdelosporangium aridum subsp. aridum fragment 1; 10, Kibdelosporangium aridum subsp. aridum fragment 2; 11, Streptoalloteichus hindustanus; 12, Kitasatospora setae fragment 1; 13, Kitasatospora setae fragment 2; 14, Streptomyces coelicolor A3 (2) chiF (DNA accession no. AB017012); 15, Streptomyces coelicolor A3(2) chiG (AB017013); 16, Streptomyces coelescens ISP5421 fragment 1 (AB031749); 17, Streptomyces coelescens ISP5421 fragment 2 (AB031750); 18, Streptomyces griseus HUT6037 chiC (AB009289); 19, Streptomyces ipomoeae MAFF4023 (AB031751); 20, Streptomyces lavendulae (AF127374); 21, Streptomyces lividans 66 fragment 1 (AB031746); 22, Streptomyces lividans 66 fragment 2 (AB031747); 23, Streptomyces lividans 66 fragment 3 (AB031748); 24, Streptomyces olivaceoviridis chi30 (AJ133186); 25, Streptomyces prasinopilosus fragment 1 (AB031752); 26, Streptomyces prasinopilosus fragment 2 (AB031753); 27, Streptomyces sp. strain AJ9463 chiIS (AB104621); 28, Streptomyces sp. strain S15 (AB031754); 29, Streptomyces sp. strain S84 (AB031755); 30, Streptomyces sp. strain S100 (AB031756); 31, Streptomyces sp. strain S159 (AB031757); 32, Streptomyces thermoviolaceus OPC-250 chi25 (AB016843); 33, Streptomyces thermoviolaceus OPC-250 chi35 (AB016842); 34, Nocardiopsis prasina OPC-131(AB086832); 35, Nonomuraea spiralis; 36, Planobispora rosea fragment 1; 37, Planobispora rosea fragment 2; 38, Planomonospora parontospora subsp. antibiotica fragment 1; 39, Planomonospora parontospora subsp. antibiotica fragment 2.
Distribution and phylogenetic relationship of the genes encoding family 19 chitinases in Actinobacteria.
As summarized in Table 1, the strains tested in these experiments could be divided into three categories, (i) strains which did not show chitinase activity in the agar plate assay (chitinase-negative strains), (ii) strains which showed chitinase activity but for which no family 19 chitinase gene was detected (family 19-negative strains), and (iii) strains which had a family 19 chitinase gene (family 19-positive strains). Family 19-positive strains were found only in the six suborders of the order Actinomycetales, the Pseudonocardineae, Micromonosporineae, Streptosporangineae, Glycomycineae, Streptomycineae, and Micrococcineae. In other orders, not even a chitinase-positive strain was detected. All three types of strains, chitinase-negative, family 19-negative, and family 19-positive strains, were found in four suborders, the Micrococcineae, Pseudonocardineae, Streptosporangineae, and Micromonosporineae. The strains belonging to the suborders Actinomycineae, Corynebacterineae, Propionibacterineae, and Frankineae did not show chitinase activity. These results demonstrated that family 19 chitinase genes are broadly dispersed in the order Actinomycetales, which comprises the greatest part of the Actinobacteria.
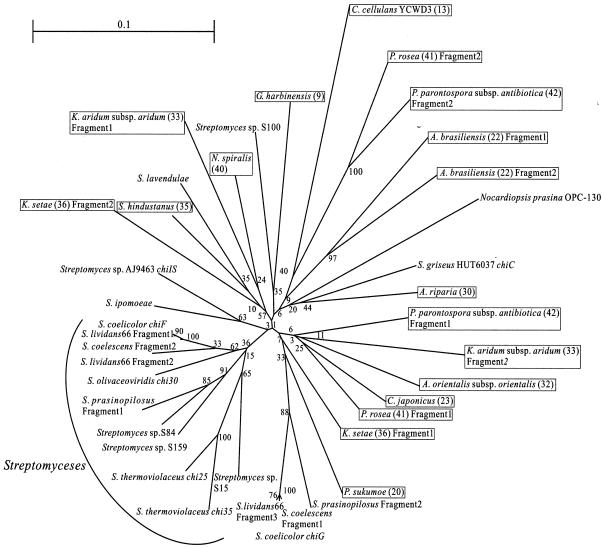
To visualize the relationship among family 19 chitinases found in Actinobacteria, a phylogenetic tree was constructed by the neighbor-joining method (27) by using the nucleotide sequences of the amplified fragments of family 19 chitinase genes and the corresponding sequences of Streptomyces species determined previously (40), as shown in Fig. 4. Bootstrap values, which were an index of the accuracy of the phylogenetic tree, were higher outside the tree but lower inside the tree. Because of the low bootstrap values observed inside the tree, which were probably due to the low relative diversity of all sequences used in the analysis, it may not be appropriate to discuss the evolutionary relationships of all family 19 chitinase genes in the tree.
FIG. 4.
Phylogenetic relationships among family 19 chitinase genes of Actinobacteria. An unrooted phylogenetic tree was calculated based on an alignment of conserved regions of family 19 chitinase genes from Actinobacteria. Organisms in which family 19 chitinase genes were found in this study are enclosed in boxes, and the numbers in parentheses are the strain numbers shown in Table 1. The numbers at the nodes are percentages which indicate the levels of bootstrap support, based on a neighbor-joining analysis of 1,000 resampled data sets.
However, it is noteworthy that approximately one-half of the family 19 chitinase genes of Streptomyces species formed a stable cluster in the tree, suggesting that there is a close evolutionary relationship at least among these genes. Interestingly, the other half of the Streptomyces family 19 chitinase genes were dispersed in the tree with the genes found in other Actinobacteria.
Comparison of amino acid sequences with the sequences of other family 19 chitinases.
Previously, the presence of family 19 chitinase genes has been reported in viruses, bacteria, nematodes, protozoans, and higher plants. Figure 5 shows an alignment of the amino acid sequences of some family 19 chitinases. These sequences were selected to represent sequence variations of all family 19 chitinases and categories of source organisms. The sequences of S. griseus ChiC and Streptomyces coelicolor ChiF are included as representatives of the Actinobacteria family 19 chitinases and are located in different clusters in Fig. 4. The alignment was first constructed with the Clustal X program and then was modified manually by referring to the 3D structures of plant class II chitinases from barley (10) and jack bean (9) and other information. Four conserved regions shown in Fig. 5, deduced in this study, were found in all sequences. Two Glu residues, which are catalytic amino acids of family 19 chitinases, were both conserved in all sequences. Asn residues corresponding to Asn124 of the barley class II chitinase, which has been suggested to participate in the catalytic reaction (1), were also well conserved. In jack bean class II chitinase, the presence of a water molecule held by hydrogen bonds to the carboxyl group of Glu90 and the hydroxyl group of Thr119 has been reported (9). This water molecule is proposed to participate as a nucleophile in the single-displacement inverting catalytic reaction (2), which is widely accepted as the catalytic mechanism of family 19 chitinases. The amino acid residues at the position corresponding to Thr119 were either Thr or Ser in all family 19 chitinases. This suggests that Ser may play the same role as Thr in the catalytic reaction as the hydroxyl amino acid. All family 19 chitinases from Actinobacteria have Ser residues at this position. The relative positions of four amino acid resides (two catalytic Glu residues, Asn, and Thr or Ser) in the 3D structures of the chitinases from jack bean and barley and S. griseus ChiC were very similar to each other.
FIG. 5.
Alignment of the amino acid sequences of the catalytic domains of family 19 chitinases. Residues conserved in all sequences are indicated by white type on a black background, while residues conserved in >80 and >60% of the proteins examined are indicated by white type on a dark gray background and by black type on a light gray background, respectively. The two catalytic amino acid residues of family 19 chitinases are indicated by solid circles. The open circle indicates amino acid residues related to the activity. Amino acid residues proposed to hold a water molecule are indicated by a solid triangle. SS1, SS2, and SS3 indicate the positions of disulfide bonds. Conserved regions of family 19 chitinases are indicated by the solid bars (C1, C2, C3, and C4). barley I, barley class I chitinase (accession no. Q42839); Osati I, O. sativa class I chitinase (Q42992); barley II, barley class II chitinase (P11955); jbean II, jack bean class II chitinase (O81934); Osati IV, O. sativa class IV chitinase (O04138); ORF_C08B6.4, Caenorhabditis elegans chitinase (Q17816); ChiB_Bglad, B. gladioli CHB101 ChiB (BAA92252); ChiF_Scoel, S. coelicolor A3 (2) ChiF (Q9Z9M6); ChiC_Sgris, S. griseus HUT6037 ChiC (O50152); ORF_N16961, V. cholerae El Tor N16961 chitinase (Q9KTW1); ORF_PAO1, Pseudomonas aeruginosa PAO1 chitinase (BAA83168); ORF_gp8, mycobacteriophage Bxb1 chitinase (AAG59713). The sequence regions used for the phylogenetic analysis of family 19 chitinases shown in Fig. 6 are indicated by arrows.
Three disulfide bonds have been identified in the 3D structures of the class II chitinases from barley (10) and jack bean (9) at the corresponding positions in the two chitinases. Cys residues, which could form the three disulfide bonds (SS1, SS2, and SS3), were found to be conserved in the amino acid sequences of all family 19 chitinases from higher plants. As shown in Fig. 5, family 19 chitinases from nematodes have all six conserved Cys residues at the corresponding positions. In the sequences of Actinobacteria strains, Burkholderia gladioli CHB101, and Vibrio cholerae El Tor N16961, Cys residues which could form SS2 and SS3 were found, but Cys residues which could form SS1 were not found. On the other hand, family 19 chitinases from other bacteria do not have any corresponding Cys residues.
In terms of the positions of insertions or deletions, the similarity of conserved regions, and the number of the deduced disulfide bonds, family 19 chitinases from Actinobacteria strains and B. gladioli CHB101 are more similar to plant family 19 chitinases than to family 19 chitinases from other bacteria.
Phylogenetic analysis of family 19 chitinases.
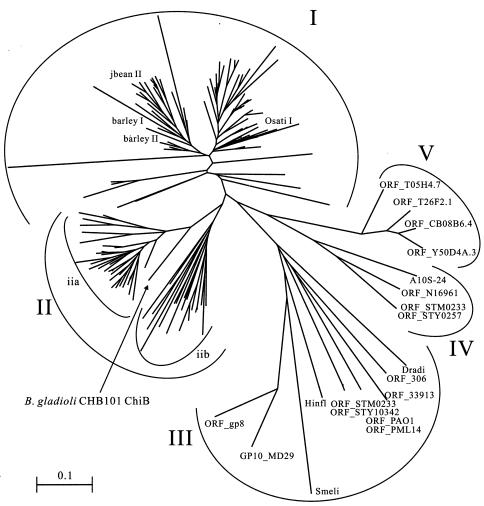
To study the phylogenetic relationships of family 19 chitinases found in Actinobacteria and other organisms, a multiple alignment of the amino acid sequences corresponding to the sequence regions shown in Fig. 3 of all family 19 chitinases available from CAZy (http://afmb.cnrs-mrs.fr/CAZY/) was prepared. Then the highly variable regions containing many gaps were manually removed from the alignment, and a phylogenetic tree was constructed by the neighbor-joining method (27) based on this modified alignment. Due to the extensive diversity of the sequences, inclusion of the highly valuable regions was impractical for construction of a reliable alignment. As shown in Fig. 6, family 19 chitinases were separated into five clusters on the phylogenetic tree. Plant family 19 chitinases are located in the two clusters, clusters I and II. Cluster I consists solely of plant class I and II chitinases. On the other hand, cluster II consists of Actinobacteria chitinases (subcluster iia in Fig. 6) and plant class IV chitinases (subcluster iib). Cluster III mainly consists of the chitinases of Proteobacteria and viruses. Cluster IV consists of family 19 chitinases found in V. cholerae El Tor N16961, Aeromonas sp. strain 10S-24, Salmonella enterica subsp. enterica serovar Typhi CT18, and Salmonella enterica serovar Typhimurium LT2, and cluster V consists solely of Nematoda chitinases. The major group of family 19 chitinases from Proteobacteria, which formed cluster III, is most distantly related to the other family 19 chitinases found in various organisms. Interestingly, one of the two family 19 chitinases found in S. enterica subsp. enterica serovar Typhi CT18 and S. enterica serovar Typhimurium LT2 is located in cluster III for each strain, and the other is located in cluster IV.
FIG. 6.
Phylogenetic relationship of family 19 chitinases of Actinobacteria and other organisms available from a database. An unrooted phylogenetic tree was calculated based on an alignment of partial amino acid sequences of all family 19 chitinases by using the neighbor-joining method implemented in the Clustal X program. The sequence regions used for the alignment are indicated in Fig. 5. Cluster I consists of plant class I and II chitinases. Cluster II consists of Actinobacteria chitinases (subcluster iia) and plant class IV chitinases (subcluster iib). Cluster III mainly consists of chitinases of Proteobacteria and viruses. Cluster IV consists of chitinases of Proteobacteria. Cluster V consists of chitinases of Nematoda. barley I, barley class I chitinase (accession no. Q42839); Osati I, O. sativa class I chitinase (Q42992); jbean II, jack bean class II chitinase (O81934); barley II, barley class II chitinase (P11955); ORF_STM0233, S. enterica serovar Typhimurium LT2 chitinase (Q8ZQH4); ORF_STY10342, S. enterica subsp. enterica serovar Typhi CT18 chitinase (CAD05435); Dradi, Deinococcus radiodurans R1 chitinase (Q9RZ37); Smeli, Sinorhizobium meliloti 1021 chitinase (Q92W46); ORF_306, Xanthomonas axonopodis pv. citri strain 306 chitinase (AAM35357); ORF_gp8, mycobacteriophage Bxb1 chitinase (AAG59713); GP10_MD29, gene 10 protein of mycobacteriophage D29 (O64203); Hinfl, Haemophilus influenzae Rd chitinase (P44187); ORF_33913, Xanthomonas campestris pv. campestris strain ATCC 33913 chitinase (AAM42250); ORF_PAO1, Pseudomonas aeruginosa PAO1 chitinase (BAA83168); ORF_PML14, Pseudomonas aeruginosa PML14 chitinase (BAA83137); ORF_STY0257, S. enterica subsp. enterica serovar Typhi CT18 chitinase (CAD08692); ORF_STM0233, S. enterica serovar Typhimurium LT2 chitinase (Q8ZRN8); ORF_N16961, V. cholerae El Tor N16961 chitinase (Q9KTW1); A10S-24, Aeromonas sp. strain 10S-24 chitinase (BAA76716); ORF_Y50D4A.3, ORF_T05H4.7, ORF_T26F2.1, and ORF_C08B6.4, Caenorhabditis elegans chitinases (AAK68504, O16512, P92013, and Q17816).
All family 19 chitinases found in Actinobacteria formed the same cluster with plant class IV chitinases, and a chitinase from B. gladioli CHB101 exhibited a close evolutionary relationship to these chitinases. On the other hand, the family 19 chitinases of other bacteria belong to the other clusters, including cluster III, which is most distantly related to the rest of the family 19 chitinases. Therefore, the family 19 chitinases of Actinobacteria strains are phylogenetically unique among the family 19 chitinases of prokaryotic organisms.
We previously proposed the hypothesis that family 19 chitinases of Streptomyces species were acquired from plants by horizontal gene transfer (40). The results obtained in this study support the contention that this hypothesis could be expanded to the family 19 chitinases of Actinobacteria. The general occurrence of family 19 chitinase genes in Streptomyces species and the high levels of sequence similarity among the genes found in Actinobacteria suggest that the family 19 chitinase gene was first acquired by an ancestor of Streptomyces species and spread among the Actinobacteria through horizontal gene transfer. However, detailed analysis of the family 19 chitinases of other prokaryotic organisms from phylogenetic and biochemical viewpoints is necessary for further discussions.
Acknowledgments
We thank Yoko Nishizawa, National Institute of Agrobiological Sciences, for offering a plasmid containing the RCC2 gene. We thank Osamu Shida, Higeta Shoyu Co., Ltd., and Koji Ohnishi, Faculty of Science, Niigata University, for their valuable advice regarding phylogenetic analysis.
This work was supported in part by grant-in-aid for scientific research 14560059 from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
REFERENCES
- 1.Andersen, M. D., A. Jensen, J. D. Robertus, R. Leah, and K. Skriver. 1997. Heterologous expression and characterization of wild-type and mutant forms of a 26 kDa endochitinase from barley (Hordeum vulgare L.). Biochem. J. 322:815-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brameld, K. A., and W. A. Goddard III. 1998. The role of enzyme distortion in the single displacement mechanism of family 19 chitinases. Proc. Natl. Acad. Sci. USA 95:4276-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broglie, K., I. Chet, M. Holliday, R. Cressman, P. Biddle, S. Knowlton, C. J. Mauvais, and R. Broglie. 1991. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254:1194-1197. [DOI] [PubMed] [Google Scholar]
- 4.Collinge, D. B., K. M. Kragh, J. D. Mikkelsen, K. K. Nielsen, U. Rasmussen, and K. Vad. 1993. Plant chitinases. Plant J. 3:31-40. [DOI] [PubMed] [Google Scholar]
- 5.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, Jr., L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, Jr., E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, Jr., A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 6.Doi, K., and A. Doi. 1986. Cloning and expression in Escherichia coli of the gene for an Arthrobacter beta-(1-3)-glucanase. J. Bacteriol. 168:1272-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. G. Sutton, W. FitzHugh, C. A. Fields, J. D. Gocayne, J. D. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 9.Hahn, M., M. Hennig, B. Schlesier, and W. Höhne. 2000. Structure of jack bean chitinase. Acta Crystallogr. D 56:1096-1099. [DOI] [PubMed] [Google Scholar]
- 10.Hart, P. J., H. D. Pfluger, A. F. Monzingo, T. Hollis, and J. D. Robertus. 1995. The refined crystal structure of an endochitinase from Hordeum vulgare L. seeds at 1.8 A resolution. J. Mol. Biol. 248:402-413. [PubMed] [Google Scholar]
- 11.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heitz, T., S. Segond, S. Kauffmann, P. Geoffroy, V. Prasad, F. Brunner, B. Fritig, and M. Legrand. 1994. Molecular characterization of a novel tobacco pathogenesis-related (PR) protein: a new plant chitinase/lysozyme. Mol. Gen. Genet. 245:246-254. [DOI] [PubMed] [Google Scholar]
- 13.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. L. Lydiate, C. P. Smith, J. M. Ward, and H. S. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. John Innes Foundation, Norwich, United Kingdom.
- 16.Iseli, B., T. Boller, and J. M. Neuhaus. 1993. The N-terminal cysteine-rich domain of tobacco class I chitinase is essential for chitin binding but not for catalytic or antifungal activity. Plant Physiol. 103:221-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeuniaux, C. 1966. Chitinase. Methods Enzymol. 8:644-650. [Google Scholar]
- 18.Jung, J. L., B. Fritig, and G. Hahne. 1993. Sunflower (Helianthus annuus L.) pathogenesis-related proteins (induction by aspirin (acetylsalicylic acid) and characterization). Plant Physiol. 101:873-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang, M. K., K. S. Park, and D. Choi. 1998. Coordinated expression of defense-related genes by TMV infection or salicylic acid treatment in tobacco. Mol. Cell 8:388-392. [PubMed] [Google Scholar]
- 20.Leah, R., H. Tommerup, I. Svendsen, and J. Mundy. 1991. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 266:1564-1573. [PubMed] [Google Scholar]
- 21.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 22.Mediavilla, J., S. Jain, J. Kriakov, M. E. Ford, R. L. Duda, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2000. Genome organization and characterization of mycobacteriophage Bxb1. Mol. Microbiol. 38:955-970. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. Analysis and visualization of genetic variation. EMBnet News 4:14. [Google Scholar]
- 24.Nishizawa, Y., N. Kishimoto, T. Saito, and A. Hibi. 1993. Sequence variation, differential expression and chromosomal location of rice chitinase genes. Mol. Gen. Genet. 241:1-10. [DOI] [PubMed] [Google Scholar]
- 25.Ohno, T., S. Armand, T. Hata, N. Nikaidou, B. Henrissat, M. Mitsutomi, and T. Watanabe. 1996. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J. Bacteriol. 178:5065-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. G. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 27.Perrakis, A., I. Tews, Z. Dauter, A. B. Oppenheim, I. Chet, K. S. Wilson, and C. E. Vorgias. 1994. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure 2:1169-1180. [DOI] [PubMed] [Google Scholar]
- 28.Saito, A., T. Fujii, T. Yoneyama, M. Redenbach, T. Ohno, T. Watanabe, and K. Miyashita. 1999. High-multiplicity of chitinase genes in Streptomyces coelicolor A3(2). Biosci. Biotechnol. Biochem. 63:710-718. [DOI] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schlumbaum, A., F. Mauch, U. Vögeli, and T. Boller. 1986. Plant chitinases are potent inhibitors of fungal growth. Nature 324:365-367. [Google Scholar]
- 32.Shimosaka, M., Y. Fukumori, T. Narita, X. Zhang, R. Kodaira, M. Nogawa, and M. Okazaki. 2001. The bacterium Burkholderia gladioli strain CHB101 produces two different kinds of chitinases belonging to families 18 and 19 of the glycosyl hydrolases. J. Biosci. Bioeng. 91:103-105. [DOI] [PubMed] [Google Scholar]
- 33.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 35.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hanchock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 36.Terwisscha van Scheltinga, A. C., K. H. Kalk, J. J. Beintema, and B. W. Dijkstra. 1994. Crystal structures of hevamine, a plant defence protein with chitinase and lysozyme activity, and its complex with an inhibitor. Structure 2:1181-1189. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsujibo, H., T. Kubota, M. Yamamoto, K. Miyamoto, and Y. Inamori. 2003. Characterization of chitinase genes from an alkaliphilic actinomycete, Nocardiopsis prasina OPC-131. Appl. Environ. Microbiol. 69:894-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda, M., M. Kojima, T. Yoshikawa, N. Mitsuda, K. Araki, T. Kawaguchi, K. Miyatake, M. Arai, and T. Fukamizo. 2003. A novel type of family 19 chitinase from Aeromonas sp. no. 10S-24. Eur. J. Biochem. 270:2513-2520. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe, T., R. Kanai, T. Kawase, T. Tanabe, M. Mitsutomi, S. Sakuda, and K. Miyashita. 1999. Family 19 chitinases of Streptomyces species: characterization and distribution. Microbiology 145:3353-3363. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe, T., K. Kimura, T. Sumiya, N. Nikaidou, K. Suzuki, M. Suzuki, M. Taiyoji, S. Ferrer, and M. Regue. 1997. Genetic analysis of the chitinase system of Serratia marcescens 2170. J. Bacteriol. 179:7111-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe, T., K. Suzuki, W. Oyanagi, K. Ohnishi, and H. Tanaka. 1990. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J. Biol. Chem. 265:15659-15665. [PubMed] [Google Scholar]
- 43.Waterston, R. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. The C. elegans Sequencing Consortium. Science 282:2012-2018. [DOI] [PubMed] [Google Scholar]
- 44.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, C. M. Fraser, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]