Abstract
The assimilable organic carbon (AOC) test is a standardized measure of the bacterial growth potential of treated water. We describe the design and initial development of an AOC assay that uses bioluminescent derivatives of AOC test bacteria. Our assay is based on the observation that bioluminescence peaks at full cell yield just prior to the onset of the stationary phase during growth in a water sample. Pseudomonas fluorescens P-17 and Spirillum sp. strain NOX bacteria were mutagenized with luxCDABE operon fusion and inducible transposons and were selected on minimal medium. Independent mutants were screened for high luminescence activity and predicted AOC assay sensitivity. All mutants tested were able to grow in tap water under AOC assay conditions. Strains P-17 I5 (with p-aminosalicylate inducer) and NOX I3 were chosen for use in the bioluminescence AOC test. Peak bioluminescence and plate count AOC were linearly related for both test bacteria, though data suggest that the P-17 bioluminescence assay requires more consistent luminescence monitoring. Bioluminescence results were obtained 2 or 3 days postinoculation, compared with 5 days for the ATP luminescence AOC assay and 8 days for the plate count assay. Plate count AOC assay results for nonmutant and bioluminescent bacteria from 36 water samples showed insignificant differences, indicating that the luminescent bacteria retained a full range of AOC measurement capability. This bioluminescence method is amenable to automation with a microplate format with programmable reagent injection.
Levels of the most general class of dissolved organic compounds, called natural organic matter, are usually highest in surface water sources and very low in groundwater. The fraction of natural organic matter known as biodegradable organic matter (BOM) can be used by indigenous microorganisms for growth. The carbon component of BOM, biodegradable organic carbon (BDOC), is commonly measured as a reduction in total organic carbon in the presence of a bacterial biofilm immobilized on sand (5a). One problem with measurement of BDOC is that biodegradation is specific for local flora (14). Assays of assimilable organic carbon (AOC) offer a standardized alternative for measurements of bacterial growth potential of treated water.
As a component of BOM, AOC is best understood as the fraction most readily used by bacteria for multiplication and therefore of greatest interest to water utilities. The AOC fraction is generally less than 1,000 in molecular weight (5) and can include such compounds as sugars, fatty acids, amino acids, and peptides. BDOC and AOC are both increased by conventional water treatment processes and ozonation (3, 16). High organic carbon levels, in the context of low disinfectant residual and warm ambient temperatures, are related to water distribution problems such as iron pipe corrosion, biofilm growth on pipes, and bacterial regrowth in distributed water (6, 8, 14, 15). While BDOC monitoring is not standardized and requires expensive analytical instrumentation, routine AOC testing has the potential to be a valuable treatment plant diagnostic tool for assessing finished water quality.
The AOC test was originally developed by van der Kooij (12, 13). Two bacterial strains, Pseudomonas fluorescens P-17 and Spirillum sp. strain NOX, are separately inoculated into a water sample. Bacteria utilize the AOC in the water sample to grow to full yield. In this way AOC is converted to biomass, and biomass at full yield is proportional to AOC. Cell yield at the stationary growth phase, measured from 3 to 5 days postinoculation, is related to an equivalent yield of the same bacteria grown in a known concentration of acetate carbon. AOC values are therefore reported as the sum of separate P-17 and NOX determinations with acetate carbon equivalents measured in micrograms per liter. It should be remembered that, when nutrients other than carbon sources limit the growth of heterotrophic organisms, AOC results measure the limiting nutrient.
AOC assay methods are dependent upon reliable quantitation of bacterial cell numbers once the test organisms have grown to full yield. The van der Kooij plate count assay relies upon enumeration of CFU for yield determination; assay results cannot be fully analyzed until 8 days postinoculation. A more rapid AOC assay, which measures cell yield as the ATP content of filter-concentrated cells, was introduced in 1993 (7). This method reduces assay time by 3 days because the need for bacterial colony growth from a single cell is eliminated. A bioluminescence AOC assay is currently being marketed by CheckLight Ltd. (Qiryat-Tiv’on, Israel; http://www.checklight.co.il). This assay employs lyophilized preparations of nonstandard AOC bacteria and reportedly produces results within 2 h. The relationship of this test to P-17 and NOX AOC is not known. In this report we describe the initial characterization of a redesigned, simpler, and less expensive AOC assay based on bioluminescence of genetically modified AOC test bacteria.
MATERIALS AND METHODS
Bacteria, media, culture conditions, and diluent.
P. fluorescens strain P-17 and Spirillum sp. strain NOX were obtained from D. van der Kooij and were stored at −70°C in 20% glycerol-2% peptone as described earlier (7). Bacteria were recovered by streaking onto R2A medium (Difco Laboratories, Detroit, Mich.) and by incubating at room temperature, 20 to 25°C, for 3 to 5 days. Single colonies were inoculated into medium containing 2 mg of sodium acetate per liter (7) and were allowed to grow for 3 to 7 days at room temperature to 30°C. The cultures were either used immediately or stored at 4°C up to 40 days for inoculation of water samples. Luminescent derivatives of P-17 and NOX were screened for high luminescence in tryptic soy broth (TSB; Difco) or nutrient broth (Difco). Agar was added to 1.5% (wt/vol) for growth on solid medium. Escherichia coli strains were routinely cultured in TSB with or without agar at room temperature to 37°C.
Culture diluent was a magnesium chloride/potassium phosphate buffer. This was made as a 10× stock from separate stocks of anhydrous potassium phosphate (3.40 g/100 ml) and magnesium chloride hexahydrate (8.11 g/100 ml). Twelve and a half milliliters of potassium phosphate stock and 50.0 ml of magnesium chloride stock were added to 800 ml of water. The pH was adjusted to 7.2, and the volume was made to 1 liter for the 10× stock. 1× buffer was used as the diluent for all experiments.
Construction of bioluminescent AOC assay strains.
E. coli strains Sm10 λ pir (pUTminiTn5Kmlux [4]) and S17-1 λ pir no. 75 (1) were used to create luminescent derivatives of P-17 and NOX bacteria. Both strains delivered the genes required for cellular luminescence as luxCDABE transposons linked to suicide plasmid vectors. Mini-Tn5Kmlux created transcriptional fusion mutations to the luxCDABE operon. The resulting mutants were designated P-17 or NOX GFX, where GF indicates a gene fusion and X is a number. The S17-1 strain transposon contains a luxCDABE operon under nominal negative regulation from a linked repressor gene; induction is provided by sodium salicylate or p-aminosalicylate (PAS; B. M. Applegate, personal communication). Similarly, these mutants were designated P-17 or NOX IX, where I indicates inducible luminescence. Evidence presented in the next section suggests that negative luminescence regulation is not fully functional for the inducible genetic constructs in AOC test strain genetic backgrounds. Both transposons carry the selectable marker kanamycin resistance (Kmr), and both vectors encode transposase and ampicillin resistance (Apr) as β-lactamase.
Mutagenesis of P-17 was performed as follows. Sm10 λ pir (pUTminiTn5Kmlux), S17-1 λ pir no. 75, and P-17 were grown to full yield in TSB. Aliquots of the two E. coli strains were separately added to equal volumes of P-17. A portion (0.1 ml) of each mixture was spread onto tryptic soy agar plates for conjugal transfer of the luciferase genes during growth at 30°C for 24 h. Lawn growth from these plates was spread as a barely visible film onto M9 minimal salt-noble agar medium with 0.5% sodium acetate and 25 μg of kanamycin sulfate (Sigma M6030, A5431, and K4000)/ml. These plates' contents were incubated 48 h at 30°C. Growth was streaked for purity on the same selection medium, and one isolate from each original mating mix was saved for further analysis. Mating controls, consisting of conjugal donor and recipient plated separately on selective medium, showed no growth. NOX mutagenesis was performed similarly, except that nutrient broth was used for growing parental strains and that nutrient agar was used for the growth and mating steps. NOX mutants were selected on M9 minimal salt-noble agar with 2% sodium oxalate and kanamycin.
Characterization of Lux+ P-17 and NOX mutants.
Luminescent derivatives of AOC test bacteria were screened for minimal disruption of cellular physiology by assays of the logarithmic-phase growth rate in rich broth at 30°C with aeration. The logarithm of optical density at 620 nm (OD620) as measured with a Hach DR/4000U spectrophotometer was plotted versus time, and the linear regression slope was divided into the logarithm of 2 for doubling time. Six to eight point growth curves were used to calculate doubling times. Logarithmic-phase luminescence per cell was described by luminescence activity, which is 1,000 multiplied by the slope of the regression line from plots of log (luminescence) versus log (OD620).
Estimated AOC assay sensitivity for each isolate was calculated as follows. Luminescence readings were normalized to 90 s and were divided by OD620. This value was multiplied by the appropriate van der Kooij cell yield/AOC relationship (see below) and the following empirically determined relationships between OD620 and cell density: 1 OD620 = 5.61 × 1010 P-17 CFU/ml and 1 OD620 = 4.89 × 1010 NOX CFU/ml. The reciprocal of the resulting value had units of AOC in microgram-per-liter acetate equivalents per single relative light unit above background. Lower values indicated greater assay sensitivity.
All isolates were tested for growth under AOC assay conditions in dechlorinated tap water with or without acetate, and all were able to multiply under these conditions. Finally, the isolates were tested for transposon vector absence by assay of vector-encoded β-lactamase activity with the chromogenic substrate nitrocefin (Calbiochem, San Diego, Calif. [9]). Enzyme activity was not detected in any of the isolates.
AOC assays.
Finished water samples from Ohio, Georgia, California, Missouri, North Carolina, Virginia, and Arizona were tested by both the plate count and bioluminescence AOC assays. Nine samples of Milli-Q laboratory water spiked with sodium acetate were also tested. Samples were collected in AOC-free glass bottles that had been muffled at 625°C for 6 h. The samples were dechlorinated with sodium thiosulfate when necessary and were pasteurized for 30 min at 65°C as described earlier (7). Each pasteurized sample was dispensed into nine precleaned 20-ml subsample vials (2000 class no. 276720; Scientific Specialties, Inc.). The manufacturer's procedure for cleaning the vial components was washing in a nonphosphate detergent, rinsing three times with tap water, rinsing three times with ASTM International type 1 deionized water, and oven drying. The vials were assembled in a carbon-free environment.
Method blank subsamples received no treatment beyond pasteurization. Matrix spikes were inoculated with 50 μl of refrigerated stock P-17 or NOX to about 104 viable cells per ml. Matrix spikes also received sodium acetate to 100 μg/liter. P-17, P-17 I5 + PAS, NOX, and NOX I3 vials were also prepared by inoculation with the appropriate bacteria and PAS when needed to 0.05% (wt/vol) for the AOC assay. PAS (Sigma A-3380) was made as a 20% stock, filter sterilized, and refrigerated. One-tenth-milliliter aliquots of all vials were removed for dilution, duplicate spread plating on R2A, and enumeration of viable counts at 3, 4, and 5 days postinoculation (7). Plate count AOC values were expressed as the mean of at least two of these values converted to acetate carbon equivalents with the van der Kooij conversion factors: 4.1 × 103 P-17 CFU/ml equals 1 μg of P-17 AOC/liter and 1.2 × 104 NOX CFU/ml equals 1 μg of NOX AOC/liter (7, 12, 13).
Separate subsample vials of method blanks, P-17 I5 + PAS, and NOX I3 were prepared for the bioluminescence AOC assay. Vials were inoculated with refrigerated stocks of P-17 I5, NOX I3 and PAS as appropriate. An initial luminescence test was performed just after inoculation; these uniformly produced low luminescence with values similar to sample method blank values. Subsequent luminescence measurements were made twice daily for approximately 3 days. Each luminescence assay used 1 ml of culture assayed in an UltraSource Inspector tube luminometer for 90 s with 0.1 ml of injected 0.2% (vol/vol) n-decanal in denatured ethanol. This instrument measured light converted to an electric current and expressed cumulative read time luminescence as relative light units. Relative light units are defined as the integral of the current-versus-time curve and are considered “relative” because measurement time may be modified.
Statistical analyses.
Least-squares linear regression, single-factor analysis of variance, and two-tailed, two-sample t tests (assuming equal variances) were performed with Excel 2000 (Microsoft, Inc.) and Axum 6.0 (MathSoft, Inc.) software packages. The significance level for all statistical tests was α of 0.05. Coefficients of variation for plate count AOC assays were defined as (triplicate standard deviation ÷ triplicate mean) × 100%.
RESULTS AND DISCUSSION
Bioluminescent mutant characterization.
The AOC test bacteria P. fluorescens P-17 and Spirillum sp. strain NOX were subjected to transposon mutagenesis for stable insertion of luxCDABE luminescence operons into their genomes. Forty independent isolates of mutated P-17 and NOX, 10 gene fusions and 10 inducible mutants of each strain, were screened for desirable AOC test properties. Criteria used were physiological similarity to parental strains as measured by doubling time, high luminescence activity, and high estimated AOC assay sensitivity. Characteristics of the four optimal isolates are presented in Table 1.
TABLE 1.
Characterization of AOC test organism luminescent mutantsa
| Strain | Mean doubling time (min) | Mean luminescence activity | Estimated AOC assay sensitivity (μg of acetate C equivalents · liter−1 per 90-s RLU) |
|---|---|---|---|
| P-17 | 78.8 ± 10 | N/A | N/A |
| P-17 GF8 | 88.6 ± 3 | 1,108 ± 82 | 0.67 |
| P-17 I5 | 88.6 ± 3 | 1,911 ± 151 | ND |
| NOX | 124 ± 3 | N/A | N/A |
| NOX GF3 | 109 ± 21 | 692 ± 127 | 0.012 |
| NOX I3 | 125 ± 6 | 1,092 ± 338 | ND |
N/A, not applicable; ND, not determined; RLU, relative light unit. Values shown are mean ± standard deviations from three or four replicates. All values were determined in rich media without luminescence inducers.
Table 1 shows that doubling time measurements were variable but not significantly different between parents and luminescence mutant strains. The inducible mutants produced more logarithmic-phase luminescence than their gene fusion counterparts. These strains are therefore better candidates for an AOC bioluminescence assay employing low-cost, photoelectric instrument-based luminometry. Doubling times and luminescence activity are not directly comparable between Pseudomonas and Spirillum sp. strains due to the use of different growth media and luminescence reaction times. However, comparison of estimated assay sensitivity for the gene fusion strains showed that NOX GF3 had an assay sensitivity predicted to be >10-fold greater than that of P-17 GF8. This trend was consistent among all the gene fusion mutants (data not shown).
Bioluminescence induction and the bioluminescence reaction.
Sodium salicylate at 0.5 mg/liter induced extremely high but rapidly decaying luminescence in the P-17 inducible mutants (data not shown). However, it also halted growth and proved toxic for both AOC assay strains. Addition of salicylate to AOC test cultures that had already reached the stationary phase did not induce luminescence. Therefore, use of the inducer sodium salicylate was discontinued. An alternate inducer, PAS, was found to promote high P-17 I5 luminescence; inducer was not necessary for NOX I3 luminescence. This fact and expression of adequate luminescence at high cell density without inducer (Table 1) suggest that repression encoded by the no. 75 inducible construct functioned only partially or not at all in P-17 and NOX genetic backgrounds.
The low cell densities encountered during AOC assays required the addition of luciferase substrate n-decylaldehyde directly to cell suspensions to achieve adequate luminescence during the test. Decanal luminescence reactions from these organisms followed a biphasic first-order decay curve, with approximately 52% of 90-s luminescence expressed during the first, “flash” phase (mean half-life, 2.6 s) and 48% expressed during the second, “glow” phase (mean half-life, 18.5 s).
Bioluminescence expression during AOC assay.
Expression of bioluminescence is known to be closely related to cellular physiology (2, 4, 10, 11). We therefore reasoned that peak luminescence, just prior to the onset of the stationary phase after exhaustion of AOC in a water sample, would be an early physiological indicator of full cell yield. Peak luminescence should also directly relate to AOC as measured by test organism stationary phase plate counts, since the latter is a viability-based test for full cell yield.
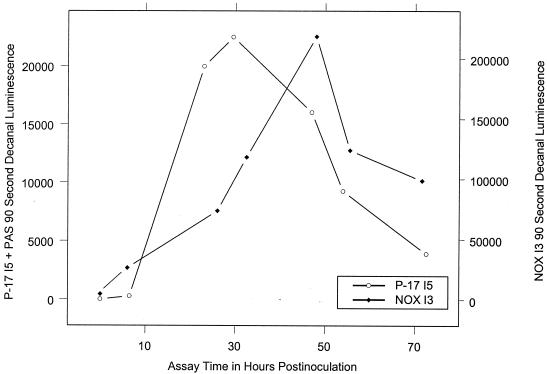
Figure 1 shows typical luminescence time courses for P-17 I5 + PAS and NOX I3 grown in dechlorinated tap water during AOC assays. Luminescence in both cases peaked less than 50 h postinoculation; this represented a significant time reduction from the approximately 72 to 120 h needed for a complete assay by other established methods.
FIG. 1.
AOC bioluminescence assay luminescence time course.
Relationship of peak luminescence to AOC.
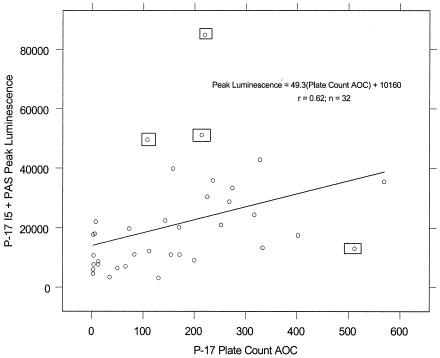
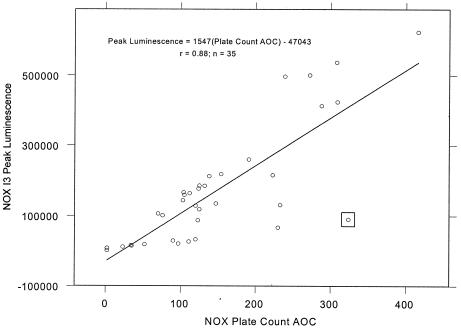
Linear plots of peak bioluminescence-versus-plate count AOC, shown in Fig. 2 and 3, portray a variable linear fit for P-17 I5 + PAS and a more consistent linear fit for NOX I3. P-17 strain variability could be a consequence of the 13% variation observed for P-17 doubling time (Table 1). Measurement of P-17 I5 peak bioluminescence will likely become more accurate with the use of a photon-counting luminometer and a microplate assay format with automated reagent injection. Significance testing of the Fig. 3 y intercept against a null hypothesis of zero suggested a marginally insignificant deviation from the expected value (P = 0.07). This suggests that extremely low values of NOX AOC (<30 μg of acetate carbon equivalents per liter) may be undetectable in this assay. When the plot slopes were compared and normalized to cell number, NOX I3 bioluminescence per cell was 10.7-fold higher than that of P-17 I5 + PAS.
FIG. 2.
Relationship of P-17 I5 peak bioluminescence to P-17 plate count AOC. The four boxed points were excluded from regression calculations.
FIG. 3.
Relationship of NOX I3 peak bioluminescence to NOX plate count AOC. The boxed point was excluded from regression calculations.
Mean luminescence peak times with standard deviations in hours postinoculation were 34 ± 12 for P-17 I5 + PAS and 38 ± 17 for NOX I3. Peak luminescence followed first-order decay kinetics for both assay strains and produced nearly identical mean half-lives of 16 ± 7 h for the Pseudomonas test and 15 ± 10 h for the Spirillum test. These results allow considerable latitude for the design of a luminescence monitoring regimen during the bioluminescence AOC assay. If we assume an average half-life of 15.5 h for peak luminescence decay, an automated assay at 5-h intervals postinoculation is expected to produce variation approximating the 20% observed with P-17 plate count assays (n = 134) and 23% for NOX assays (n = 133).
Bioluminescence method validation.
In order to prove the utility of the bioluminescence method as an AOC assay technique, it is necessary to show that the modified test strains can function as well as the original strains in plate count assays. This was accomplished through a concurrent plate count assay of the bioluminescence tests shown in Fig. 2 and 3. Standard t tests of parent and bioluminescent mutant plate count AOC values revealed no significant differences for either Pseudomonas or Spirillum bacteria (P = 0.69 or 0.61, respectively). Taken together, these results show that an AOC assay based on luminescence of test organisms is time saving and, with more consistent monitoring, would likely be at least as accurate as the original van der Kooij plate count assay.
Conclusions.
The results presented here describe the initial development of an AOC assay based on bioluminescence of AOC test organisms. They also provide detailed functional characterization of two suitable bioluminescent strains with a simple luminometer and an inexpensive aldehyde substrate. Future work will evaluate this bioluminescence test with more sophisticated instrumentation in efforts to increase accuracy, sensitivity, and ease of use through automated measurement. Further method refinement will allow AOC assay incorporation into the water treatment laboratory as a general measure of finished water quality.
Acknowledgments
We acknowledge with gratitude the gifts of E. coli strains Sm10 λ pir (pUTminiTn5Kmlux) and S17-1 λ pir no. 75 from John Coote and Bruce Applegate, respectively.
This work was funded by the utility subsidiaries of American Water, Voorhees, N.J.
REFERENCES
- 1.Applegate, B. M., S. R. Kehrmeyer, and G. S. Sayler. 1998. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethylbenzene, and xylene (BTEX) sensing. Appl. Environ. Microbiol. 64:2730-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorn, J. G., R. J. Frye, and R. M. Maier. 2003. Effect of temperature, pH, and initial cell number on luxCDABE and nah gene expression during napthalene and salicylate catabolism in the bioreporter organism Pseudomonas putida RB1353. Appl. Environ. Microbiol. 69:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escobar, E. C., and A. A. Randall. 2001. Case study: ozonation and distribution system biostability. J. Am. Water Works Assoc. 93:77-89. [Google Scholar]
- 4.Forde, C. B., R. Parton, and J. G. Coote. 1998. Bioluminescence as a reporter of intracellular survival of Bordetella bronchiseptica in murine phagocytes. Infect. Immun. 66:3198-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hem, L. J., and H. Efraimsen. 2001. Assimilable organic carbon in molecular weight fractions of organic matter. Water Res. 35:1106-1110. [DOI] [PubMed] [Google Scholar]
- 5a.Joret, C., Y. Levi, and M. Gilbert. 1989. The measurement of biodegradable organic carbon (BDOC): a tool in water treatment. Water Supply 7:41-45. [Google Scholar]
- 6.LeChevallier, M. W., C. D. Lowry, R. G. Lee, and D. L. Gibbon. 1993. Examining the relationship between iron corrosion and the disinfection of biofilm bacteria. J. Am. Water Works Assoc. 85:111-124. [Google Scholar]
- 7.LeChevallier, M. W., N. E. Shaw, L. A. Kaplan, and T. L. Bott. 1993. Development of a rapid assimilable organic carbon method for water. Appl. Environ. Microbiol. 59:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeChevallier, M. W., N. J. Welch, and D. B. Smith. 1996. Full-scale studies of factors related to coliform regrowth in drinking water. Appl. Environ. Microbiol. 62:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shingler. 1972. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie, J. M., G. R. Campbell, J. Shepherd, Y. Beaton, D. Jones, K. Killham, and R. R. E. Artz. 2003. A stable bioluminescent construct of Escherichia coli O157:H7 for hazard assessments of long-term survival in the environment. Appl. Environ. Microbiol. 69:3359-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unge, A., R. Tombolini, L. Mølbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Kooij, D. 1990. Assimilable organic carbon (AOC) in drinking water, p. 57-87. In G. A. McFeters (ed.), Drinking water microbiology. Springer-Verlag, New York, N.Y.
- 13.van der Kooij, D. 1992. Assimilable organic carbon as an indicator of bacterial regrowth. J. Am. Water Works Assoc. 84:57-65. [Google Scholar]
- 14.Volk, C. J., and M. W. LeChevallier. 1999. Impacts of the reduction of nutrient levels on bacterial water quality in distribution systems. Appl. Environ. Microbiol. 65:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volk, C. J., and M. W. LeChevallier. 2000. Assessing biodegradable organic matter. J. Am. Water Works Assoc. 92:64-76. [Google Scholar]
- 16.Volk, C. J., and M. W. LeChevallier. 2002. Effects of conventional treatment on AOC and BDOC levels. J. Am. Water Works Assoc. 94:112-123. [Google Scholar]