Determination of the orientation of the enterovirus 71 virions in the crystal required the calculation of a locked rotation function that included only icosahedral threefold and fivefold symmetry axes. Otherwise, misleading high rotation-function values were produced by accidental alignment of icosahedral and crystallographic twofold axes.
Keywords: enterovirus 71, viruses, rotation function, molecular replacement
Abstract
Enterovirus 71 is a picornavirus that causes hand, foot and mouth disease but may induce fatal neurological illness in infants and young children. Enterovirus 71 crystallized in a body-centered orthorhombic space group with two particles in general orientations in the crystallographic asymmetric unit. Determination of the particle orientations required that the locked rotation function excluded the twofold symmetry axes from the set of icosahedral symmetry operators. This avoided the occurrence of misleading high rotation-function values produced by the alignment of icosahedral and crystallographic twofold axes. Once the orientations and positions of the particles had been established, the structure was solved by molecular replacement and phase extension.
1. Introduction
Enterovirus 71 (EV71) is a picornavirus causing hand, foot and mouth disease (Chan et al., 2000 ▶). In infants and small children the disease may progress to serious neurological implications (Alexander et al., 1994 ▶).
Picornaviruses are small non-enveloped animal viruses with positive-sense RNA genomes. Picornavirus capsids contain 60 copies of each of the four viral proteins VP1, VP2, VP3 and VP4, which form an ∼300 Å diameter quasi-T = 3 icosahedral shell. VP1, VP2 and VP3 each have the β-sandwich ‘jelly-roll’ fold common to many capsid proteins of icosahedral viruses. Each copy of the small VP4 is attached to the inner surface of the capsid and extends from an icosahedral fivefold axis towards a threefold axis. Most enteroviruses have a depression on the viral surface around the icosahedral fivefold axes of symmetry called the ‘canyon’ (Rossmann et al., 1985 ▶; Rossmann, 1989 ▶). The canyon is the site of binding of receptors that have an immunoglobulin-like fold (Belnap et al., 2000 ▶; Colonno et al., 1988 ▶; Olson et al., 1993 ▶). The binding of these receptors into the canyon results in the expulsion of a ‘pocket factor’ from a pocket immediately below the floor of the canyon. The shape of the pocket and the mostly hydrophobic side chains of the residues forming the pocket suggest that the pocket factor could be a lipid (Rossmann, 1994 ▶; Smyth et al., 2003 ▶; Filman et al., 1989 ▶; Muckelbauer et al., 1995 ▶). Release of the pocket factor presumably destabilizes the virions (Rossmann, 1994 ▶), which is a likely prelude to genome release.
The structure of EV71 and its implications for the design of pocket-factor-replacing antiviral drugs have been described elsewhere (Wang et al., 2012 ▶; Plevka et al., 2012 ▶). Here, we report the determination of the EV71 structure by X-ray crystallography.
2. Materials and methods
2.1. Virus growth and purification
Rhabdomyosarcoma cells were grown on 150 mm diameter plates to a density of 105 cells ml−1. 100 plates were infected with EV71 strain MY104-9-SAR-97 (GenBank DQ341368.1; obtained from Jane Cardosa, University of Malaysia, Sarawak, Malaysia; McMinn et al., 2001 ▶) at a multiplicity of infection of 3 and incubated at 310 K. Both cells and virus-containing supernatant were harvested 4 d post-infection. The cells were pelleted by centrifugation at 4500 rev min−1 for 10 min in a Beckman JA-10 rotor (Beckman Coulter Inc., Brea, California, USA) and lysed by freezing and thawing several times followed by Dounce homogenization. Cell debris was removed by centrifugation at 10 000 rev min−1 for 10 min (Beckman JA-10) and the virus-containing supernatant was pooled with the previously harvested virus-containing supernatant. The virus was pelleted by centrifugation at 45 000 rev min−1 in a Beckman Ti50.2 rotor at 277 K for 2 h. The pelleted virus was resuspended in buffer A (0.25 M HEPES, 0.25 M NaCl pH 7.5) and treated with MgCl2 (0.005 M), DNase (0.01 mg ml−1) and RNase (7.5 mg ml−1) for 30 min at room temperature followed by trypsin (0.8 mg ml−1) digestion for 10 min at 308 K. Subsequently, 0.015 M EDTA and 1% sarcosine were added, the solution was centrifuged at 4500 rev min−1 for 10 min (Beckman JA-10) and the supernatants were collected. The virus was pelleted by centrifugation at 45 000 rev min−1 in a Beckman Ti50.2 rotor for 2 h. The pellets were resuspended in buffer A and spun through a 10–40% potassium tartrate gradient at 36 000 rev min−1 in a Beckman SW41 Ti rotor at 277 K for 90 min. The virus band was collected, diluted in buffer A and centrifuged at 45 000 rev min−1 for 2 h (Beckman Ti50.2 rotor) to remove the potassium tartrate. The purified virus pellet was resuspended in buffer A and the concentration of the virus was measured with a spectrophotometer using an absorption coefficient of 7.7 mg ml−1 cm−1 at 260 nm wavelength.
2.2. Crystallization and data collection
Crystals of EV71 were obtained using the hanging-drop technique with a well solution consisting of 2.5%(w/w) PEG 800, 2.5%(w/w) glycerol, 600 mM NaCl, 150 mM CaCl2, 100 mM Tris pH 8.0. The drops were prepared by mixing 1.5 µl well solution with an equal volume of 10 mg ml−1 virus solution in buffer A. Crystals formed within one week. For data collection, crystals were soaked for 1–2 min in mother liquor containing 30%(w/w) PEG 400 and immediately cooled in liquid nitrogen. Single-crystal diffraction data were collected at 100 K using a MAR CCD 225 detector on BioCARS beamline 14B at the Advanced Photon Source synchrotron. An oscillation range of 0.2° was used during data collection. The diffraction pattern extended to 3.8 Å resolution. The diffraction images were processed and scaled using the HKL-2000 package (Otwinowski & Minor, 1997 ▶). Quality indicators of the diffraction data are listed in Table 1 ▶.
Table 1. Scaling and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Space group | I212121 |
| Unit-cell parameters () | a = 600.2, b = 610.6, c = 851.8 |
| Matthews coefficient (3Da1) | 3.44 |
| Resolution limits () | 503.8 (3.973.80) |
| Completeness (%) | 30.4 (11.5) |
| R merge † | 0.24 (0.79) |
| Average multiplicity | 1.7 (1.1) |
| I/(I) | 2.41 (0.88) |
| Reciprocal-space correlation coefficient of F obs and F calc after map convergence | 0.82 (0.47) |
| R factor | 0.279 (0.386) |
| Average B factor (2) | 79.2 |
| Ramachandran plot‡ | |
| Outliers (%) | 0.97 |
| Most favored regions (%) | 87 |
| Rotamer outliers (%) | 0.85 |
| R.m.s.d., bonds () | 0.003 |
| R.m.s.d., angles () | 0.93 |
| No. of unique reflections | 459113 |
R
merge = 
 .
.
According to MolProbity (Chen et al., 2010 ▶).
2.3. Data deposition
The EV71 coordinates together with the observed structure amplitudes and phases calculated by phase extension have been deposited in the Protein Data Bank (accession code 4aed).
3. Results and discussion
3.1. X-ray structure determination of EV71
The highest symmetry space groups consistent with the EV71 diffraction pattern were I422 or I4122. However, scaling the integrated intensities in the 50–10 Å resolution range gave an R merge of 0.39 when assuming a fourfold symmetry axis along c. In contrast, the R merge was 0.12 when assuming only twofold symmetry along c. It was therefore concluded that the virions of EV71 crystallized in a body-centered orthorhombic space group. The unit-cell parameters determined by post-refinement in an orthorhombic space group were a = 600.2, b = 610.6, c = 851.8 Å. The two possible space groups I222 and I212121 cannot be differentiated by systematic absences of reflections. Quality indicators of the diffraction data, which have been published previously (Plevka et al., 2012 ▶), are reprinted in Table 1 ▶ for the completeness of the present discussion. The Matthews coefficient indicated that there were the equivalent of two virus particles in the crystallographic asymmetric unit. Plots of fivefold, threefold and twofold self-rotation functions did not show any obvious sets of icosahedrally related peaks (Figs. 1 ▶ a, 1 ▶ b and 1 ▶ c; Tong & Rossmann, 1997 ▶). A more powerful procedure for determining the orientation of a high-symmetry object is to search simultaneously, with a locked rotation function, for all of the symmetry axes assuming their known relative orientations (Rossmann et al., 1972 ▶; Tong & Rossmann, 1997 ▶). However, a locked rotation function failed to establish the orientations of the particles in the crystal because it was dominated by peaks resulting from the alignment of icosahedral twofold symmetry axes with crystallographic twofold axes. Thus, a modified locked rotation function was calculated in which only the icosahedral fivefold and threefold symmetry axes were included. The search was performed over polar angles 0 < ψ < 90°, 0 < ϕ < 90°, 0 < κ < 90° (Table 2 ▶). This range covers several icosahedral asymmetric units. This procedure identified two independent particle orientations ϕ = 42.5, ψ = 68.5, κ = 45.5° for particle 1 and ϕ = 24.5, ψ = 87.5, κ = 77.0° for particle 2 with respect to the standard icosahedral orientation as defined by Arnold et al. (1984 ▶) and the XYK polar angle convention (Tong & Rossmann, 1997 ▶). None of the icosahedral twofold axes can be aligned with the crystallographic twofold axes. Therefore, the crystallographic asymmetric unit must contain two particles in general orientations. However, the arrangement of the crystallographic symmetry axes in space group I222 with the current unit-cell parameters does not permit the positioning of two particles in general orientations. Therefore, the space group must be I212121. The positions of all of the noncrystallographic symmetry (NCS) operators of both independent particles and their crystallographic symmetry-related copies are shown in Fig. 1 ▶(d). The high density and accidental near-coincidence of the NCS operators explains why the original rotation functions failed to show sets of icosahedrally related peaks. However, there remained strong twofold peaks such as at ψ = 45, ϕ = 0° in the rotation function that are not part of the icosahedral symmetries. These peaks would be consistent with a tetragonal 422 point group and indeed there is a moderately high fourfold peak in the rotation function coincident with the c axis.
Figure 1.
Rotation function of EV71 crystal diffraction data. Stereographic plots of (a) fivefold, (b) threefold and (c) twofold rotation functions were calculated using 18–13 Å resolution data and a 150 Å radius of integration. The plots are contoured starting from 0.5σ in 0.5σ increments. There are no obvious sets of icosahedrally related peaks. (d) Stereographic diagram showing the positions of icosahedral symmetry elements for all particles in the EV71 crystals. Fivefold axes are shown as red pentagons, threefold axes as green triangles and twofold axes as blue ovals.
Table 2. List of peaks from the EV71 locked rotation function that included only fivefold and threefold symmetry elements.
| Search angle (polar XYK) | |||||
|---|---|---|---|---|---|
| Peak No. | () | () | () | Height/ | Particle orientation† |
| 1 | 39 | 60 | 35 | 8.6 | 1 |
| 228‡ | 090 | 0 | 91 | 7.7 | St 90 |
| 29 | 0 | 91 | 91 | 6.6 | St 90 |
| 30 | 46 | 56 | 46 | 6.6 | St 90 |
| 31 | 42 | 67 | 46 | 6.1 | 1 |
| 32 | 56 | 53 | 53 | 5.8 | 1 |
| 33 | 42 | 42 | 49 | 5.6 | 1 |
| 34 | 25 | 88 | 77 | 5.6 | 2 |
| 35 | 14 | 4 | 84 | 5.5 | 1 |
| 36 | 7 | 88 | 88 | 4.8 | 1 |
| 37 | 32 | 77 | 11 | 4.3 | 2 |
| 38 | 91 | 91 | 91 | 4.1 | St 90 |
| 39 | 14 | 32 | 67 | 3.6 | 2 |
| 40 | 67 | 91 | 60 | 3.0 | Wrong solution |
| 41 | 88 | 35 | 77 | 3.0 | Wrong solution |
| 42 | 53 | 46 | 39 | 3.0 | Wrong solution |
| 43 | 91 | 88 | 46 | 2.7 | Wrong solution |
| 44 | 81 | 84 | 60 | 2.7 | Wrong solution |
| 45 | 18 | 67 | 91 | 2.6 | Wrong solution |
| 46 | 84 | 28 | 91 | 2.5 | Wrong solution |
Particle orientation corresponds to particle orientation 1 or 2 as described in the text or to the standard orientation rotated by 90 about the x axis (St 90). Note that peaks with a height of 3 or lower correspond to incorrect solutions.
Peaks 228 correspond to the same effective rotation since is zero.
The positions of particle centers were identified with the fast translation function in the program Phaser (McCoy et al., 2007 ▶) using correctly oriented particles of echovirus 7 as search models (Plevka et al., 2010 ▶). The center of particle 1 was located at fractional coordinates x = 0.2501, y = 0.2527, z = 0.2479 and that of particle 2 at x = 0.5020, y = 0.4969, z = 0.5001. This arrangement of particles in the crystal corresponds roughly to I4122 symmetry (Figs. 1 ▶ d and 2 ▶). Therefore, EV71 may have crystallized in space group I4122, in which there is only one independent particle, and then lost the fourfold symmetry owing to strain applied to the crystal during vitrification or other crystal handling.
Figure 2.
Arrangement of the two crystallographically independent EV71 virions (red and blue) within the I212121 crystal viewed along the c axis. The two particles are related by roughly a fourfold rotation, producing an approximate I4122 symmetry. Corresponding edges of each of the two icosahedrons are marked by arrows to highlight the approximate 90° rotation relating the two particles. The boundaries of the unit cell and the positions of symmetry elements in space group I212121 are shown in black. The putative fourfold screw axis which replaces one of the twofold screw axes is shown in green.
Correctly placed echovirus 7 particles (PDB entry 2x5i; Plevka et al., 2010 ▶) were used to calculate initial phases for reflections with resolution lower than 10 Å using the program CNS (Brünger et al., 1998 ▶). The phases were refined by 15 cycles of real-space averaging with the program AVE using 120-fold NCS (Kleywegt & Read, 1997 ▶). The mask defining the volume of electron density to be averaged was derived from the echovirus 7 atomic model by including all grid points within 5 Å of each atom. Grid points outside the capsid were set to the average value of the density outside the capsid. Phase information for reflections immediately outside the current resolution limit was obtained by extending the resolution by (1/c) Å−1 followed by three cycles of averaging. This procedure was repeated until 3.8 Å resolution had been reached. Particle orientations, positions and relative unit-cell lengths were refined by changing these in small steps and searching for the highest correlation between observed and calculated structure-factor amplitudes. Fourier inversion of the averaged electron-density map determined the calculated amplitudes. The refined particle orientations and positions were found to be ψ = 42.8, ϕ = 68.8, κ = 45.9°, x = 0.2501, y = 0.2530, z = 0.2479 for particle 1 and ψ = 24.8, ϕ = 87.4, κ = 77.4°, x = 0.5020, y = 0.4967, z = 0.5001 for particle 2.
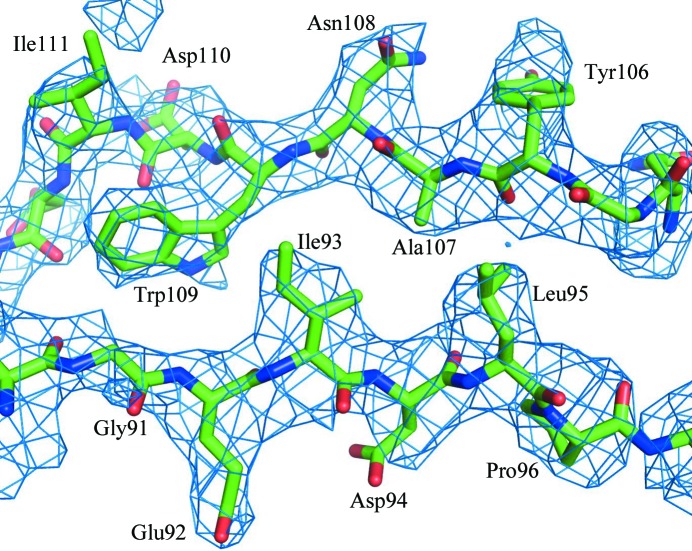
To enhance the high-resolution information in the absence of better than 3.8 Å resolution data, the structure amplitudes for map calculation were modified by application of a −125 Å2 B factor (Fig. 3 ▶). The final map was calculated with phases from the phase extension. The structure was built using the programs O (Jones et al., 1991 ▶) and Coot (Emsley & Cowtan, 2004 ▶) starting with a homology model of EV71 based on the echovirus 7 structure generated with the program Phyre2 (Kelley & Sternberg, 2009 ▶). The structure was built manually and subjected to coordinate and B-factor refinement using the program CNS. The refinement target was the crystallographic residual. Atoms from each residue were constrained to have the same B value because of the limited resolution of the diffraction data. The structure refinement utilized NCS constraints. Other calculations used the CCP4 suite of programs (Winn et al., 2011 ▶). Water molecules were not included in the model because of the limited resolution of the diffraction data. Had it been calculated, the R free would have been very similar to R work because of the 120-fold NCS (Arnold & Rossmann, 1990 ▶; Kleywegt & Brünger, 1996 ▶). Therefore, all measured reflections were used in the structure refinement. Basic structure-quality indicators are listed in Table 1 ▶.
Figure 3.
Electron-density map of EV71 showing strands B and C on opposite sides of the jelly-roll β-barrel of VP1.
Although the completeness of the data is only 11.5% in the highest resolution shell because of the exceptionally high NCS, the effective completeness is 1380%. Hence, the quality of the map (Fig. 3 ▶) was sufficient to build the capsid structure except for residue 1 of VP1, residues 1–9 and 135–143 of VP2, and residues 1–12 of VP4. The structure of the icosahedral asymmetric unit consists of 831 amino-acid residues and five putative divalent cations that were modeled as calcium ions. There is an elongated and fragmented density within the VP1 pocket. This density was modeled as lauric acid, although in reality the density probably represents an average of different fatty acids.
3.2. EV71 capsid structure
The root-mean-square deviations between equivalent Cα atoms of EV71 and other enteroviruses (Table 3 ▶) show that EV71 is most similar to bovine enterovirus (Smyth et al., 1995 ▶), to which it also has the highest sequence identity. Prominent structural differences among these picornaviruses are found in the loop regions exposed on the capsid surface, which are also the most important neutralizing immunogenic sites (Filman et al., 1989 ▶; Rossmann et al., 1985 ▶; Sherry et al., 1986 ▶; Icenogle et al., 1986 ▶). The most variable regions of VP1 are the loops located around the fivefold axes. Because of the smaller size of the EV71 VP1 loops, the canyon of EV71 is shallower than in most other picornaviruses. The prominent and most structurally variable surface features of VP2 and VP3 are called ‘puff’ and ‘knob’, respectively. These regions are involved in binding to non-immunoglobulin-like receptors in some picornaviruses (Hafenstein et al., 2007 ▶) and may have the same function in EV71 (Miyamura et al., 2011 ▶).
Table 3. Sequence similarity and structural similarity in the icosahedral asymmetric units of picornaviruses.
Lower left portion of table: root-mean-square deviations (r.m.s.d.s; ) of superimposed C atoms of the respective three-dimensional structures. The second number (following the slash) indicates the percentage of equivalent amino acids used for calculating the r.m.s.d. with respect to the number of residues in the smaller of the two structures being compared. The icosahedral asymmetric unit consisting of subunits VP1, VP2, VP3 and VP4 was used as a rigid body in all cases. The program O (Jones et al., 1990 ▶) was used for superposition of the molecules. The cutoff for inclusion of residues in the r.m.s.d. calculation was 3.8. Upper right portion of table: percentage of equivalent residues that are identical.
| EV71 | Bovine enterovirus (BEV) | Coxsackievirus B3 (CVB3) | HRV14 | HRV1a | Echovirus 7 | Poliovirus 1 | |
|---|---|---|---|---|---|---|---|
| EV71 | 49 | 43 | 42 | 40 | 41 | 41 | |
| BEV | 1.0/94 | 45 | 44 | 43 | 45 | 43 | |
| CVB3 | 1.1/90 | 1.0/94 | 48 | 46 | 72 | 54 | |
| HRV14 | 1.2/92 | 1.1/93 | 1.0/94 | 49 | 46 | 48 | |
| HRV1a | 1.1/89 | 1.1/91 | 0.9/92 | 1.0/92 | 45 | 44 | |
| Echovirus 7 | 1.1/89 | 1.1/94 | 0.7/98 | 1.1/95 | 1.0/93 | 53 | |
| Poliovirus 1 | 1.1/88 | 1.1/93 | 1.0/95 | 1.0/95 | 1.1/93 | 1.0/95 |
The largest differences in the capsid structure between EV71 and other enteroviruses are located in VP4 and the N-terminal arm of VP1, which are attached to the inner face of the capsid. Each main chain of VP4 forms an elliptical loop with a major axis of about 20 Å and a minor axis of about 10 Å positioned next to the fivefold axes (Fig. 4 ▶ a). In contrast, in most other enteroviruses the N-termini of VP4 form a five-pointed star-like β-structure around the fivefold axes (Fig. 4 ▶ b). The structure of the N-terminus of EV71 VP1 was ordered from residue 2 onwards. It has previously been shown that after picornaviruses have bound to a receptor, VP4 and the N-terminus of VP1 become externalized from the capsid, possibly forming a transmembrane channel for release of the genome into the cytoplasm of the host cell (Lin et al., 2011 ▶). It was suggested that the N-terminus of VP1 forms a transmembrane helix as a part of the pore for genome release. It may therefore be relevant that in EV71 there is a short α-helix formed by residues 5–9 of VP1. Similarly, residues 5–11 in the N-terminus of human rhinovirus serotype 1A (HRV1A) VP1 form a short α-helix although they differ in sequence and location in the capsid compared with EV71.
Figure 4.
Comparison of the N-terminal arms of VP4 in (a) EV71 and (b) poliovirus 1. In each case one of the polypeptide chains is rainbow-coloured from blue at the N-terminus to yellow.
Supplementary Material
PDB reference: enterovirus 71, 4aed
Acknowledgments
We thank Vukica Srajer, Robert Henning and other staff of the Advanced Photon Source (Argonne National Laboratory) BioCARS beamline 14 and Robert Fischetti of GM/CA beamline 23 for help with data collection. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources under grant No. RR007707. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-06CH11357. We thank Paul Chipman for his negative-stain assistance on the electron microscope. We thank Sheryl Kelly for help with the preparation of the manuscript. This study was supported by grant AI11219 from the National Institutes of Health awarded to MGR.
References
- Alexander, J. P., Baden, L., Pallansch, M. A. & Anderson, L. J. (1994). J. Infect. Dis. 169, 905–908. [DOI] [PubMed]
- Arnold, E., Erickson, J. W., Fout, G. S., Frankenberger, E. A., Hecht, H.-J., Luo, M., Rossman, M. G. & Rueckert, R. R. (1984). J. Mol. Biol. 177, 417–430. [DOI] [PubMed]
- Arnold, E. & Rossmann, M. G. (1990). J. Mol. Biol. 211, 763–801. [DOI] [PubMed]
- Belnap, D. M., McDermott, B. M., Filman, D. J., Cheng, N., Trus, B. L., Zuccola, H. J., Racaniello, V. R., Hogle, J. M. & Steven, A. C. (2000). Proc. Natl Acad. Sci. USA, 97, 73–78. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Chan, L. G., Parashar, U. D., Lye, M. S., Ong, F. G., Zaki, S. R., Alexander, J. P., Ho, K. K., Han, L. L., Pallansch, M. A., Suleiman, A. B., Jegathesan, M. & Anderson, L. J. (2000). Clin. Infect. Dis. 31, 678–683. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Colonno, R. J., Condra, J. H., Mizutani, S., Callahan, P. L., Davies, M. E. & Murcko, M. A. (1988). Proc. Natl Acad. Sci. USA, 85, 5449–5453. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Filman, D. J., Syed, R., Chow, M., Macadam, A. J., Minor, P. D. & Hogle, J. M. (1989). EMBO J. 8, 1567–1579. [DOI] [PMC free article] [PubMed]
- Hafenstein, S., Bowman, V. D., Chipman, P. R., Bator Kelly, C. M., Lin, F., Medof, M. E. & Rossmann, M. G. (2007). J. Virol. 81, 12927–12935. [DOI] [PMC free article] [PubMed]
- Icenogle, J. P., Minor, P. D., Ferguson, M. & Hogle, J. M. (1986). J. Virol. 60, 297–301. [DOI] [PMC free article] [PubMed]
- Jones, T. A., Bergdoll, M. & Kjeldgaard, M. (1990). Crystallographic and Modeling Methods in Molecular Design, edited by C. Bugg & S. Ealick, pp. 189–195. New York: Springer-Verlag.
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kelley, L. A. & Sternberg, M. J. (2009). Nature Protoc. 4, 363–371. [DOI] [PubMed]
- Kleywegt, G. J. & Brünger, A. T. (1996). Structure, 4, 897–904. [DOI] [PubMed]
- Kleywegt, G. J. & Read, R. J. (1997). Structure, 5, 1557–1569. [DOI] [PubMed]
- Lin, J., Cheng, N., Chow, M., Filman, D. J., Steven, A. C., Hogle, J. M. & Belnap, D. M. (2011). J. Virol. 85, 9974–9983. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McMinn, P., Lindsay, K., Perera, D., Chan, H. M., Chan, K. P. & Cardosa, M. J. (2001). J. Virol. 75, 7732–7738. [DOI] [PMC free article] [PubMed]
- Miyamura, K., Nishimura, Y., Abo, M., Wakita, T. & Shimizu, H. (2011). J. Gen. Virol. 92, 287–291. [DOI] [PMC free article] [PubMed]
- Muckelbauer, J. K., Kremer, M., Minor, I., Diana, G., Dutko, F. J., Groarke, J., Pevear, D. C. & Rossmann, M. G. (1995). Structure, 3, 653–667. [DOI] [PubMed]
- Olson, N. H., Kolatkar, P. R., Oliveira, M. A., Cheng, R. H., Greve, J. M., McClelland, A., Baker, T. S. & Rossmann, M. G. (1993). Proc. Natl Acad. Sci. USA, 90, 507–511. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Plevka, P., Hafenstein, S., Harris, K. G., Cifuente, J. O., Zhang, Y., Bowman, V. D., Chipman, P. R., Bator, C. M., Lin, F., Medof, M. E. & Rossmann, M. G. (2010). J. Virol. 84, 12665–12674. [DOI] [PMC free article] [PubMed]
- Plevka, P., Perera, R., Cardosa, J., Kuhn, R. J. & Rossmann, M. G. (2012). Science, 336, 1274. [DOI] [PMC free article] [PubMed]
- Rossmann, M. G. (1989). J. Biol. Chem. 264, 14587–14590. [PubMed]
- Rossmann, M. G. (1994). Protein Sci. 3, 1712–1725. [DOI] [PMC free article] [PubMed]
- Rossmann, M. G., Arnold, E., Erickson, J. W., Frankenberger, E. A., Griffith, J. P., Hecht, H.-J., Johnson, J. E., Kamer, G., Luo, M., Mosser, A. G., Rueckert, R. R., Sherry, B. & Vriend, G. (1985). Nature (London), 317, 145–153. [DOI] [PubMed]
- Rossmann, M. G., Ford, G. C., Watson, H. C. & Banaszak, L. J. (1972). J. Mol. Biol. 64, 237–245. [DOI] [PubMed]
- Sherry, B., Mosser, A. G., Colonno, R. J. & Rueckert, R. R. (1986). J. Virol. 57, 246–257. [DOI] [PMC free article] [PubMed]
- Smyth, M., Pettitt, T., Symonds, A. & Martin, J. (2003). Arch. Virol. 148, 1225–1233. [DOI] [PubMed]
- Smyth, M., Tate, J., Hoey, E., Lyons, C., Martin, S. & Stuart, D. (1995). Nature Struct. Biol. 2, 224–231. [DOI] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol. 276, 594–611. [PubMed]
- Wang, X. et al. (2012). Nature Struct. Mol. Biol. 19, 424–429. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: enterovirus 71, 4aed