Abstract
Microorganisms have been reported to induce settlement and metamorphosis in a wide range of marine invertebrate species. However, the primary cue reported for metamorphosis of coral larvae is calcareous coralline algae (CCA). Herein we report the community structure of developing coral reef biofilms and the potential role they play in triggering the metamorphosis of a scleractinian coral. Two-week-old biofilms induced metamorphosis in less than 10% of larvae, whereas metamorphosis increased significantly on older biofilms, with a maximum of 41% occurring on 8-week-old microbial films. There was a significant influence of depth in 4- and 8-week biofilms, with greater levels of metamorphosis occurring in response to shallow-water communities. Importantly, larvae were found to settle and metamorphose in response to microbial biofilms lacking CCA from both shallow and deep treatments, indicating that microorganisms not associated with CCA may play a significant role in coral metamorphosis. A polyphasic approach consisting of scanning electron microscopy, fluorescence in situ hybridization (FISH), and denaturing gradient gel electrophoresis (DGGE) revealed that coral reef biofilms were comprised of complex bacterial and microalgal communities which were distinct at each depth and time. Principal-component analysis of FISH data showed that the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Cytophaga-Flavobacterium of Bacteroidetes had the largest influence on overall community composition. A low abundance of Archaea was detected in almost all biofilms, providing the first report of Archaea associated with coral reef biofilms. No differences in the relative densities of each subdivision of Proteobacteria were observed between slides that induced larval metamorphosis and those that did not. Comparative cluster analysis of bacterial DGGE patterns also revealed that there were clear age and depth distinctions in biofilm community structure; however, no difference was detected in banding profiles between biofilms which induced larval metamorphosis and those where no metamorphosis occurred. This investigation demonstrates that complex microbial communities can induce coral metamorphosis in the absence of CCA.
Most scleractinian coral species on the Great Barrier Reef reproduce during annual mass spawning events, where gametes are synchronously released into the seawater for external fertilization and dispersal (11). The developing larvae usually become competent to attach and metamorphose into juvenile polyps within a week of fertilization (2). Larvae of many coral species actively select a site of permanent attachment using external chemical cues that induce metamorphosis (13, 25-27). The primary source of chemical morphogens described for coral larvae are various species of nongeniculate calcareous coralline algae (CCA). These are thought to produce cell-wall-bound, high-molecular-mass polysaccharides that are recognized by chemoreceptors on the planula (25, 27). The skeleton of the coral Goniastrea sp. and coral rubble were also found to induce metamorphosis in corals, indicating that either multiple inducers for metamorphosis exist or specific inducers may originate from a variety of natural sources (13).
Biofilm formation on a marine substratum modifies the surface chemistry, thereby potentially affecting recruitment of larval invertebrates (4). Bacterial biofilms are present on the surface of various CCA species (9, 10, 16, 22) and may be primarily responsible for inducing larval metamorphosis of some invertebrates. A single strain of Pseudoalteromonas, isolated from the CCA Hydrolithon onkodes, was able to induce significant levels of metamorphosis of coral larvae in laboratory assays (30). However, the biological relevance of this interaction is not yet known, as sterile H. onkodes was still able to induce metamorphosis. Metamorphosis of other marine invertebrate larvae is also triggered by microorganisms associated with CCA. Johnson and Sutton (17) showed that the removal of bacteria from the surface of CCA using antibiotic treatments resulted in a reduction of metamorphosis of crown-of-thorns starfish larvae (Acanthaster planci). This demonstrated that bacteria on the surface of CCA produce morphogenic substances.
Marine biofilms alone have been reported to induce the metamorphosis in several classes of cnidarians, including Anthozoa (hard and soft corals) (12, 26), Scyphozoa (jellyfish) (3), and Hydrozoa (20). The species composition of natural coral reef biofilms that play a role in coral metamorphosis have as yet not been determined. In this study we utilized a variety of molecular techniques to examine the community structure of developing coral reef biofilms and their ability to induce the metamorphosis of coral larvae.
MATERIALS AND METHODS
Biofilm establishment.
Glass microscope slides (pathology grade; Livingstone International, Sydney, Australia) were cleaned (washed in 70% ethanol and rinsed three times in sterile distilled water) and placed in square polyvinyl chloride frames (112 per grid), allowing the top surface of the slides to be exposed to the seawater. The frames were deployed at Davies Reef, part of the Great Barrier Reef, and secured to the natural substratum using plastic cable ties. Two sites were randomly chosen in areas that allowed the secure placement of the frames (site 1, 18°49.77′S, 147°37.50′E; site 2, 18°49.64′S, 147°37.52′E). Frames were deployed at depths of 4 m (shallow) and 10 m (deep) within each site. To allow biofilm development, slides were maintained at site 1 for 2, 4, and 8 weeks and at site 2 for 8 weeks prior to the November 2000 mass coral spawning. The frames were collected; each treatment set was placed in separate aquaria with flowthrough seawater and transferred to the laboratory within 2 h. One set of clean negative-control slides were immersed in aquaria and transported as per treatments. Slides were used immediately in larval metamorphosis assays, with a minimum of 20 replicate biofilm slides randomly selected for each treatment. To determine biofilm community composition, an additional 30 slides were randomly selected and frozen at −80°C for DNA extraction, 30 slides were fixed in 4% paraformaldehyde for fluorescence in situ hybridization (FISH), and 10 slides were fixed in 2.5% glutaraldehyde for scanning electron microscopy (SEM).
Coral collection.
Coral larvae used in the metamorphosis assays were raised from gametes collected from live colonies of the reef-building coral Acropora microphthalma (Verrill, 1859). Corals were collected by snorkeling from a depth of 6 to 8 m on a fringing reef of Pelorus Island, Great Barrier Reef (18°32′S, 146°29′E), placed in 80-liter seawater tubs, and transported to flowthrough aquaria at the nearby Orpheus Island Research Station within 30 min of collection. Synchronous spawning occurred on 17 November 2000, and the released gametes, in the form of buoyant egg-sperm bundles, were collected in 250-ml containers from the water surface by gentle suction. Gametes from all colonies were cross-fertilized for 1 h in a single 500-liter plastic tank. The eggs, which formed a monolayer on the water surface, were then transferred to a 700-liter tank for primary rearing (29). At 36 h postfertilization, the ciliated larvae were transferred to 100-liter flowthrough aquaria and raised until competent. Water temperature was monitored continuously with an in situ data logger and fluctuated daily between 25 and 30°C.
Metamorphosis assays.
Larval metamorphosis assays were performed in 10-cm-diameter sterile plastic petri dishes maintained at 28°C. Eight-day-old coral larvae (n = 10 to 20) were introduced to each dish containing a single biofilm slide to be tested and filtered (pore size, 0.2 μm) seawater to a final volume of 30 ml. Two controls were established, the first being slides that were transported from the reef back to the laboratory in aquarium seawater (control A) and the second being sterile glass slides (control B). A minimum of 20 replicate slides were assayed for each treatment. Larvae were defined as metamorphosed (13) when they had changed from either free swimming or casually attached pear-shaped forms to squat, firmly attached disk-shaped structures with pronounced flattening of the oral-aboral axis and typically obvious septal mesenteries radiating from the central mouth region. Twenty-four hours was chosen as the endpoint for scoring early-stage metamorphosis (13). Settlement and metamorphosis were assessed by direct counting of all larvae and newly metamorphosed polyps in each well using a dissecting microscope. Digital photographs of metamorphosed larvae and relevant biofilm communities were taken for each treatment. Slides were sorted according to whether or not they induced larval metamorphosis, and replicate slides were then processed for SEM, FISH, and denaturing gradient gel electrophoresis (DGGE) as outlined below.
Community analysis. (iii) SEM.
Individual slides were fixed in 0.1 M sodium cacodylate buffer, pH 7.4, prepared in artificial seawater (Instant Ocean; Aquarium Systems) and containing 2.5% (vol/vol) glutaraldehyde for 20 h. Fixed slides were removed, placed in fresh 0.1 M sodium cacodylate buffer, and stored at 4°C until further processing. Fixed slides were placed in a 1% (wt/vol) osmium tetroxide solution and 1.5% potassium ferricyanide for 2.5 h and subsequently dehydrated in a graded ethanol series (15, 35, 55, 75, 85, 95, and 100% [vol/vol] ethanol). Slides were placed in hexamethyldisilazane for 1 h and allowed to dry overnight. Biofilms were sputter coated with gold for 180 s at 11 mA. Samples were visualized by SEM in a JEOL 6300F scanning electron microscope operated at 8 kV.
(ii) FISH.
Biofilm slides were fixed in 4% paraformaldehyde suspended in phosphate-buffered saline for 8 h at 4°C and transferred to 50% ethanol-50% phosphate-buffered saline at −20°C until further processing. All probes were labeled with either fluorescein or the indocarbocyanine fluorochromes Cy3 or Cy5 (Table 1) and synthesized by MWG AG Biotech (Ebersberg, Germany). Replicate wells (n = 8) were created on each slide using a wax circle. Hybridization solution (8 μl of 0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.01% sodium dodecyl sulfate [SDS], 20% [vol/vol] formamide) was mixed with 1 μl of the appropriate fluorescently labeled oligonucleotide and applied to each well of the slide. Slides were incubated in 50-ml polypropylene tubes at 46°C for 3 h. After hybridization, slides were carefully removed and rinsed immediately in prewarmed wash buffer (20 mM Tris-HCl, 0.01% SDS, 0.225 M NaCl) at 48°C for 10 min. Slides were rinsed in fresh water to remove excess salts, air dried, and mounted in the antifading glycerol medium (Citifluor; Citifluor Ltd, London, United Kingdom). FISH preparations were viewed and imaged on a Bio-Rad MRC-1024 confocal laser scanning microscope (CLSM). The illumination source was a 15-mW argon-krypton laser (American Laser Corporation) with excitation peaks at 488 nm (blue), 568 nm (green), and 647 nm (red). The images were captured in three different photomultiplier tubes. A 560-nm long-pass emission filter separated the green signal (fluorescein isothiocyanate) from the red signal, and a 640-nm short-pass emission filter separated the far-red signal (from Cy5) from the near-red signal (from Cy3). The CLSM was controlled by an OS-2 PC running the Bio-Rad LaserSharp Software package. Images were collected and final image evaluation was done in Adobe PhotoShop. The green emission was presented as the green channel of the color image, the red emission was presented as the red channel, and by convention, the far-red emission (Cy5) was presented as the blue channel.
TABLE 1.
Sequences of oligonucleotide probes used for FISH
| Probe | Sequence | Specificity | % Formamide | Reference |
|---|---|---|---|---|
| EUB338c | GCTGCCTCCCGTAGGAGT | Most bacteria | 20 | 1 |
| ALFIba | CGTTCGYTCTGAGCCA | Alphaproteobacteria | 20 | 23 |
| BET42ab | GCCTTCCCACTTCGTTT | Betaproteobacteria | 35 | 23 |
| GAM42ab | GCCTTCCCACATCGTTT | Gammaproteobacteria | 35 | 23 |
| HGC69ab | TATAGTTACCACCGCCGT | Actinobacteria | 25 | 33 |
| LGC354b | YSGAAGATTCCCTACTGC | Some Firmicutes | 35 | 24 |
| CF319ab | TGGTCCGTGTCTCAGTAC | Cytophaga-Flavobacterium of Bacteroidetes | 35 | 23 |
| PLA46b | GACTTGCATGCCTAATCC | Planctomycetales | 30 | 28 |
| ARC915b | GTGCTCCCCCGCCAATTCCT | Archaea | 20 | 34 |
| NonEub338c | ACTCCTACGGGAGGCAGC | Negative control | 20 | 1 |
Fluorochrome, fluorescein.
Fluorochrome, Cy3.
Fluorochrome, Cy5.
(iii) DGGE.
DNA was extracted from individual slides by scraping the biofilms into Eppendorf tubes using sterile scalpels and 0.5 ml of grinding buffer per sample (2 ml of 1 M Tris, 4 ml of 0.5 M EDTA, 2 ml of 10% SDS, 400 μl of 5 M NaCl, and 11.6 ml of distilled water). Tubes were immersed in liquid nitrogen, and their contents were ground with plastic pestles. Samples were incubated at 65°C for 60 min prior to addition of 187.5 μl of 5 M potassium acetate. Samples were incubated on ice for 30 min and centrifuged at 5,900 × g for 15 min. The supernatants were transferred to fresh tubes, and DNA was precipitated with a 0.8 volume of isopropanol. The 16S rRNA genes were amplified by PCR with bacterial and eukaryotic primers. For bacterial PCR, the forward primer 1055f (5′-ATG GCT GTC GTC AGC T-3′) and the reverse primer 1392r, incorporating a 40-base GC clamp (5′-CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC CAC GGG CGG TGT GTA C-3′), were used (7). For eukaryotic PCR, the forward primer NS1f (5′-GTA GTC ATA TGC TTG TCT C-3′) and reverse primer NS2r, incorporating a 40-base GC clamp (5′-CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC CGG CTG CTG GCA CCA GAC TTG C-3′), were used (38). Products from triplicate PCRs were combined, and 15 μl was applied to a 6.5% (wt/vol) polyacrylamide gel containing a 40 to 60% (bacterial) or 10 to 70% (eukaryotic) denaturing gradient of formamide and urea. Gels were electrophoresed at 60°C for 17 h in 1× Tris acetate-EDTA buffer at a constant voltage of 50 V. Gels were stained with ethidium bromide (0.5 μg ml−1) and visualized with a transilluminator.
Data analyses.
The importance of three factors (depth, time, and CCA presence) in inducing larval metamorphosis was investigated in an unbalanced repeated-measures analysis of variance (type IV SS; SPSS version 11). The percentage of metamorphosis was transformed [arcsin√(percent metamorphosis/100)] and expressed as radians to satisfy the analysis of variance assumption of homogeneity of variances (37). Data from site 2 and week 2 were removed from the analysis due to lack of replication for some treatments.
A principal-component analysis (PCA) was used to summarize biofilm community composition using FISH data (S-Plus release 3). Depth of biofilm formation and exposure time of slides were the variables. To assist in the interpretation, biplots of the response variables were overlaid on the PCA plots as vectors. These identify the bacteria primarily responsible for differences between treatments (S-Plus release 3). Ninety-five percent confidence limits were drawn around group means and are shown on the plots.
Cluster analysis was used to identify replicates that generated similar DGGE profiles. Computation of DGGE and treatment parameter matrices used binary Euclidean distances, with between-group linkage as the clustering method, and software from the Statistical Package for Social Science (SPSS version 11).
RESULTS
Metamorphosis assays.
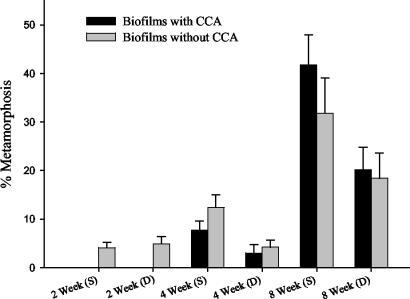
Glass slides were deployed on the Great Barrier Reef, and the biofilms that formed on them were tested for their ability to induce coral metamorphosis in laboratory assays. No metamorphosis was observed on control slides (data not shown), indicating that biofilms which develop within 12 h of submersion in aquarium seawater do not induce coral metamorphosis. Two-week-old biofilms induced metamorphosis in less than 10% of larvae (Fig. 1). The level of metamorphosis increased significantly between 4 and 8 weeks of biofilm development (F = 36.5; df = 1, 21; P < 0.01), with a maximum of 41% of larvae induced to metamorphose in response to biofilms developed in shallow depths (Fig. 1). There was a significant influence of depth at both 4 and 8 weeks, with greater levels of metamorphosis occurring on shallow-water biofilms than deep-water films (F = 6.5; df = 1, 21; P < 0.02). CCA, tentatively identified as the early-recruiting species including Titanaderma protoypum (L. Harrington, personal communication), appeared on some slides after 4 weeks of biofilm development. The presence of CCA on 4- and 8-week-old biofilms established at both depths made no significant difference to the ability of biofilms to induce larval metamorphosis (F = 0.1; df = 1, 21; P > 0.50). Two-week-old biofilms were not included in the statistical analysis as they did not contain CCA.
FIG. 1.
Percentage of metamorphosis of A. microphthalma larvae on 2-, 4-, and 8-week-old biofilms. Biofilms were developed at site 1 at two depths, 4 m (S) and 12 m (D). Twenty replicate slides were assessed in each treatment (bars = 1 standard error).
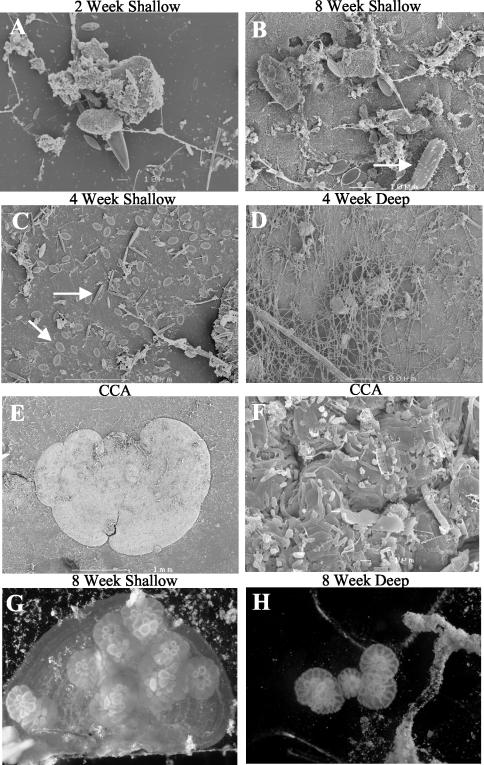
Larvae were found to settle and metamorphose in response to microbial biofilms lacking CCA from both shallow and deep treatments (Fig. 1 and 2), indicating that microorganisms not associated with CCA may also play a significant role in triggering metamorphosis. In biofilms where CCA was present, larvae showed a preference for settling directly upon the surface of the CCA (Fig. 2G). However, where larvae metamorphosed in the absence of CCA, they settled on clean regions of the glass slide away from other macro-members of the biofilm community (Fig. 2H).
FIG. 2.
Scanning electron micrographs of marine biofilms over time and depth (A to D) and on CCA (E and F), with diatoms indicated by arrows. Early stage metamorphosis of coral larvae in response to reef biofilms developed on glass slides in the presence (G) and absence (H) of CCA is shown, with individual corals indicated by arrows.
Community analysis.
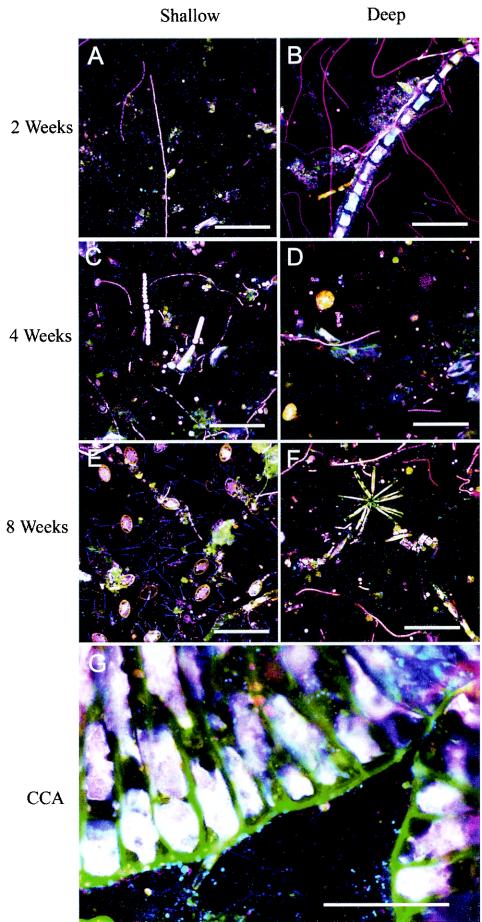
SEM, FISH, and DGGE each revealed that coral reef biofilms were comprised of a wide diversity of bacterial and microalgal species (Fig. 2 to 6). The density of microbial colonization increased with biofilm age (Fig. 2A to C and 3A to F). The microbial biomass also tended to be greater in shallow-water biofilms after 2 and 4 weeks but appeared to be similar at all depths after 8 weeks (Fig. 2 and 3). The community composition varied with biofilm depth for the microalgal component (Fig. 2C and D and 3A to F). In particular, shallow-water biofilms exhibited a higher density of diatoms than the deep-water biofilms (Fig. 2C and D). In all biofilms analyzed, FISH detected a number of bacteria which hybridized to the bacterial probe but did not hybridize to probes designed to target Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Actinobacteria, Firmicutes, the Cytophaga-Flavobacterium of Bacteroidetes, or the Planctomycetales (Fig. 3).
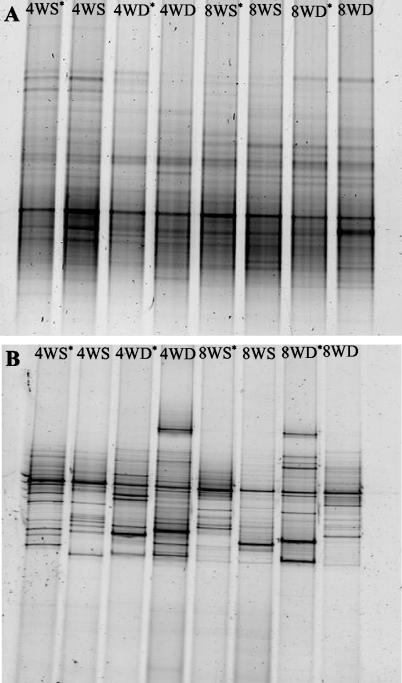
FIG. 6.
The large diversity of bacteria (A) and eukarya (B) associated with marine biofilms was detected using DGGE. All treatments were from site 1. Lanes: 4WS and 4WD, shallow- and deep-water biofilms, respectively, developed over 4 weeks; 8WS and 8WD, shallow- and deep-water biofilms, respectively, developed over 8 weeks. *, induced metamorphosis.
FIG. 3.
FISH images of marine biofilms with the Cy5-labeled bacterium-specific probe (EUB338), the Cy3-labeled GAM42a (for Gammaproteobacteria), the fluorescein-labeled ALF1b (for Alphaproteobacteria), and the Cy3-labeled Arch915 (for Archaea). Cells which appear magenta are Gammaproteobacteria, cyan cells are Alphaproteobacteria, blue cells are other bacteria, and red cells are Archaea. The scale bar on all images is 100 μm. Average bacterial counts ± standard errors per microscopic field of view (n = 60) with the Cy5-labeled bacterium-specific probe EUB338 were 160 ± 4.4 (2 weeks, shallow), 146 ± 4.0 (2 weeks, deep), 176 ± 4.7 (4 weeks, shallow), 166 ± 4.3 (4 weeks, deep), 185 ± 5.1 (8 weeks, shallow), and 184 ± 5.3 (8 weeks, deep).
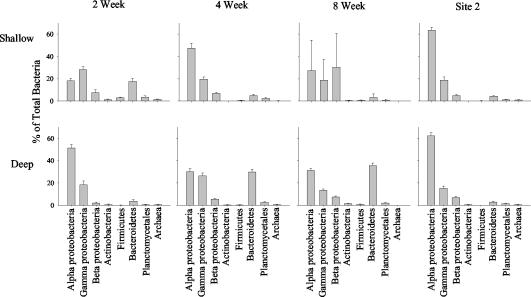
Relative densities of specific probe-targeted bacteria and archaea were estimated (Fig. 3 and 4). In the 2-week-old biofilms, deep films were dominated by Alphaproteobacteria and contained fewer Gammaproteobacteria and low levels of Betaproteobacteria (Fig. 4). The deep biofilms also contained very low numbers of Actinobacteria, Firmicutes, Cytophaga-Flavobacterium of Bacteroidetes, Planctomycetales, and Archaea (Fig. 4) and high densities of algae (Fig. 3B). In comparison, the shallow biofilms were dominated by Gammaproteobacteria and had fewer Alphaproteobacteria; a moderate density of Cytophaga-Flavobacterium of Bacteroidetes; and very low numbers of Actinobacteria, Firmicutes, Planctomycetales, and Archaea (Fig. 4). Differences in community composition were not as evident in the 4-week biofilms. Shallow-water biofilms after 4 weeks were dominated by Alphaproteobacteria and had a moderate abundance of Gammaproteobacteria; low numbers of Betaproteobacteria; and very low numbers of Actinobacteria, Firmicutes, Cytophaga-Flavobacterium of Bacteroidetes, Planctomycetales, and Archaea (Fig. 4). Deep-water biofilms after 4 weeks also exhibited high numbers of Alpha- and Gammaproteobacteria; low numbers of Betaproteobacteria; and very low numbers of Actinobacteria, Firmicutes, Cytophaga-Flavobacterium of Bacteroidetes, Planctomycetales, and Archaea. The primary difference between the two depths after 4 weeks was the high number of Cytophaga-Flavobacterium of Bacteroidetes (approximately 30%) in the deep films compared with only 5% in the shallow films (Fig. 4). The 8-week biofilms from both depths also had similar numbers of Alpha- and Gammaproteobacteria; however, the shallow communities were dominated by Betaproteobacteria and the deep communities had very high numbers of Cytophaga-Flavobacterium of Bacteroidetes. Both depths had large diatom populations (Fig. 3E and F) and very low numbers of Actinobacteria, Firmicutes, and Planctomycetales (Fig. 4). Interestingly, no archaea could be detected in any of these 8-week biofilms. In contrast to site 1, the community compositions were very similar for both depths at site 2. The biofilms were each dominated by Alphaproteobacteria, with fewer Gammaproteobacteria; small populations of Betaproteobacteria; and very few Actinobacteria, Firmicutes, Cytophaga-Flavobacterium of Bacteroidetes, and Planctomycetales (Fig. 4). Biofilms grown at both depths at site 2 contained very few Archaea.
FIG. 4.
Quantitative estimates of biofilm community composition using group-specific FISH probes. All densities are expressed as a percentage of total bacterial numbers obtained using dual hybridization reactions with the bacterium-specific probe EUB338. Site 2 data were collected after 8 weeks of biofilm development. Error bars, standard errors.
No differences in the relative densities of each subdivision of the Proteobacteria were observed between slides that induced larval metamorphosis and those that did not (Table 2). Also, the presence of CCA did not influence the relative subdivision densities. CCA was primarily observed on biofilm slides at 4 and 8 weeks and hosted a complex microbial community including diatoms, microalgae, and Alpha-, Beta-, and Gammaproteobacteria on their surfaces (Fig. 2E and F and 3G).
TABLE 2.
Community composition of Proteobacteria in 8-week-old biofilms identified by FISH and CLSM
| Depth and metamorphosis | CCA | Avg % composition ± SE
|
||
|---|---|---|---|---|
| Alphaproteo- bacteria | Betaproteo- bacteria | Gammaproteo- bacteria | ||
| Shallow | ||||
| Induced | − | 23.9 ± 3.9 | 33.7 ± 4.6 | 18.3 ± 2.4 |
| Induced | + | 19.9 ± 1.9 | 24.0 ± 2.7 | 21.4 ± 1.9 |
| Not induced | − | 30.6 ± 4.0 | 27.0 ± 2.6 | 19.1 ± 2.9 |
| Not induced | + | 32.3 ± 3.2 | 24.5 ± 2.7 | 20.3 ± 1.7 |
| Deep | ||||
| Induced | − | 55.8 ± 3.0 | 7.0 ± 0.8 | 15.8 ± 2.0 |
| Induced | + | 51.6 ± 2.4 | 8.1 ± 0.9 | 22.7 ± 1.9 |
| Not induced | − | 47.4 ± 2.6 | 7.4 ± 0.9 | 20.4 ± 1.8 |
| Not induced | + | 48.2 ± 2.5 | 7.9 ± 0.8 | 18.3 ± 0.9 |
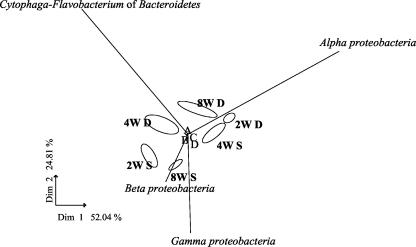
Each treatment (depth and time) combination had distinct biofilm communities, which clearly separated on a PCA plot (Fig. 5). While shallow-water biofilms had relatively high abundances of Beta- and Gammaproteobacteria compared to deeper-water films, there was no clear successional pattern over the three time periods examined. The lengths of the vector lines indicated that the Alphaproteobacteria and Cytophaga-Flavobacterium had the largest influence on overall community composition. This is particularly evident with the 2-, 4-, and 8-week deep-water films and the 4-week shallow-water biofilms. The Beta- and Gammaproteobacteria have considerable influence on biofilm community composition in the 2- and 8-week biofilms at shallow-water sites. In contrast, Actinobacteria, Firmicutes, Planctomycetales, and Archaea had little effect on the biofilm communities, potentially due to their relatively low abundance and also due to the minimal variability they displayed over depth and time.
FIG. 5.
PCA of biofilm community composition at site 1 using FISH counts to describe the community structure of reef biofilms. Abbreviations: 2W S and 2W D, shallow- and deep-water biofilms, respectively, developed over 2 weeks; 4W S and 4W D, shallow- and deep-water biofilms, respectively, developed over 4 weeks; 8W S and 8W D, shallow- and deep-water biofilms, respectively, developed over 8 weeks. Biplots of the response variables were overlaid on the PCA plots as vectors. A, Actinobacteria; B, Planctomycetales; C, Firmicutes; D, Archaea.
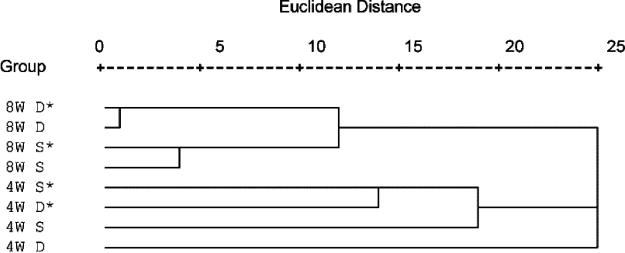
Most of the DGGE bands for both the bacterial (Fig. 6A) and eukaryotic (Fig. 6B) biofilms were common to all treatments irrespective of biofilm age or depth. The DGGE analysis of the bacterial community resulted in numerous bands on the DGGE gel throughout the gradient, indicating a very diverse microbial assemblage. The DGGE analysis of the eukaryotic community revealed much less diversity. However, there appeared to be more variability between biofilms in the eukaryotic community composition than in the bacterial communities (Fig. 6). It was not possible to detect any bands that were exclusively and consistently present in biofilms that induced coral metamorphosis (Fig. 6). Additionally, there were no bands exclusively present in biofilms where no coral metamorphosis occurred. Comparative analysis of bacterial DGGE patterns using cluster analysis revealed that there is little difference in banding patterns between biofilms which induced larval metamorphosis and those where no metamorphosis occurred (Fig. 7). With the exception of 4-week deep-water biofilms (which formed their own lineage), clustering of bacterial DGGE patterns was observed between the 4- and 8-week-old films. Considerably more variability (larger Euclidean distances) was evident in the films established over 4 weeks (Fig. 7). In the 8-week-old samples, clear distinctions in DGGE patterns were observed between shallow- and deep-water biofilms (Fig. 7); however, this trend was not obvious in 4-week-old samples. No ecological patterns could be ascertained from cluster analysis of eukaryotic DGGE patterns (data not shown).
FIG. 7.
Cluster analysis of DGGE banding profiles using binary Euclidean distances and between-group linkage. Abbreviations: S, shallow; D, deep; 4W, 4-week biofilms; 8W, 8-week biofilms. *, induced metamorphosis.
DISCUSSION
Although it has been clearly demonstrated that many species of coral larvae settle and metamorphose in response to CCA (13, 25-27), there is mounting evidence for a variety of potential sources of chemical inducers for coral larval metamorphosis (13, 30). This study reveals for the first time the complex nature of coral reef biofilms as they develop over 8 weeks and how these biofilms have the potential to induce coral metamorphosis in the absence of CCA.
The coral reef biofilms established in the present study induced up to 41% metamorphosis in A. microphthalma larvae. This builds upon previous research that demonstrated a strain of Pseudoalteromonas isolated from CCA was able to induce significant metamorphosis of Acropora willisae larvae (30). However, the larvae in that experiment were not exposed to a natural biofilm containing Pseudoalteromonas sp., and thus the environmental relevance of the result could not be confirmed. The larvae of the soft coral Heteroxenia fuscenscens were also shown to undergo metamorphosis in response to two strains of unidentified bacteria isolated from a coral skeleton (12). Other research has shown that scleractinian coral larvae undergo metamorphosis in response to indeterminate microalgal and bacterial biofilms (26), but the present study is the first attempt to describe the composition of naturally developed biofilms and their potential effects on coral metamorphosis.
In the present study, metamorphosis of A. microphthalma larvae was induced equally by microbial biofilms in both the presence and absence of CCA. This indicates that recruitment of corals onto reefs may be influenced as much by early-stage biofilms as they are by the presence of CCA. High bacterial densities have been visualized on the surface of CCA in previous electron microscopy studies; however, no attempts were made to describe the phylogenetic affiliation of these bacterial communities (9, 10, 16, 22). The present study revealed that CCA hosts a complex and dense microbial community on its surface, including numerous microalgae, diatom species, and members of the Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria. Interestingly, no differences in the relative densities of Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria could be detected between biofilms that did and did not induce coral metamorphosis and between biofilms that did and did not contain CCA. This strongly suggests that the metamorphic cues are associated not with bacterial density but most probably with specific microbial species that colonize the biofilms after 2 weeks. Unabia and Hadfield (36) found a wide phylogenetic range of bacteria capable of inducing settlement of the polychaete Hydroides elegans, suggesting either that multiple cues existed in different bacterial species or that the settlement cue was common to many bacterial species (36). In a separate study, the metamorphosis-inducing bacteria isolated from marine biofilms belonged to the Gammaproteobacteria, Cytophaga-Flavobacterium of Bacteroidetes, and gram-positive organisms, and different species within the same bacterial genus were found to vary considerably in their ability to induce settlement of H. elegans (19).
Coral larvae did show a preference for metamorphosing on older biofilms and films that were established in shallow water. This preference is likely to be due to differences in microbial density and microbial community composition within the different biofilm treatments. The density of microbial biofilms increased markedly with time and tended to be greater in shallow-water than in deeper-water films. Our results for tropical biofilm development agree with several previous studies on temperate marine biofilms that showed marine biofilms to be heterogenous in composition and highly variable over time and that larvae of marine invertebrates are able to differentiate between biofilms of various compositions, densities, ages, and origins during metamorphosis (31, 32, 39, 40). Many invertebrate species also preferentially settle and metamorphose on multispecies biofilms (18, 32).
In this study three techniques were applied to examine the effects of microbial biofilms on coral metamorphosis. By employing a polyphasic approach a deeper understanding of the relationship between the microbial biofilm and the coral was gained. The results indicate that after 2 weeks of biofilm development there was considerable disparity in community composition between shallow- and deep-water samples. Shallow-water biofilms were dominated by Gammaproteobacteria, with large numbers of diatoms, whereas deeper-water films were dominated by Alphaproteobacteria, with increased densities of microalgae. These differences in community composition probably reflect the early successional stage of the biofilm, and differences between depths may be due to differing ambient light levels. To date, little research effort has focused on succession in marine microbial biofilms, particularly in reef ecosystems. In the salt marsh environment, members of the α subdivision of the Proteobacteria were found to be rapid colonizers of most marine surfaces, although a few species of Gammaproteobacteria (primarily Alteromonas spp.) were also involved in very early colonization (5). Early stages of surface colonization were very dynamic (5), which we also found with our observations of early succession in coral reef biofilms. A separate study examining the phylogenetic diversity of free-living and aggregate-attached marine bacteria showed that Alphaproteobacteria were the most-abundant group of free-living bacterioplankton in coastal and open-ocean habitats. In contrast, Gammaproteobacteria, Cytophaga-Flavobacterium of Bacteroidetes, and the Planctomycetales were the most abundant aggregate-attached organisms (6).
After 4 weeks the reef biofilm community composition was more stable, similar between depths, and dominated by Alphaproteobacteria with moderate densities of Gammaproteobacteria. After 8 weeks a successional shift was observed, with both depths containing moderate abundances of Alpha- and Gammaproteobacteria; however, the shallow-water community was dominated by Betaproteobacteria while Cytophaga-Flavobacterium of Bacteroidetes was much more prevalent in deep-water biofilm communities. In contrast, 8-week biofilms from site 2 had very similar community compositions between depths, with all biofilms being entirely dominated by Alphaproteobacteria. No other studies have examined the community composition of reef biofilms at this late successional stage. The presence of low numbers of Archaea in all treatments except 8-week films from site 1 was interesting. This is the first description of archaea from coral reef biofilms and extends the known ecological niche for this group of microorganisms. These results emphasize the fine spatial scale variability that may occur in microbial community composition of coral reef ecosystems, and this may affect the patterns of coral recruitment on a reef.
PCA clusters samples with similar patterns and generates new variables (or principal components) which can be used to explain the highest dispersion of the data (8). PCA of FISH data showed that microbial community structure is very distinct at each depth, with no common trajectories of the communities through time. The Alphaproteobacteria and Cytophaga-Flavobacterium of Bacteroidetes had the largest influence on the overall biofilm community composition, with the Beta- and Gammaproteobacteria having considerable influence on biofilm community structure in shallow-water biofilms.
Broad-scale visual techniques such as SEM and FISH are ideal tools for describing overall community structure and architecture but could not elucidate the role of individual microorganisms in the process of coral settlement and metamorphosis. DGGE was employed as a fingerprinting technique to characterize bacterial community structures. DGGE showed a division in community composition between biofilms established for 4 and 8 weeks and a clear distinction in community composition between shallow and deep communities in 8-week-old biofilms. However, cluster analysis of DGGE banding patterns and FISH data revealed no differences in bacterial community structure between biofilms which induced metamorphosis and those that did not. No bands were exclusively present in metamorphosis-inducing biofilms (metamorphosis stimulators). Nor did DGGE analysis reveal bands that were exclusively present in biofilms where no metamorphosis occurred (metamorphosis inhibitors). The microorganisms involved in this process may be a small fraction of a highly diverse community that we cannot detect using the techniques employed. Furthermore, the microorganisms may be part of a complex interaction between a number of species, and it is this combination that produces cues for larval development. These results further illustrate the complexity of the relationship between coral larvae and marine biofilms.
Although it has been repeatedly demonstrated that coral larvae of many species are induced to settle and metamorphose in response to external chemicals present in CCA (13, 25-27), there is mounting evidence that microbial biofilms play a primary role in the synthesis of these external morphogens, as is the case in other cnidarians (15, 21, 30, 35). The results presented here demonstrate that complex microbial biofilms more than 2 weeks old are able to induce metamorphosis both in the presence and absence of CCA and highlight the potential for biofilms to influence coral recruitment patterns following mass spawning events. These findings may be applied to enhance the artificial rehabilitation of reefs degraded by natural climatic phenomena, crown-of-thorns starfish, or coral bleaching (14). Timely mechanical preparation of the seabed and subsequent development of appropriate microbial biofilms, prior to predictable mass spawning events, may enhance coral recruitment. These results also suggest that the timing of disturbance, in relation to annual recruitment pulses, may have major consequences for the nature of reef recovery.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babcock, R. C., and A. J. Heyward. 1986. Larval development of certain gamete-spawning scleractinian corals. Coral Reefs 5:111-116. [Google Scholar]
- 3.Brewer, R. H. 1976. Larval settling behaviour in Cyanea capillata (Cnidaria: Scyphozoa). Biol. Bull. 150:183-199. [Google Scholar]
- 4.Characklis, W. G., and K. E. Cooksy. 1983. Biofilms and microbial fouling, p. 93-108. In A. I. Laskin (ed.), Applied microbiology. Academic Press, New York, N.Y.
- 5.Dang, H., and C. R. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLong, E. F. 1993. Single-cell identification using fluorescently labeled, ribosomal RNA-specific probes, p. 285-294. In P. S. Kemp, B. E. Sherr, and J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, New York, N.Y.
- 7.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 9.Galbary, D., and C. J. Veltkamp. 1980. Observations on Mesophyllum lichenoides (Corallinaceae, Rhodophyta) with scanning electron microscope. Phycologia 19:49-53. [Google Scholar]
- 10.Garland, C. D., S. L. Cooke, J. F. Grant, and T. A. McMeekin. 1985. Ingestion of the bacteria on and the cuticle of crustose (non-articulated) coralline algae by post-larval and juvenile abalone (Haliotis ruber Leach) from Tasmanian waters. J. Exp. Mar. Biol. Ecol. 91:137-149. [Google Scholar]
- 11.Harrison, P. L., and C. C. Wallace. 1990. Reproduction, dispersal and recruitment of scleractinian corals, p. 133-207. In Z. Dubinsky (ed.), Coral reefs. Ecosystems of the world, vol. 25. Elsevier Science Publishing Company, New York, N.Y.
- 12.Henning, G., Y. Benayahu, and D. K. Hofmann. 1991. Natural substrates, marine bacteria and a phorbol ester induce metamorphosis of the soft coral Heteroxenia fuscenscens (Anthozoa: Octocorallina). Verh. Dtsch. Zool. Ges. 84:486-487. [Google Scholar]
- 13.Heyward, A. J., and A. P. Negri. 1999. Natural inducers for coral larval metamorphosis. Coral Reefs 18:273-279. [Google Scholar]
- 14.Heyward, A. J., L. D. Smith, M. Rees, and S. N. Field. 2002. Enhancement of coral recruitment by in situ mass culture of coral larvae. Mar. Ecol. Prog. Ser. 230:113-118. [Google Scholar]
- 15.Hofmann, D. K., and U. Brand. 1987. Induction of metamorphosis in the symbiotic scyphozoan Cassiopea andromeda: role of marine bacteria and of biochemicals. Symbiosis 4:99-116. [Google Scholar]
- 16.Johnson, C. R., D. G. Muir, and A.-L. Reysenbach. 1991. Characteristic bacteria associated with the surfaces of coralline algae: a hypotheses for bacterial induction of marine invertebrate larvae. Mar. Ecol. Prog. Ser. 74:281-294. [Google Scholar]
- 17.Johnson, C. R., and D. C. Sutton. 1994. Bacteria on the surface of crustose coralline algae induce metamorphosis of the crown of thorns starfish Acanthaster planci. Mar. Biol. 120:305-310. [Google Scholar]
- 18.Kirchmann, D., S. Graham, D. Reish, and R. Mitchell. 1982. Bacteria induce settlement and metamorphosis of Janua (Dexiospira) brasiliensis Grube (Polychaeta: Spirorbidae). J. Exp. Mar. Biol. Ecol. 56:153-163. [Google Scholar]
- 19.Lau, S. C. K., K. K. W. Mak, F. Chen, and P.-Y. Qian. 2002. Bioactivity of bacterial strains isolated from marine biofilms in Hong Kong waters for the induction of larval settlement in the marine polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 226:301-310. [Google Scholar]
- 20.Leitz, T. 1997. Induction of settlement and metamorphosis of Cnidarian larvae: signals and signal transduction. Invertebr. Reprod. Dev. 31:109-122. [Google Scholar]
- 21.Leitz, T., and T. Wagner. 1993. The marine bacterium Alteromonmas espejiana induces metamorphosis of the hydroid Hydractinia echinate. Mar. Biol. 115:173-178. [Google Scholar]
- 22.Lewis, T. E., C. D. Garland, and T. A. McMeekin. 1985. The bacterial biota on crustose (nonarticulated) coralline algae from Tasmanian waters. Microb. Ecol. 11:221-230. [DOI] [PubMed] [Google Scholar]
- 23.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 24.Meier, H., R. Amann, W. Ludwig, and K.-H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of Gram-positive bacteria with low DNA G + C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 25.Morse, A. N. C., K. Iwao, M. Baba, K. Shimoike, T. Hayashibara, and M. Omori. 1996. An ancient chemosensory mechanism brings new life to coral reefs. Biol. Bull. 191:149-154. [DOI] [PubMed] [Google Scholar]
- 26.Morse, D. E., N. Hooker, A. N. C. Morse, and R. A. Jensen. 1988. Control of larval metamorphosis and recruitment in sympatric agariciid corals. J. Exp. Mar. Biol. Ecol. 116:193-217. [Google Scholar]
- 27.Morse, D. E., Morse, A. N., Raimondi, P. T., and Hooker, N. 1994. Morphogen-based chemical flypaper for Agaricia humilis coral larvae. Biol. Bull. 186:172-181. [DOI] [PubMed] [Google Scholar]
- 28.Neef, A., R. Amann, H. Schlesner, and K.-H. Schleifer. 1998. Monitoring a widespread group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 29.Negri, A. P., L. D. Smith, N. S. Webster, and A. J. Heyward. 2002. Understanding ship-grounding impacts on a coral reef: potential effects of anti-foulant paint contamination on coral recruitment. Mar. Pollut. Bull. 44:111-117. [DOI] [PubMed] [Google Scholar]
- 30.Negri, A. P., N. S. Webster, R. T. Hill, and A. J. Heyward. 2001. Metamorphosis of broadcast spawning corals in response to bacteria isolated from crustose algae. Mar. Ecol. Prog. Ser. 223:121-131. [Google Scholar]
- 31.Parsons, G. J., M. J. Dadswell, and J. C. Roff. 1993. Influence of biofilm on settlement of sea scallop, Placopecten magellanicus (Gmelin, 1791), in Passamaquoddy Bay, New Brunswick, Canada. J. Shellfish Res. 12:279-283. [Google Scholar]
- 32.Patel, P., M. E. Callow, I. Joint, and J. A. Callow. 2003. Specificity in the settlement-modifying response of bacterial biofilms towards zoospores of the marine alga Enteromorpha. Environ. Microbiol. 5:338-349. [DOI] [PubMed] [Google Scholar]
- 33.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of Gram-positive bacteria with high DNA G + C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 34.Stahl, D. A., R. I. Amann, L. K. Poulsen, L. Raskin, and W. C. Capman. 1995. Archaea, a laboratory manual of methanogens. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 35.Szewzyk, U., C. Holmström, M. Wranstadh, M. O. Samuelsson, J. S. Maki, and S. Kjelleberg. 1991. Relevance of the exopolysaccharide of marine Pseudomonas sp., strain S9 for the attachment of Ciona intestinalis larvae. Mar. Ecol. Prog. Ser. 75:259-265. [Google Scholar]
- 36.Unabia, C. R. C., and M. G. Hadfield. 1999. Role of bacteria in larval settlement and metamorphosis of the polychaete Hydroides elegans. Mar. Biol. 133:55-64. [Google Scholar]
- 37.Underwood, A. J. 1999. Experiments in ecology: their logical design and interpretation using analysis of variance. Cambridge University Press, Cambridge, United Kingdom.
- 38.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, N.Y.
- 39.Wieczorek, S. K., A. S. Clare, and C. D. Todd. 1995. Inhibitory and facilitatory effects of microbial films on settlement of Balanus amphitrite larvae. Mar. Ecol. Prog. Ser. 119:221-228. [Google Scholar]
- 40.Wieczorek, S. K., and C. D. Todd. 1998. Inhibition and facilitation of settlement of epifaunal marine invertebrate larvae by microbial biofilm cues. Biofouling 12:81-118. [Google Scholar]