Abstract
Ammonium oxidation by autotrophic ammonia-oxidizing bacteria (AOB) is a key process in agricultural and natural ecosystems and has a large global impact. In the past, the ecology and physiology of AOB were not well understood because these organisms are notoriously difficult to culture. Recent applications of molecular techniques have advanced our knowledge of AOB, but the necessity of using PCR-based techniques has made quantitative measurements difficult. A quantitative real-time PCR assay targeting part of the ammonia-monooxygenase gene (amoA) was developed to estimate AOB population size in soil. This assay has a detection limit of 1.3 × 105 cells/g of dry soil. The effect of the ammonium concentration on AOB population density was measured in soil microcosms by applying 0, 1.5, or 7.5 mM ammonium sulfate. AOB population size and ammonium and nitrate concentrations were monitored for 28 days after (NH4)2SO4 application. AOB populations in amended treatments increased from an initial density of approximately 4 × 106 cells/g of dry soil to peak values (day 7) of 35 × 106 and 66 × 106 cells/g of dry soil in the 1.5 and 7.5 mM treatments, respectively. The population size of total bacteria (quantified by real-time PCR with a universal bacterial probe) remained between 0.7 × 109 and 2.2 × 109 cells/g of soil, regardless of the ammonia concentration. A fertilization experiment was conducted in a tomato field plot to test whether the changes in AOB density observed in microcosms could also be detected in the field. AOB population size increased from 8.9 × 106 to 38.0 × 106 cells/g of soil by day 39. Generation times were 28 and 52 h in the 1.5 and 7.5 mM treatments, respectively, in the microcosm experiment and 373 h in the ammonium treatment in the field study. Estimated oxidation rates per cell ranged initially from 0.5 to 25.0 fmol of NH4+ h−1 cell−1 and decreased with time in both microcosms and the field. Growth yields were 5.6 × 106, 17.5 × 106, and 1.7 × 106 cells/mol of NH4+ in the 1.5 and 7.5 mM microcosm treatments and the field study, respectively. In a second field experiment, AOB population size was significantly greater in annually fertilized versus unfertilized soil, even though the last ammonium application occurred 8 months prior to measurement, suggesting a long-term effect of ammonium fertilization on AOB population size.
Ammonium oxidation by autotrophic ammonia-oxidizing bacteria (AOB) is a key process in agricultural and natural ecosystems, with a large global impact. The product of this process, nitrite, is immediately oxidized by nitrite-oxidizing bacteria to nitrate, a nitrogen form susceptible to leaching. Nitrogen leaching can lead to groundwater pollution and surface and groundwater eutrophication. Nitrous oxide and nitric oxide, by-products of ammonia oxidation, contribute to the greenhouse effect and ozone layer depletion. On a local scale, loss of nitrate to groundwater and nitrous oxide and nitric oxide to the atmosphere reduces the amount of nitrogen available to plants in soil.
Until recently, the ecology of AOB communities largely has been extrapolated from pure-culture studies on Nitrosomonas europaea, the AOB most easily grown in laboratory cultures (30). Recent advances in DNA-based techniques for direct whole microbial community analysis have revealed that Nitrosospira, rather than Nitrosomonas, bacteria are ubiquitous in many environments (nutrient-poor and nutrient-enriched water and soil) and are the dominant AOB in soil (14, 18, 24; M. A. Bruns, J. Stephen, J. I. Prosser, and E. A. Paul, Abstr. 96th Gen. Meet. Am. Soc. Microbiol., abstr. N-101, p. 339, 1996). In the last decade, it has become possible to study terrestrial AOB communities without culturing by using PCR primers to specifically amplify 16S rRNA sequences from AOB cells (34). Also, PCR primers have been designed to target amoA, a functional gene coding for the active subunit of ammonia monooxygenase (31), a key enzyme in ammonia oxidation by AOB.
The need to quantify microbial populations is a pressing one in many areas of microbial ecology. In recent years, fluorescence in situ hybridization (FISH) techniques were developed to detect AOB in different environments (2, 6, 7, 17). However FISH is time-consuming and difficult to use in soil and requires physiologically active cells. More recently, Hesselsoe et al. (17) quantified soil AOB by separating AOB cells from soil by density gradient centrifugation, incubating the cells on a membrane for microcolony formation, and then staining and counting the cells. However, this approach may underestimate AOB density because it is not possible to separate all AOB from soil particles by centrifugation and not all cells can form microcolonies under laboratory incubation conditions.
Quantitative PCR methods, such as competitive and real-time PCR assays, are now being applied in investigations of AOB. Competitive PCR quantification includes a competitive sequence that serves as an internal control in each reaction. Competitive PCR requires time- and resource-consuming post-PCR analyses (8). Also, the primers used may not have the same affinity for the target and competitive sequences, which can lead to erroneous results. Nevertheless, competitive PCR has been successfully used to quantify AOB in groundwater (21), wastewater (H. M. Dionisi, G. Harms, A. Layton, S. Hawkins, I. Gregory, A. Meyers, K. Robinson, and G. Sayler, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. Q-320, p. 433, 2002), compost (23), activated sludge (3), and soil (24, 28, 33).
The quantitative real-time PCR method uses a fluorescence-labeled probe and measures fluorescence emitted continuously during the amplification reaction. The method does not require the use of a competitive sequence in the reaction, and it does not require post-PCR manipulation. In contrast to end point detection PCR, quantitative real-time PCR calculations are based on the initial exponential phase of the PCR. More detail on the theoretical background of quantitative real-time PCR is described by Heid et al. (15). Real-time PCR has been used to quantify the 16S rRNA gene (rDNA) from AOB (16; A. E. Bernhard, A. Schramm, and D. A. Stahl, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. N-118, p. 325-326, 2002; G. Harms, H. Dionisi, I. R. Gregory, A. C. Layton, K. G. Robinson, and G. S. Sayler, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. Q-90, p. 601, 2001) and amoA in aquatic environments (11; Dionisi et al., Abstr. 102nd Gen. Meet. Am. Soc. Microbiol.; M. A. Guerrero, and C. D. Sinigalliano, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. I-41, p. 253, 2002).
The objective of this research was to quantify soil AOB population dynamics in response to ammonium fertilizer additions. To achieve this goal, a real-time PCR assay targeting the amoA gene was developed and applied in laboratory and field experiments. A functional gene, amoA, rather than a 16S rDNA associated with AOB was targeted to provide quantitative information about the process of ammonia oxidation. We also estimated total bacterial population size by using a universal eubacterial 16S rDNA probe designed by Suzuki et al. (35). Therefore, changes in the relative proportions of AOB to the total bacterial density could be monitored following ammonium addition.
MATERIALS AND METHODS
Bacterial cultures.
N. europaea ATCC 19718 was purchased from the American Type Culture Collection (ATCC) and cultured in ATCC liquid medium 221. Strains of Nitrosospira multiformis, Nitrosospira tenuis, and Nitrosospira sp. strain NpAV were obtained from Jeanette Norton, Utah State University, and cultured in ATCC liquid medium 929. All strains were cultured at 28°C in the dark with periodic pH adjustment.
DNA extraction from soil.
DNA was extracted from 0.5 g (dry weight) of soil by using a Bio 101 FastDNA SPIN kit (for soil) as described by the manufacturer (Bio 101, Inc., Vista, Calif.). Extracted DNA was dissolved in 50 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]) and quantified by measuring its absorbance at 260 nm with a spectrophotometer. The quality of the extracted DNA was analyzed by electrophoresis on a 1.0% agarose gel.
To test whether PCR inhibitors were present in DNA samples extracted from soil, DNA samples from pure N. europaea cultures were mixed with an equal volume of soil DNA. Results of triplicate amoA real-time PCRs with and without addition of soil DNA were compared.
DGGE with amoA primers.
For denaturing gradient gel electrophoresis (DGGE) analysis, a GC clamp was attached to primer AmoA-1F (5′-GGGGTTTCTACTGGTGGT-3′) (31). Reverse primer amoA-2R-TG (5′-CCCCTCTGGAAAGCCTTCTTC-3′) was designed to be a perfect match for the N. tenuis amoA-specific sequence. PCRs were performed in a PE thermocycler with a 1:10 dilution of the DNA extracts as a template in a total volume of 50 μl with Ex Taq DNA polymerase (Takara Shuzo, Shiga, Japan) in the supplied buffer. After initial denaturation for 3 min at 95°C, amplification proceeded for 35 cycles consisting of 1 min at 95°C, 1 min at 55°C, and 45 s at 72°C, followed by a final 5-min extension at 72°C. amoA PCR products were subjected to DGGE as described previously (26) and visualized by UV transillumination after staining with SYBR green. Excised bands were cloned as described previously (20). Selected clones were sequenced with an ABI Prism automatic sequencer (Applied Biosystems) at Davis Sequencing Inc., Davis, Calif. Sequence identities were determined by BLAST and RDP analyses.
Extraction efficiency.
To test efficiency of DNA extraction from soil, a known number of amoA genes was added to soil. The number of N. europaea cells in liquid medium was determined after 4′,6′-diamidino-2-phenylindole (DAPI) staining with a fluorescence microscope (4), and the amoA concentration was calculated on the basis of the assumption that N. europaea has two amoA copies per cell (5, 27). A total of 1.5 ×108 ± 1.0 ×108 copy numbers of amoA was added to 0.5 mg of soil and vortexed for 10 s, and then DNA was extracted in triplicate as described above. The results of the real-time PCR assays with and without target DNA addition were compared.
TaqMan primers and probe design.
The real-time quantitative PCR method, also known as TaqMan PCR, requires an internal probe that hybridizes to the target sequence. This probe is labeled by two fluorescent dyes: a reporter and a quencher. The two fluorescent dyes interact whenever the probe is intact, causing the quencher to quench the reporter's fluorescence. During PCR amplification, Taq DNA polymerase cleaves the 5′ end of the probe, so the quencher is no longer associated with the reporter, and fluorescence is emitted. Fluorescence intensity is correlated with the number of accumulated PCR amplicons.
Copy numbers of the amoA gene were quantified with the probe and primer sequences shown in Table 1. The A189 forward primer was designed by Holmes et al. (19). The amoA-2R′ reverse primer, which is a modification of primer AmoA-2R designed by Rotthauwe et al. (31), and the A337 probe were designed in this study with Primer Express 1.0 software (Applied Biosystems, Foster City, Calif.) by aligning the amoA sequences of 14 different strains obtained from GenBank. The number of total bacteria was estimated by real-time PCR with 16S rDNA-based primers and a probe as described previously (35). TaqMan probes were dually labeled with the fluorescent dyes 6-carboxyfluorescein and 6-carboxytetramethyl rhodamine at the 5′ and 3′ ends, respectively. All of the primers and probes used in this study were synthesized by BioSource International (Camarillo, Calif.).
TABLE 1.
PCR primers and probe for the amoA gene
| Name | Sequence (5′-3′) | Positiona | Concn (nM) | Reference |
|---|---|---|---|---|
| Primer A189 | GGHGACTGGGAYTTCTGG | 151-168 | 300 | 19 |
| Primer amoA-2R′ | CCTCKGSAAAGCCTTCTTC | 802-820 | 900 | This study |
| Probe A337 | TTCTACTGGTGGTCRCACTACCCCATCAACT | 337-367 | 100 | This study |
| Primer amoA-2R-TG | CCCCTCTGGAAAGCCTTCTTC | 800-820 | 400 | This study |
Position is based on the open reading frame of amoA of N. europaea.
Real-time PCR.
Real-time PCR was performed in a 25-μl reaction mixture that consisted of 5 ng of template DNA, 12.5 μl of TaqMan Universal Master Mix (a 2×-concentrated mixture of AmpliTaq Gold DNA polymerase, uracil-N-glycosylase, deoxynucleoside triphosphates with UTP, passive reference dye, and optimized buffer [Applied Biosystems]), and primers and probe at the concentrations shown in Table 1. MicroAmp optical caps and tubes (Applied Biosystems) were used. The PCR protocol for amoA quantification was as follows: 2 min at 50°C, 10 min at 95°C, and then 40 cycles consisting of 45 s at 95°C, 1 min at 55°C, and 45 s at 72°C. The PCR protocol for bacterial 16S rDNA quantification was as follows: 2 min at 50°C, 10 min at 95°C, and then 40 cycles consisting of 15 s at 95°C and 1 min at 56°C. Reactions were carried out in an ABI Prism 7700 sequence detector (Applied Biosystems). The fluorescence signal was normalized by dividing the reporter dye (6-carboxyfluorescein) emission by the passive reference dye emission. The parameter CT (threshold cycle) is the cycle number at which the fluorescence emission crosses a threshold within the logarithmic increase phase. The threshold was defined as 10 times the standard deviation around the average intensity of background fluorescence from nontemplate controls. For signals giving CT values of less than 15, the calculation of the threshold was adjusted to 3 to 10 cycles. The greater the amount of initial template DNA, the earlier the fluorescence crosses the threshold and the smaller the CT value will be. Data were analyzed with Sequence Detector software, version 1.7.
Optimization of primer concentrations.
Primer concentrations were optimized in accordance with the Applied Biosystems guidelines. Template DNA and TaqMan probe concentrations were not changed, while PCR primer concentrations were systematically varied in all pairwise combinations (150, 300, 600, and 900 nM) of both the forward and reverse primers. The combination of 300 nM forward primer and 900 nM reverse primer was determined to be optimum and used in all reactions.
Standard curve for amoA quantification.
An external standard curve that shows the relationship between amoA copy numbers and CT values was generated with serial dilutions of a known copy number of the amoA gene. PCR primers A189 and amoA-2R′ were used to amplify the amoA gene from the total DNA extracted from N. europaea ATCC 19718. The PCR product was cloned with a TOPO TA cloning kit (Invitrogen). Plasmid DNA was extracted with a Plasmid Mini Kit (Qiagen), and the plasmid concentration (nanograms per microliter) was measured with a spectrophotometer. Since the sequences of the vector and PCR insert are known (5), we calculated the copy numbers of amoA directly from the concentration of extracted plasmid DNA.
The standard curve for bacterial 16S rDNA was made with genomic DNA extracted from Escherichia coli K-12. The 16S rDNA copy number was estimated on the basis of the genome size (4.64 Mbp) and seven copies of the 16S rDNA per E. coli genome (37).
Quantification of amoA and the 16S rDNA in soil samples.
Extracted DNA was diluted to 1 ng/μl, and 5 μl of this dilution was used in the 25-μl reaction mixtures of the real-time PCR. Triplicate PCR samples for each soil sample were used to calculate amoA copy numbers. AOB population size was estimated by dividing the amoA copy number by 2.5 on the basis of the knowledge that each AOB cell carries two or three nearly identical copies of the amoA gene (27). We assumed that the average copy number of 16S rDNAs per cell was seven on the basis of the knowledge that 16S rDNA copy numbers range from 1 to 13 in all described bacteria (9) and E. coli has seven 16S rDNA copies per genome (37). The AOB and total bacterial population sizes presented in this paper were not corrected for efficiency of extraction from soil.
Microcosm experiment: soil sampling.
In November 2001, soil cores from depths of 0 to 15 cm were taken from an agricultural plot farmed in accordance with organic practices (plot identification no. 8-8) at the Long-Term Research on Agricultural Systems (LTRAS) facility (http://LTRAS.ucdavis.edu/). A tomato crop had been harvested in September, and no vegetation was present at the time of sampling. Soil at this site is classified as a mixture of Rincon silty clay loam and Yolo silt loam (12). The soil was air dried, passed through a 2-mm sieve, and stored at 4°C.
Microcosm preparation.
Soil preparation was conducted in a cold (4°C) room to prevent ammonia oxidization. Ammonium sulfate was added to the soil as described by Venterea and Rolston (36) to minimize spatial stratification of ammonium sulfate. Soil was flushed with an aqueous solution designed to approximate soil solution concentrations for a Sacramento Valley agricultural soil (38) and amended with different concentrations of (NH4)2SO4. For the 0 mM ammonium sulfate treatment, the flushing solution contained 2.5 mM CaCl2, 2.5 mM CaSO4, 1.25 mM MgSO4, and 8.125 mM K2SO4. For the 1.5 mM ammonium sulfate treatment, the flushing solution contained 2.5 mM CaCl2, 2.5 mM CaSO4, 1.25 mM MgSO4, 6.68 mM K2SO4, and 1.5 mM (NH4)2SO4. For the 7.5 mM ammonium sulfate treatment, the flushing solution contained 2.5 mM CaCl2, 2.5 mM CaSO4, 1.25 mM MgSO4, 0.68 mM K2SO4, and 7.5 mM (NH4)2SO4. Soils were flushed slowly with their respective solutions until effluent ammonium concentrations were equal to influent levels and then were drained with a pressure plate to equilibrate to 0.05 MPa. Added concentrations of 1.5 and 7.5 mM ammonium sulfate in soil solution were equivalent to approximately 80 and 400 kg of ammonium sulfate per hectare, respectively, assuming that all of the N was in the soil solution (18% moisture content and bulk density of 1.5 g/cm3). This estimate also assumed that the ammonium sulfate is evenly mixed into the top 15-cm layer of the field soil. The rate of 400 kg of ammonium sulfate (84 kg of N) per hectare is about half the amount of fertilizer routinely applied to wheat in California. The measured initial concentrations of ammonium in soil were 103 and 548 μg of N/g of dry soil for the 1.5 and 7.5 mM ammonium sulfate treatments, respectively.
A total of 60 g of soil was placed in each of 50 300-ml mason jar microcosms for each ammonium concentration. After preparation, microcosms were incubated at ∼20°C. At 0, 1, 3, 7, 14, and 28 days, four jars of each ammonium concentration were extracted with 2 M KCl to measure ammonium and nitrate concentrations and 5 g of soil was taken from each microcosm and stored at −80°C for DNA extraction.
Ammonium and nitrate concentration measurements in microcosm soil.
A total of 200 ml of 2 M KCl was added to 60 g of microcosm soil, shaken on a reciprocating shaker, and centrifuged at 4,100 × g for 5 min at 5°C. The supernatant was collected and stored at −20°C until analysis. Ammonium and nitrate-plus-nitrite concentrations (referred to as nitrate concentrations in this study) in the extracts were determined colorimetrically with a Lachat Quick Chem II Flow Injection Analyzer (Lachat 8000; Zellweger Analytical, Milwaukee, Wis.).
Field experiment 1.
The effect of ammonium fertilization on AOB population size was measured in an organically managed corn (Zea mays)-tomato (Lycopersicum esculentum) rotation plot at LTRAS (plot identification no. 2-3). The study was conducted after the tomato harvest, from 15 November to 24 December 2002. Two fertilized and two unfertilized control plots, each measuring 0.5 by 2 m, were randomly chosen. A total of 40 g of ammonium sulfate per plot (400 kg/ha) was applied to the fertilized treatments and mixed thoroughly into the top 15 cm of soil with a shovel. Soils were sampled immediately before and 1, 3, 7, 13, and 39 days after fertilization to quantify ammonium and nitrate concentrations and AOB population size. Three soil cores from the 0- to 15-cm depth were taken at three different locations in each plot. Soil samples were air dried for approximately 24 h, sieved (2-mm maximum particle size), and then mixed thoroughly. Immediately after sieving, the soil were extracted with a KCl solution (described below) and a second soil subsample was frozen for further DNA analysis.
Field experiment 2.
The long-term effect of ammonium fertilization on AOB populations was studied by collecting soil samples from six plots located in conventionally managed wheat-fallow plots at LTRAS in November 2002, several months after wheat was harvested and at least 8 months after the last fertilizer application. At LTRAS, treatments were randomly assigned to plots. Three of the plots had, since 1993, annually received preplanting fertilization (112 kg of N [as ammonium nitrate]/ha in November) and in-season fertilization (56 kg of N [as ammonium nitrate]/ha in March) during the wheat year of the rotation. These plots were compared to three unfertilized plots. Soil samples were collected and analyzed as described previously.
Ammonium and nitrate concentration measurements in field samples.
Five milliliters of 1 M KCl was added to 1g of soil, shaken for 30 min, and centrifuged (5,200 × g for 10 min). The supernatant was collected and kept at −20°C until analysis. Ammonium and nitrate concentrations were measured colorimetrically as described by Foster (10). By this method, nitrate cannot be distinguished from nitrite.
Data analysis.
For each field replicate, ammonium and nitrate concentrations were measured once. Triplicate results of real-time PCR measurements were averaged, and the standard error was calculated. When the standard error (in units of the CT value) for a soil sample was greater than 0.5, the real-time PCR assay of that sample was repeated.
Ammonium and nitrate concentrations and AOB population sizes were averaged among locations within each plot. Means and standard errors were calculated with the following sample sizes: n = 4 in the microcosm experiment, n = 2 in field experiment 1, and n = 3 in field experiment 2.
Kinetic parameters.
The generation time, growth yield, and oxidation rate per AOB cell were calculated for the 1.5 and 7.5 mM ammonium treatments of the microcosm experiment and for the ammonium treatment in field experiment 1. The generation time (g) is the time required for a population to double in size. This parameter was calculated from the logarithmic growth equation Nt = N0 × eμt, where Nt is the population size at time t, N0 is the population size at time zero, and μ is the growth rate constant. The μ (assumed constant) was calculated from the slope of AOB population increase during the period of exponential growth. The generation time is calculated from g = (ln 2)/μ, an expression that is derived from the logarithmic-growth equation.
The rate of oxidation per cell was estimated by dividing the rate of ammonium disappearance between two sampling time points (assuming all ammonium consumption is by AOB) by the mean population size during the time period. The growth yield was estimated by dividing the population growth rate by the ammonium disappearance rate between sampling time points. These estimates make the assumption that rates of net nitrogen mineralization are equal to net immobilization rates, which is reasonable given the soil type and its previous agricultural management.
RESULTS
Real-time PCR assay. (i) TaqMan probe design and specificity.
The newly designed TaqMan probe perfectly matched the target amoA sequence of N. europaea ATCC 19718. N. multiformis, N. tenuis, and Nitrosospira sp. strain NpAV, on the other hand, had sequences containing 1, 3, and 1 mismatches with the probe, respectively (as shown in Table 2). To test the hypothesis that the designed TaqMan probe could also detect strains with mismatches in their target sequences, we compared calibration curves generated with dilutions of genomic DNAs from N. europaea, N. tenuis, and Nitrosospira sp. strain NpAV. There was no significant difference in the slopes and intercepts for N. europaea and Nitrosospira sp. strain NpAV (one mismatch, P = 0.75) over a DNA concentration range of 10 to 104 fmol. The slope and intercept for the N. tenuis (three mismatches with the probe) standard curve were significantly different from those of the other two nitrospiras (P = 0.02).
TABLE 2.
Probe and target sequences
| Name | Sequenceb |
|---|---|
| A337 (TaqMan probe) | 5′TTCTACTGGTGGTCRCACTACCCCATCAACT3′ |
| Targets | |
| Nitrosomonas europaea | 3′AAGATGACCACCAGTGTGATGGGGTAGTTGA5′ |
| Nitrosospira tenuis | 3′AAAATGACCACCAGCGTGATGGGGTACTCGA5′ |
| Nitrosospira multiformis | 3′AAGATGACCACCAGCGTGATGGGGTACTTGA5′ |
| Nitrosospira sp. strain NpAV | 3′AAGATGACCACCAGCGTGATGGGCTAGTTGA5′ |
| Nitrosospira sp. strain 39-19 | 3′AAGATGACCACCAGCGTGTTGGGGTAGTCGA5′ |
| Nitrosospira sp. strain 24C | 3′AAGATGACCACCAGCGTGATGGGGTAGTTGA5′ |
| Nitrosospira briensis | 3′AAGATGACCACCAGCGTGATGGGGTAGTTGA5′ |
| Methlyococcus capsulatusa | 3′AAGACCCCGACCTGGATGAAGGGCTAGTTGA5′ |
| Methylosinus trichosporium OB3ba | 3′AAGACCCCGACCTGGATAAAGGGCTAGTCGG5′ |
| Methylocystis sp. strain Ma | 3′AAGACCCCGACCTGGATGAAGGGCTAGTCGG5′ |
Methane-oxidizing bacterium.
Underlined bases are mismatches with the probe.
To determine whether N. tenuis (or other strains having three mismatches with probe A337) was a member of the AOB community in our soil, we performed DGGE analysis of the soil communities with amoA-specific primers. Analysis of soil microcosms with amoA primers (amoA-2R-GT, targeting N. tenuis) revealed three distinctive bands (data not shown). Sequence analysis indicated that the three sequences were most similar to an amoA gene from an unidentified strain, Nitrosospira sp. strain Nsp1, and Nitrosospira sp. strain NpAV (GenBank accession numbers AY445617 to AY445619). TaqMan probe A337 is a 100% match for the amoA genes from those three uncultured bacteria. The specificity of our real-time PCR assay was confirmed by sequence analysis of PCR products derived from soil DNA samples. Each yielded the expected bacterial amoA sequence (data not shown).
(ii) Standard curve and detection limit of amoA real-time PCR assay.
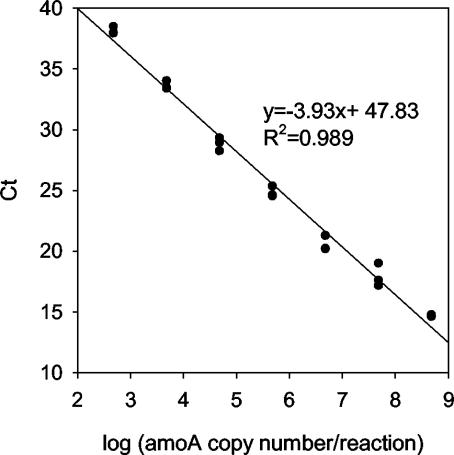
A standard curve relating the CT value and AOB was generated with a 10-fold dilution series of a plasmid containing the amoA gene. There was a strong linear (R2 = 0.99) inverse relationship between CT and the log10 number of amoA copies over 7 orders of magnitude (Fig. 1). The equation describing the relationship is CT = −3.93 × log10 (amoA) + 47.8. The detection limit was 98.3 copies per 5 μl of DNA sample used in a reaction mixture, corresponding to 3.9 × 104 cells/g of dry soil, with the assumption that the DNA extraction efficiency was 100%.
FIG. 1.
Standard curve for amoA real-time PCR.
The standard curve based on plasmid DNA (Fig. 1) was compared to a genomic DNA standard with extracted N. europaea cells (no mismatch with the TaqMan probe). The slopes and intercepts of the calibration curves for the genomic and plasmid-based standards agreed within 0.8%.
(iii) DNA extraction efficiency and effect of PCR inhibitors in soil.
A known number of N. europaea cells was added to soil, and the AOB population size was measured by real-time PCR. The calculated population size, based on DAPI counts, was 1.5 × 108 ± 1.0 × 108 AOB cells/g of soil. The measured population size was 7.2 × 107 ± 2.0 × 107 AOB cells/g of soil. Therefore, the extraction efficiency of the Bio 101 kit per gram of soil was 31.0%. On the basis of this extraction efficiency obtained, the detection limit of the real-time PCR assay was calculated as 1.3 × 105 AOB cells/g of soil.
To test for the potential presence of PCR inhibitors in soil DNA samples, the amoA copy numbers in pure-culture DNA, with and without addition of soil DNA, were quantified by real-time PCR (Table 3). There was no significant difference in the quantified amoA genes between the samples with and without soil DNA addition (two-tailed t test, n = 3, t = 1.99, P = 0.12). We concluded, therefore, that soil PCR inhibitors did not interfere with the real-time PCR assay.
TABLE 3.
Quantification of amoA copy numbers with and without addition of soil DNA
| PCR template | amoA copy no./PCR (mean ± SE, n = 3) |
|---|---|
| Soil DNA | 3.9 × 102 ± 32.2 |
| Nitrosomonas europaea DNA | 3.0 × 105 ± 3.7 × 103 |
| N. europaea DNA + soil DNA | 3.6 × 105 ± 2.7 × 104 |
(iv) Real-time PCR standard curve for bacterial 16S rDNA quantification.
A calibration curve was generated for bacterial 16S rDNAs, similar to what was described for amoA quantification. Standards ranged from 102 to 108 genome copies of E. coli. The calibration curve for 16S rDNA copies was linear (R2 = 0.99) over 7 orders of magnitude (CT = −3.53 × log10 [16S rRNA] + 42.4; data not shown).
Microcosm experiment. (i) Ammonium and nitrate concentrations.
Ammonium concentration changes were measured after adding ammonium sulfate (0, 1.5, and 7.5 mM) to the laboratory microcosms (Fig. 2A). Two weeks after addition, ammonium decreased to below the detection limit in the 1.5 mM treatment. In the 7.5 mM treatment, ammonium decreased to approximately 60% of the added concentration by the end of 1 month. Nitrate concentrations remained at <3.5 μg of N/g of dry soil and changed little over time in the control soils (Fig. 2B), whereas in the 1.5 mM treatment, nitrate concentrations increased substantially up to day 14 and then decreased to a little over half the peak value after 1 month. In the 7.5 mM treatment, the nitrate concentration increased over the 1-month incubation period.
FIG. 2.
Changes in ammonium concentration, nitrate concentration, AOB population size, and total bacterial population size after application of ammonium to microcosms. Dashed, dotted, and solid lines are 0, 1.5, and 7.5 mM ammonium sulfate treatments, respectively.
(ii) AOB and total bacterial population sizes estimated by real-time PCR assay.
In the absence of ammonium addition, the AOB population density changed little with time (Fig. 2C). In the 1.5 mM ammonium sulfate treatment, after 1 week, the AOB population density was almost eight times as high as the initial density of (4.0 ± 0.95) ×106 cells/g of dry soil. In the 7.5 mM treatment, the AOB population density was 11 times higher after 1 week. AOB population sizes were not significantly different among treatments on day 0 (one-way analysis of variance [ANOVA], P = 0.12) but were significantly different on day 28 (one-way ANOVA, P = 0.02).
The total bacterial density did not increase following ammonia application (Fig. 2D) at either concentration and remained between 0.7 × 109 and 2.2 × 109 copies/g of dry soil at all ammonium treatment levels (one-way ANOVA, P = 0.10 at day 0 and P = 0.10 at day 28).
Field experiment 1. (i) Ammonium and nitrate concentrations.
A field experiment was conducted to determine whether relationships between AOB densities and ammonium oxidation observed in the microcosms could be detected in the field under similar conditions. In the control plots, ammonium concentrations were less than 3.0 μg of N/g of dry soil and did not fluctuate much over time (Fig. 3A). In the ammonium treatment, the ammonium concentration was 267.5 ± 22.0 μg of N/g of dry soil (n = 2) at day 3 and decreased to 21.2 ± 1.2 μg of N/g of dry soil by day 39. The ammonium concentration was lower, but not statistically significantly so, on day 1 than on day 3 (one-way ANOVA, P = 0.22).
FIG. 3.
Changes in ammonium concentration, nitrate concentration, and AOB population size after application of ammonium to field plots. Solid lines are ammonium treatment. Dashed lines are control plots.
The nitrate concentrations in the control plots slowly decreased by 9.9 μg of N/g of dry soil by day 39 (Fig. 3B). In the ammonium-treated plots, the nitrate concentrations doubled by day 39 and were significantly different from those of the control plots (one-way ANOVA, P = 0.005). It should be noted that in field experiment 1, day 0 samples were taken immediately before ammonium application, whereas day 0 samples in the microcosm experiment were collected immediately after ammonium application.
(ii) AOB population size.
In the control plots, the AOB population size remained less than 9.0 × 106 cells/g of dry soil and did not change much over time (Fig. 3C). In the ammonium-treated plots, the AOB population size increased fourfold by day 39 and was significantly greater than that in control soils (one-way ANOVA, P = 0.014).
Field experiment 2.
Ammonium, nitrate, and AOB concentrations were measured in unfertilized and fertilized plots that had received fertilizer 8 and 12 months before soil sampling (Table 4). The AOB population size and nitrate concentrations were significantly higher in fertilized than in unfertilized plots (two-tailed t test, P = 0.007 and 0.002, respectively), while the difference in ammonium concentration was not statistically significant (two-tailed t test, P = 0.17).
TABLE 4.
Ammonium and nitrate concentrations and AOB population sizes in fertilized and unfertilized plots
| Agricultural soila | Mean N concn (μg/g of dry soil) ± SE
|
Mean AOB population size (no. of cells/g of dry soil) ± SE | |
|---|---|---|---|
| Ammonium | Nitrate | ||
| Unfertilized plots | 3.5 ± 1.0 | 2.0 ± 0.2 | (5.0 ± 1.8) × 106 |
| Fertilized plotsb | 5.1 ± 0.1 | 5.3 ± 0.4 | (1.5 ± 1.0) × 107 |
Samples were taken in November 2002.
Fertilized plots received ammonium nitrate in November 2001 and March 2002.
Kinetic parameters.
AOB growth generation time was calculated for the 1.5 and 7.5 mM microcosm treatments and for the field experiment 1 ammonium treatment (Table 5). The specific growth rate (μ) was calculated for days 0 to 3 in the 1.5 mM treatment, days 0 to 7 in the 7.5 mM treatment, and days 1 to 39 in the ammonium treatment. The curves yielded R2 values of 0.99, 0.94, and 0.97 and calculated generation times of 28.0, 51.7, and 372.7 h in the 1.5 mM microcosm, the 7.5 mM microcosm, and field experiment 1, respectively. All calculations assumed no cell death (e.g., due to predation by protozoa) during the experiments. The rates of oxidation per cell (based on ammonium disappearance) and growth yields were calculated (Table 5). Growth yields were 5.6 × 106, 17.5 × 106, and 1.7 × 106 cells/μmol of NH4+ in the 1.5 and 7.5 mM microcosm treatments and the field study ammonium treatment, respectively. If cell death occurred under field conditions, these growth yield measurements would be underestimated.
TABLE 5.
Kinetic parameters
| Method, environment, and organism | Generation time (h) | Oxidation rate (fmol of NH4+/h/cell) | Yield (106 cells/μmol) | Reference |
|---|---|---|---|---|
| Fluorescent-antibody staining, pure culture | ||||
| Nitrosospira | 20.1 | 4 | 8.0 | 1 |
| Nitrosolobus | 23 | 23 | 1.9 | |
| Acridine orange direct counting, pure culture, Nitrosospira spp. | 50 | 4.3 | 2.8 | 22 |
| 58 | 6.0 | 1.8 | ||
| 63 | 5.3 | 1.7 | ||
| 138 | 7.1 | 0.7 | ||
| Real-time PCR | ||||
| Microcosms, indigenous AOB, 1.5 mM NH4+ | 28 (0-3)a | 15.6 (0-1) | 5.6 (0-7) | This study |
| 1.0 (1-3) | ||||
| 0.8 (3-7) | ||||
| 0.4 (7-14) | ||||
| Microcosms, indigenous AOB, 7.5 mM NH4+ | 51.7 (0-7) | 8.5 (0-1) | 17.5 (0-7) | This study |
| 3.5 (1-3) | ||||
| 1.1 (3-7) | ||||
| 0.2 (7-14) | ||||
| 0.5 (14-28) | ||||
| Soil, indigenous AOB, field expt 1 | 372.7 (1-39) | 7.2 (3-7) | 1.7 (3-39) | This study |
| 2.3 (7-13) | ||||
| 0.5 (13-39) |
Values in parentheses are experiment durations in days.
DISCUSSION
This study is the first to develop and apply real-time PCR analysis to quantify amoA genes in soil and compare their dynamics with ammonium and nitrite concentrations. We measured net rather than gross (e.g., by stable-isotope pool dilution methods) rates of ammonium disappearance and nitrate production. The strong relationships observed among AOB densities, ammonium disappearance, and nitrate accumulation suggested that rates of net nitrogen mineralization and immobilization were similar and that denitrification was minimal in this well-aerated agricultural soil. Since the real-time PCR method allowed processing of large samples numbers, we measured population sizes at multiple time points and in response to different ammonium concentrations. This permitted us to estimate kinetic parameters describing ammonium oxidation (based on various assumptions) and provided a basis for comparison of our data with those of previous studies.
AOB population size in soil.
Under unfertilized conditions, the mean AOB population sizes were 7.1 × 106 and 5.5 × 106 cells/g of dry soil in microcosms and field samples, respectively. Our AOB population size estimates were several orders of magnitude greater than AOB population sizes in agricultural soils reported on the basis of most-probable-number (MPN) methods (13, 25, 32) and FISH with the microcolony method (17). This discrepancy was not surprising because the MPN method and the FISH-microcolony method count only culturable cells active under MPN assay conditions or on the membrane used for microcolony formation. In contrast, the real-time PCR method counted both active and dormant cells. The AOB population sizes in our study are comparable to those measured in agricultural soil (16, 24) by other PCR-based methods. With a real-time PCR assay targeting the AOB 16S rDNA, Hermansson and Lindgren (16) found 106 to 107 cells/g of dry soil. Mendum et al. (24) used a competitive PCR assay targeting both amoA and the 16S rDNA to find 105 to 108 gene copies/g of dry soil. Phillips and coworkers used a competitive PCR assay targeting the 16S rDNA and reported 105 cells of AOB/g of dry soil in fertilized soils (29). These results indicate a large, and as yet unexplained, variation in AOB population size among different agricultural soils.
Microcosm experiment.
The 0 mM ammonium sulfate treatment demonstrated that soil pretreatment by leaching reduced ammonium to <1 μg of N/g of dry soil and nitrate to <2 μg of N/g of dry soil and thus decreased interference from background levels of these ions. Very low nitrate levels were produced during incubation of the 0 mM treatment, indicating low net N mineralization and nitrification.
In the 1.5 mM treatment, AOB population growth stopped by the time most of the added ammonium was depleted. However, in the 7.5 mM microcosm, the AOB population growth reached a plateau around 6.0 × 107 cells/g of dry soil even though there was still a high ammonium concentration (330 μg of N/g of dry soil) remaining. In this case, factors other than ammonium concentration may have been limiting AOB population growth. Because microcosms were aerated every other day, oxygen was presumably not limiting. The soil pH may have decreased during incubation and limited population growth in this treatment. Unfortunately, pH was not measured in this study.
The total bacterial population size remained the same regardless of the ammonium concentration added. The proportion of AOB to total bacteria initially was 0.39%. After incubation in microcosms, this percentage increased to peak values (at day 7) of 0.6, 3.1, and 5.7% in the 0, 1.5, and 7.5 mM treatment, respectively. Thus, the absence of change in the total population size is not surprising given that AOB, even when enriched by ammonium, were still a small fraction of the total bacterial density and because the lack of carbon input prevented substantial growth of any heterotrophic bacteria.
Field experiments.
Nitrate concentrations were far lower in the field than in microcosm experiments and were lower than expected given the amount of added ammonium that disappeared (Fig. 3B). The low nitrate concentrations may have resulted from denitrification during the processing of the field soils required before KCl extraction. In the microcosm experiment, soil samples were not air dried or sieved because nitrate and ammonium concentrations were homogenized by leaching. The AOB population growth in the field ammonium treatment was first detected on day 7, contrasting with the microcosm data that showed growth beginning immediately after ammonium addition. Mendum et al. (24) reported no AOB growth within 3 days after ammonium application. One possible explanation for the longer lag period observed in the field was that ammonium may not have been as uniformly distributed and available to AOB in the field as in the laboratory soil. Moisture content, pH, and competition with heterotrophic bacteria for ammonium also may have differed between the microcosms and the field and contributed to differences in growth dynamics.
In the second field experiment, we examined the long-term effects of ammonium fertilizer on the AOB population and found a significant difference in AOB population size between fertilized and unfertilized plots. These differences, measured at least 8 months after the last fertilizer application, showed that differences in AOB population density were maintained independently of recent fertilizer application. The fertilized soil had received ammonium inputs every other year since 1993, whereas unfertilized plots had not been fertilized for 10 years. Differences in AOB may have been due, in part, to higher crop productivity and soil organic matter levels in the fertilized soils. In the unfertilized soils, AOB were likely maintained on ammonia that mineralized from soil organic matter.
Kinetic parameters.
Kinetic parameters obtained in this study were compared with values obtained from pure-culture studies on Nitrosospira spp. (Table 5) (1, 22). Calculated AOB generation times in the 1.5 and 7.5 mM treatments were similar to those determined in these pure-culture studies. AOB generation time in the ammonium-treated field plots was longer than that measured in the microcosm experiment, perhaps because field conditions were less favorable for AOB than those in microcosms or pure cultures.
The rates of ammonium oxidation per cell and AOB growth yields were, surprisingly, of the same order of magnitude as values obtained in pure-culture studies. The rate of ammonium oxidation per cell would be higher than expected if the AOB population size were underestimated. The fact that the measurements of ammonium oxidation per cell were in the range of reported values suggests that the real-time PCR developed in this study did not grossly underestimate the AOB population size.
Future applications.
The effects of agricultural management practices such as irrigation, tillage, cover cropping, and winter fallow on AOB population size can now be studied with the real-time PCR method developed in this study. The real-time PCR method permits rapid analysis of multiple samples and is thus useful in studies of population dynamics. The nitrogen cycle and reducing nitrogen losses from agroecosystems may be better managed with knowledge of AOB population dynamics. If the rate of ammonium oxidation per cell is relatively constant, as appears to be the case in this study, it may be possible to estimate ammonium oxidation rates from AOB population sizes.
Acknowledgments
We thank Dennis Bryant and other members of the LTRAS staff for providing field help. Also, we thank Jenny Norton for providing the Nitrosospira strains. Martin Burger's help with the design of the microcosm experiment is appreciated.
USDA-NRI Soils and Soil Biology Program grant 2000-00991 and a Kearney Foundation grant to R.F.D. helped fund this project. Financial support was also provided by the NIEHS Superfund Basic Research Program (2P42 ESO 4699), EPA Center for Ecological Health Research, and NSF Microbial Observatories (NSF-MO MCB-0137210).
REFERENCES
- 1.Belser, L. W., and E. L. Schmidt. 1978. Serological diversity within a terrestrial ammonia-oxidizing population. Appl. Environ. Microbiol. 36:589-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesterfeld, S., L. Figueroa, M. Hernandez, and P. Russell. 2001. Quantification of nitrifying bacterial populations in a full-scale nitrifying trickling filter using fluorescent in situ hybridization. Water Environ. Res. 73:329-338. [DOI] [PubMed] [Google Scholar]
- 3.Bjerrum, L., T. Kjaer, and N. B. Ramsing. 2002. Enumerating ammonia-oxidizing bacteria in environmental samples using competitive PCR. J. Microbiol. Methods 51:227-239. [DOI] [PubMed] [Google Scholar]
- 4.Bloem, J. 1995. Fluorescent staining of microbes for total direct counts, p. 1-12. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Brujin (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Chain, P., J. Lamerdin, F. Larimer, W. Regala, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daims, H., U. Purkhold, L. Bjerrum, E. Arnold, P. A. Wilderer, and M. Wagner. 2001. Nitrification in sequencing biofilm batch reactors: lessons from molecular approaches. Water Sci. Technol. 43:9-18. [PubMed] [Google Scholar]
- 7.Daims, H., N. B. Ramsing, K.-H. Schleifer, and M. Wagner. 2001. Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol. 67:5810-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diviacco, S., P. Norio, L. Zentilin, S. Menzo, M. Clementi, G. Biamonti, S. Riva, A. Falaschi, and M. Giacca. 1992. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene 122:313-320. [DOI] [PubMed] [Google Scholar]
- 9.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microbial Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 10.Foster, J. C. 1995. Soil nitrogen. p. 79-87. In K. Alef, and P. Nannipieri (ed.). Methods in applied soil microbiology and biochemistry. Academic Press, San Diego, Calif.
- 11.Harms, G., A. Layton, H. Dionisi, I. Gregory, V. Garrett, S. Hawkins, K. Robinson, and G. Sayler. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343-351. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa, H., J. M. Labavitch, A. M. McGuire, D. C. Bryant, and R. F. Denison. 1999. Testing CERES model predictions of N release from legume cover crop residue. Field Crops Res. 63:255-267. [Google Scholar]
- 13.Hashimoto, T., and T. Hattori. 1987. Length of incubation for the estimation of the most probable number of nitrifying bacteria in soil. Soil Sci. Plant Nutr. 33:507-510. [Google Scholar]
- 14.Hastings, R. C., J. R. Saunders, G. H. Hall, R. W. Pickup, and A. J. McCarthy. 1998. Application of molecular biological techniques to a seasonal study of ammonia oxidation in a eutrophic freshwater lake. Appl. Environ. Microbiol. 64:3674-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 16.Hermansson, A., and P.-E. Lindgren. 2001. Quantification of ammonia-oxidizing bacteria in arable soil by real-time PCR. Appl. Environ. Microbiol. 67:972-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hesselsoe, M., K. K. Brandt, and J. Sorensen. 2001. Quantification of ammonia oxidizing bacteria in soil using microcolony technique combined with fluorescence in situ hybridization (MCFU-FISH). FEMS Microbiol. Ecol. 38:87-95. [Google Scholar]
- 18.Hiorins, W. D., R. C. Hastings, I. M. Head, A. J. McCarthy, J. R. Saunders, R. W. Pickup, and G. H. Hall. 1995. Amplification of 16s ribosomal RNA genes of autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of nitrosospiras in the environment. Microbiology 141:2793-2800. [DOI] [PubMed] [Google Scholar]
- 19.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 20.Hristova, K., B. Gebreyesus, D. Mackay, and K. M. Scow. 2003. Naturally occurring bacteria similar to the methyl tert-butyl ether (MTBE)-degrading strain PM1 are present in MTBE-contaminated groundwater. Appl. Environ. Microbiol. 69:2616-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanova, I. A., J. R. Stephen, Y.-J. Chang, J. Bruggemann, P. E. Long, J. P. McKinley, G. A. Kowalchuk, D. C. White, and S. J. Macnaughton. 2000. A survey of 16S rRNA and amoA genes related to autotrophic ammonia-oxidizing bacteria of the beta-subdivision of the class Proteobacteria in contaminated groundwater. Can. J. Microbiol. 46:1012-1020. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, Q. Q., and L. R. Bakken. 1999. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30:171-186. [DOI] [PubMed] [Google Scholar]
- 23.Kowalchuk, G. A., Z. S. Naoumenko, P. J. L. Derikx, A. Felske, J. R. Stephen, and I. A. Arkhipchenko. 1999. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl. Environ. Microbiol. 65:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendum, T. A., R. E. Sockett, and P. R. Hirsch. 1999. Use of molecular and isotopic techniques to monitor the response of autotrophic ammonia-oxidizing populations of the β subdivision of the class Proteobacteria in arable soils to nitrogen fertilizer. Appl. Environ. Microbiol. 65:4155-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrill, L. G., and J. E. Dawson. 1967. Patterns observed for the oxidation of ammonium to nitrate by soil organisms. Soil Sci. Soc. Am. Proc. 31:757-760. [Google Scholar]
- 26.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 27.Norton, J. M., J. J. Alzerreca, Y. Suwa, and M. G. Klotz. 2002. Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch. Microbiol. 177:139-149. [DOI] [PubMed] [Google Scholar]
- 28.Phillips, C. J., D. Harris, S. L. Dollhopf, K. L. Gross, J. I. Prosser, and E. A. Paul. 2000. Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl. Environ. Microbiol. 66:5410-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips, C. J., E. A. Paul, and J. I. Prosser. 2000. Quantitative analysis of ammonia oxidising bacteria using competitive PCR. FEMS Microbiol. Ecol. 32:167-175. [DOI] [PubMed] [Google Scholar]
- 30.Prosser, J. I. 1989. Autotrophic nitrification in bacteria. Adv. Microb. Physiol. 30:125-181. [DOI] [PubMed] [Google Scholar]
- 31.Rotthauwe, J.-H., K.-P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarathchandra, S. U. 1979. A simplified method for estimating ammonium oxidising bacteria. Plant Soil 52:305-309. [Google Scholar]
- 33.Stephen, J. R., Y.-J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venterea, R. T., and D. E. Rolston. 2000. Mechanisms and kinetics of nitric and nitrous oxide production during nitrification in agricultural soil. Global Change Biol. 6:303-316. [Google Scholar]
- 37.Welch, R. A., V. Burland, G. D. I. Plunkett, P. Redford, P. Roesch, D. A. Rasko, E. L. Buckles, S.-R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. T. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolt, J. 1994. Soil solution chemistry. John Wiley & Sons, Inc., New York, N.Y.