Abstract
Background and Aims
There is currently much speculation about the role of epigenetic variation as a determinant of heritable variation in ecologically important plant traits. However, we still know very little about the phenotypic consequences of epigenetic variation, in particular with regard to more complex traits related to biotic interactions.
Methods
Here, a test was carried out to determine whether variation in DNA methylation alone can cause heritable variation in plant growth responses to jasmonic acid and salicylic acid, two key hormones involved in induction of plant defences against herbivores and pathogens. In order to be able to ascribe phenotypic differences to epigenetic variation, the hormone responses were studied of epigenetic recombinant inbred lines (epiRILs) of Arabidopsis thaliana – lines that are highly variable at the level of DNA methylation but nearly identical at the level of DNA sequence.
Key Results
Significant heritable variation was found among epiRILs both in the means of phenotypic traits, including growth rate, and in the degree to which these responded to treatment with jasmonic acid and salicylic acid. Moreover, there was a positive epigenetic correlation between the responses of different epiRILs to the two hormones, suggesting that plant responses to herbivore and pathogen attack may have a similar molecular epigenetic basis.
Conclusions
This study demonstrates that epigenetic variation alone can cause heritable variation in, and thus potentially microevolution of, plant responses to defence hormones. This suggests that part of the variation of plant defences observed in natural populations may be due to underlying epigenetic, rather than entirely genetic, variation.
Keywords: Arabidopsis thaliana, DNA methylation, epigenetic recombinant inbred line, epiRIL, growth rate, induced defence, jasmonic acid, salicylic acid
INTRODUCTION
Plants have evolved an impressive suite of mechanisms that allow them to respond plastically to changing environmental conditions and thus to persist in variable environments. One important class of such response mechanisms are induced defences, i.e. chemical or physical defences that are produced rapidly in response to pathogen or herbivore attack and that therefore, in contrast to constitutive defences, avoid the waste of valuable resources when enemies are absent or rare (e.g. Karban and Baldwin, 1997; Agrawal, 2001; Karban et al., 2002; Kempel et al., 2011). Two of the key elements in the signalling cascades of induced plant defences are the hormones jasmonic acid (JA) and salicylic acid (SA). Jasmonic acid is mainly involved in the production of defences against insect herbivores (Thaler et al., 1996; McConn et al., 1997; Baldwin, 1998; Cipollini and Redman, 1999), whereas SA is involved in plant responses to fungal, bacterial and viral pathogens (Delaney, 1997, and citations therein). Although the induction of defences against herbivores and pathogens generally appears to be controlled by separate molecular pathways (e.g. Thomma et al., 1998), their expression can sometimes be antagonistic (Kunkel and Brooks, 2002). The nature of this relationship determines whether the two kinds of defences can act, and evolve, independently, or whether there are physiological or genetic trade-offs between them.
One key question on variation in phenotypic traits, such as growth or induced defence, concerns the amount of variation that is heritable, because this heritable variation determines the evolutionary potential of traits and thus their ability to adapt to local environmental conditions. We know that there is significant heritable variation in many ecologically important plant traits, including phenotypic plasticity and induced defences, in many plant species (Linhart and Grant, 1996; Agrawal et al., 2002; Pigliucci, 2005), including Arabidopsis thaliana (Koornneef et al., 2004). The usual assumption is that such heritable variation reflects underlying variation in DNA sequence.
Recently, increasing evidence has accumulated showing that heritable phenotypic variation can also be brought about by underlying variation in epigenetic modifications of the genome, such as DNA methylation or histone modifications (Jaenisch and Bird, 2003), which themselves can be heritable and to some degree independent of DNA sequence variation (Vaughn et al., 2007; Bossdorf et al., 2008; Jablonka and Raz, 2009; Johannes et al., 2009). Heritable epigenetic variation has been found to influence flowering time, height and fitness of plants (Johannes et al., 2009; Roux et al., 2011), as well as their phenotypic plasticity in response to different nutrient (Bossdorf et al., 2010) or light conditions (Tatra et al., 2000). Nothing is known, however, about the potential role of epigenetic variation for heritable variation in induced plant defences.
Testing for epigenetic influences on phenotypic variation is not a trivial task, because in most natural systems epigenetic variation is confounded with DNA sequence variation, and it therefore becomes difficult to ascribe phenotypic differences to underlying epigenetic differences (Richards, 2006, 2008; Bossdorf et al., 2008). One elegant solution to this problem, at least for proof-of-principle, is the use of epigenetic recombinant inbred lines (epiRILs; Johannes et al., 2009; Reinders et al., 2009), which have been constructed to be highly variable at the epigenetic level, but nearly identical at the level of DNA sequence. Since the epigenetic variation among epiRILs is heritable (Johannes et al., 2009), and there is virtually no DNA sequence variation, any observed heritable phenotypic variation, i.e. significant line effects, must be of epigenetic origin.
Here, we used epiRILs of A. thaliana to address the following questions. (1) Does epigenetic variation among lines cause heritable variation in mean plant growth, phenology and reproduction? (2) Can we find epigenetic variation in plant responsiveness to JA and SA, and thus, indirectly, in induced defences, and, if yes, (3) what is the relationship between the responses of epiRILs to the two signalling hormones?
MATERIALS AND METHODS
Study system
Arabidopsis thaliana is a small annual weed in the mustard family (Brassicaceae). It is a predominantly selfing ruderal species that usually occurs in open or disturbed habitats. Arabidopsis thaliana is the model species of plant biology (ASPB, 2002) and also a popular study system in ecological and evolutionary genetics and genomics (Mitchell-Olds and Schmitt, 2006). The biochemical and physiological basis of its defence against herbivores and pathogens, as well as the ecological relevance and evolutionary dynamics of these defence mechanisms have been thoroughly studied (e.g. Wittstock and Halkier, 2002; Tian et al., 2003; Weinig et al., 2003; Kliebenstein et al., 2005). Among others, we know that JA and SA play a key role in the species' defence against herbivores and pathogens (e.g. Traw and Bergelson, 2003), and that natural variation exists in the constitutive and induced chemical defences of A. thaliana (e.g. Mauricio and Rausher, 1997; Kliebenstein et al., 2001).
In our study, we worked with epiRILs of A. thaliana, which are characterized by a large amount of heritable among-line variation in DNA methylation, but are nearly identical at the level of DNA sequence (Johannes et al., 2009). A detailed description of the creation of these lines can be found in Johannes et al. (2009). Briefly, the epiRILs are derived from a cross of the hypomethylation mutant Col-ddm1 (Kakutani et al., 1995), which shows a 70 % reduction of overall DNA methylation, with its wild type. Backcrossing the heterozygous F1 with the wild type created, through recombination, a large amount of variation in DNA methylation. About half of this artificially created epigenetic variation has been found to be inherited over eight generations (Johannes et al., 2009). For the epiRIL population, Johannes et al. (2009) selected only plants that were homozygous wild type at the DDM1 locus, and used these to create several hundred inbred lines from single seed. First phenotypic screenings demonstrated that there is also heritable variation in phenotype (growth, fitness and phenology) among the epiRILs (Johannes et al., 2009; Roux et al., 2011). In our experiment, we used 12 epiRILs of the ninth generation. Based on data from a larger screening experiment with 135 lines (Zhang et al., unpubl. res.), these 12 lines were selected to represent the greatest possible variation in plant growth (biomass production).
Experiment
In order to test for heritable variation in plant responses to JA and SA among the epiRILs, we conducted an experiment in which we subjected replicate plants of each of the 12 lines to either single or weekly application of JA or SA.
Seeds of all lines were stratified under cold (4 °C), dark conditions for 2 d, and then transferred to a short-day growth chamber (8/16 h light/dark, 21 °C). Ten days later, we transplanted seedlings individually into 5 cm pots filled with a standard potting soil and randomized these pots in the growth chamber. After another 2 weeks, we transferred the plants to a long-day growth chamber (Percival E-36L, Percival Scientific, Perry, USA) with long-day conditions (16/8 h, 24/20 °C day/night) where the experiment was conducted and the plants were grown until harvest.
There were five experimental treatments: (1) control; (2) a single application of JA, (3) repeated application of JA; (4) a single application of SA; and (5) repeated application of SA, with eight replicates for each epiRIL × treatment combination, thus a total of 480 plants. The hormones were applied by spraying rosettes with a 0·50 mm solution of JA (Sigma-Aldrich GmbH, Switzerland) or a 0·50 mm solution of SA (Sigma-Aldrich GmbH, Switzerland), respectively. We always sprayed plants from the same direction and height until their surfaces were completely wet. Plants that were treated once were sprayed when they were 6 weeks old; plants that were treated repeatedly were sprayed five times, starting at week 4. Within the growth chamber, the positions of the plants were completely randomized, and re-randomized weekly.
To estimate the growth rates of epiRILs, we measured the rosette diameters of all plants four times at weeks 4, 5, 6 and 7, fitted a power function y = abx to these data, separately for each plant individual, and used the parameter b as the measure of growth rate. In addition, we recorded the time to flowering daily of all plants during the experiment. When the plants were 11 weeks old and all had produced fruits, we measured plant height and counted the numbers of siliques on each plant as a measure of reproduction. After that, we cut the above-ground biomass, dried it at 70 °C until 48 h, and weighed it.
Statistical analyses
To test for heritable variation among epiRILs in mean phenotypes, as well as in their responses to the hormone treatments, we analysed all data with linear models that included treatment, line (=epiRIL) as well as the treatment × line interaction as fixed effects. Lines were treated as fixed effects because they were non-randomly selected from the pool of epiRILs. A significant line effect indicates epigenetically based heritable variation in mean phenotype; a significant line × treatment effect indicates epigenetically based heritable variation in plasticity. For all variables, the residuals were normally distributed, hence no data transformations were necessary. All analyses were done in JMP (JMP 9·0; SAS Institute, Cary, NC, USA).
In addition, we wanted to test whether there is a relationship between JA responses and SA responses of epiRILs. For each trait and each of the four hormone treatments, we calculated the responsiveness of each line as the percentage change from the average value in the controls relative to that in the respective treatment, and we then calculated Pearson's correlations for the four possible combinations JA1–SA1, JA1–SA5, JA5–SA1 and JA5–SA5. However, since in each of these cases spurious correlations must be expected from the shared control value in the denominator, we tested the significance of these correlations with permutation tests that removed possible epigenetic correlations between two responses while maintaining the spurious correlation. This was achieved by randomizing treatment means across epiRILs and re-calculating the JA–SA correlation, as described above, from these values. The resulting correlations then only reflect the spurious parts, and a confidence envelope based on 3000 permutations can be used to test whether the observed correlation value significantly differs from this expectation. If yes, it is considered to be evidence of a true epigenetic correlation.
RESULTS
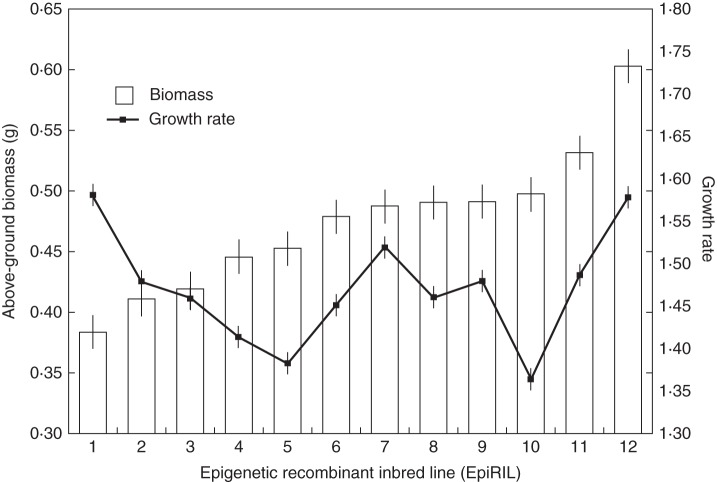
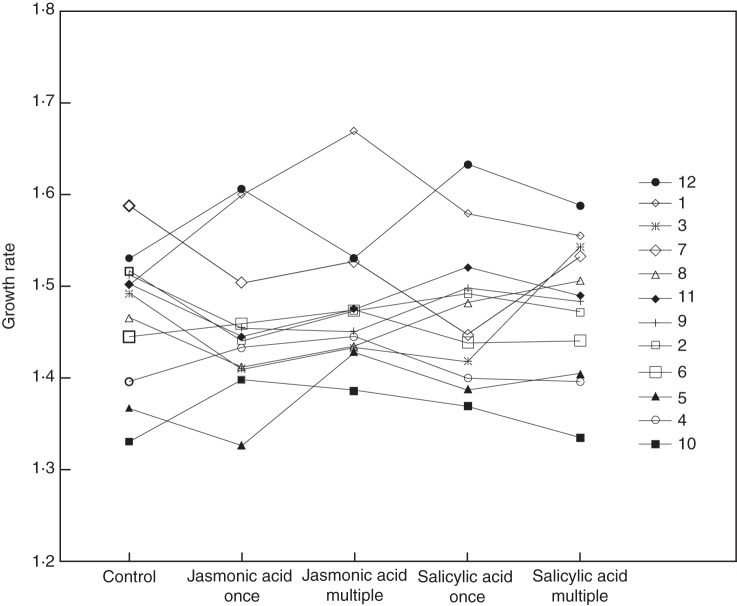
Across lines, spraying plants with JA or SA decreased plant biomass and delayed plant flowering. These effects were generally more pronounced for JA than for SA, and they were stronger when hormones were applied several times, rather than only once. We found that there was significant heritable variation among epiRILs for all of the measured traits (Tables 1 and 2). These line effects were strongest for the growth rates and biomasses of epiRILs (Fig. 1). Moreover, epiRILs also differed significantly in their growth rate responses to the experimental treatments (Table 1), indicating epigenetically based heritable variation in plant responses to defence hormones. While some lines clearly suffered from the hormone application and delayed their growth in response to JA, SA or both hormones, the growth rate of other lines increased in response to the hormone treatments (Fig. 2). Out of 20 different pairwise correlations between line responses to JA and SA, 17 were positive, and these correlations remained significant even after scrutinizing them with the permutation test that accounted for spurious correlations (Table 3).
Table 1.
Summary of analyses of variance testing for the effects of epigenetic variation, treatment of plants with defence hormones, and their interaction on plant growth, phenology and reproduction
| Growth rate |
Flowering time |
Plant height |
Biomass |
Fruit number |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Source* | d.f. | F | P | F | P | F | P | F | P | F | P |
| Line | 11 | 27·04 | <0·001 | 6·67 | <0·001 | 9·51 | <0·001 | 17·43 | <0·001 | 6·82 | <0·001 |
| Treatment | 4 | 1·21 | 0·305 | 3·81 | 0·005 | 1·02 | 0·396 | 4·10 | 0·003 | 2·06 | 0·085 |
| Line × treatment | 44 | 2·20 | <0·001 | 0·89 | 0·678 | 0·69 | 0·934 | 1·28 | 0·115 | 0·80 | 0·811 |
| Error | 404 | ||||||||||
* Line = differences between epigenetic recombinant inbred lines of Arabidopsis thaliana.
Table 2.
Flowering time, plant height and reproduction (means ± s.e.), averaged over all treatments, of the 12 epigenetic recombinant inbred lines (epiRILs) of Arabidopsis thaliana used in this study
| Line | Days to flowering | Plant height (cm) | Fruit number |
|---|---|---|---|
| 1 | 55·63 ± 0·27 | 35·72 ± 0·73 | 170·04 ± 5·92 |
| 2 | 55·93 ± 0·26 | 37·46 ± 0·71 | 182·56 ± 5·76 |
| 3 | 56·55 ± 0·26 | 37·73 ± 0·72 | 174·50 ± 5·84 |
| 4 | 56·30 ± 0·27 | 38·82 ± 0·73 | 164·71 ± 5·92 |
| 5 | 56·68 ± 0·26 | 38·73 ± 0·71 | 199·90 ± 5·76 |
| 6 | 57·06 ± 0·26 | 34·00 ± 0·72 | 169·23 ± 5·84 |
| 7 | 55·85 ± 0·26 | 33·49 ± 0·72 | 193·26 ± 5·84 |
| 8 | 54·90 ± 0·26 | 39·69 ± 0·71 | 159·18 ± 5·76 |
| 9 | 57·54 ± 0·28 | 36·48 ± 0·75 | 162·75 ± 6·11 |
| 10 | 56·41 ± 0·27 | 34·49 ± 0·73 | 173·09 ± 5·92 |
| 11 | 56·44 ± 0·27 | 34·49 ± 0·73 | 153·18 ± 5·92 |
| 12 | 56·05 ± 0·26 | 33·83 ± 0·71 | 150·88 ± 5·76 |
Fig. 1.
Biomass production and growth rates (means ± s.e.), averaged over all treatments, of the 12 epigenetic recombinant inbred lines of Arabidopsis thaliana used in this study. See Methods for definition of growth rate.
Fig. 2.
Responses in growth rate of 12 epigenetic recombinant inbred lines (epiRILs) of Arabidopsis thaliana to treatments involving single or repeated application of jasmonic acid or salicylic acid. See Methods for definition of growth rate.
Table 3.
Epigentic correlations (represented by Pearson's r) between the mean phenotypic responses (percentage change relative to control plants) of 12 epiRILs of A. thaliana to treatment with jasmonic acid (JA) vs. salicylic acid (SA), treated either once (JA1/SA1) or five times (JA5/SA5)
| Growth rate | Flowering time | Plant height | Biomass | Fruit number | |
|---|---|---|---|---|---|
| JA1–SA1 | 0·750** | 0·323 | 0·862*** | 0·684* | 0·658* |
| JA1–SA5 | 0·429 | 0·673* | 0·870*** | 0·922*** | 0·783** |
| JA5–SA1 | 0·624* | 0·576* | 0·806** | 0·769** | 0·734** |
| JA5–SA5 | 0·461 | 0·663* | 0·785** | 0·834*** | 0·643* |
To control for potential spurious correlation, significance levels were obtained with permutation tests as described in the text. *P < 0·05; **P < 0·01; ***P < 0·001.
DISCUSSION
Little is known about the phenotypic effects of heritable epigenetic variation in plants. Here we used epiRILs of A. thaliana to demonstrate that heritable variation in DNA methylation alone can cause significant variation in ecologically important plant traits, including plant growth rate and the plant responses to defence hormones.
Epigenetic variation in ecologically important traits
There was highly significant variation in mean phenotype among epiRILs for all of the traits analysed in our study. As epiRILs are almost identical at the level of DNA sequence, but harbour several orders of magnitude more variation at the level of DNA methylation, the observed variation is most probably of epigenetic origin. Our results are in accordance with previous epiRIL studies (Johannes et al., 2009; Reinders et al., 2009; Roux et al., 2011), and they add to the growing body of evidence that epigenetic variation alone can cause heritable variation in ecologically important traits.
While previous epiRIL studies have already demonstrated heritable variation in some of the measured traits (Johannes et al., 2009; Roux et al., 2011), our study is the first that examined effects on plant growth rates. Growth rate is one of the key determinants of plant life-history strategy (Grime and Hunt, 1975; Reich et al., 1992; Grime, 2001), and it is related, among others, to the defence strategies of plants (Coley, 1988) and their invasiveness (Dawson et al., 2011). Our study shows that epigenetic variation can cause heritable variation and thus potentially microevolution of this important trait.
Epigenetic variation in responses to defence hormones
Induced defences allow plants to optimize resource allocation by producing defences only when needed (Herms and Mattson, 1992; Karban and Baldwin, 1997; Agrawal et al., 1999). Heritable variation in inducibility, in turn, allows adaptation and thus fine-tuning of this ability to local herbivore regimes. Here, we found significant epigenetic variation in plant responses to JA and SA, the two key hormones involved in induced herbivore and pathogen defence, respectively. As such variation in response to JA and SA is usually associated with variation in the strength of induced defence, or with different defence strategies, our results suggest that induced plant defence can be variable, and thus potentially evolve, based on epigenetic variation. Interestingly, we found significant epigenetic variation in hormone responses only with regard to plant growth rates even though two other traits (plant biomass and flowering time) were both variable among epiRILs and generally affected by the hormone treatments.
In a few epiRILs, multiple application of a defence hormone had an opposite effect to its single application. We can only speculate about the underlying causes for this. Jasmonic acid and SA are not only involved in the induction of plant defences, but they are also known to affect many other processes in plants (Raskin, 1992; Creelman and Mullet, 1997), several of which influence the traits we measured in our study. If different pathways that respond to JA or SA have different sensitivities, this could generally create the observed patterns of opposite responses at different JA/SA intensities. Moreover, the pattern was only observed in some epiRILs, hence specific DNA methylation changes appear to have contributed to this.
Previously, Stokes et al. (2002) reported on a heritable epigenetic variant of A. thaliana with increased expression of the SA pathway and thus increased pathogen resistance, and Reinders et al. (2009) found significant variation for resistance to the bacterial pathogen Pseudomonas syringae in another population of epiRILs. Although these studies did not discriminate between constitutive and induced defences, they also suggest, as does our study, a potential for heritable epigenetic variation in plant defences.
Quantitative genetic studies of plant defence often find substantial heritable variation in both constitutive and induced plant defences within and among natural populations (e.g. Agrawal et al., 2002; Handley et al., 2005; Gols et al., 2008). It is possible that part of this variation reflects underlying epigenetic rather than genetic variation.
Another intriguing result of our study is the positive epigenetic correlation between plant responses to JA and SA, which indicates a common molecular basis of the observed epigenetic variation in plant responses to the two hormones. This is somewhat surprising because many previous studies found the two defences to involve separate molecular pathways (e.g. Thomma et al., 1998), or their expression to be antagonistic (Kunkel and Brooks, 2002). Neither seems to be the case in our data. We do not know what the molecular mechanism behind this positive correlation is, but one important implication is that, at least in this system, one should expect correlated epigenetic evolution of plant responses to JA and SA induction. Detailed molecular studies are necessary to elucidate the precise mechanisms underlying this positive correlation.
Conclusions
Our study demonstrates, for the first time, that epigenetic variation alone can cause heritable variation in, and thus potentially microevolution of, plant growth rates and responses to defence hormones. It will be interesting to see to what extent the conclusions from our proof-of-principle system also apply to real ecological systems, and how common ecologically relevant epigenetic variation, as observed here, is in natural populations. Clearly, field studies and studies of natural genotypes (e.g. Herrera and Bazaga 2010) will be needed to address this question.
ACKNOWLEDGEMENTS
We thank Vincent Colot and Frank Johannes for epiRIL seeds and advice. This work, as part of the European Science Foundation EUROCORES Programme EuroEEFG, was supported by funds from the Swiss National Science Foundation (grant number 31EE30-131171) to O.B., and an ERASMUS stipend to K.K.M.
LITERATURE CITED
- Agrawal AA. Phenotypic plasticity in the interactions and evolutions of species. Science. 2001;294:321–326. doi: 10.1126/science.1060701. [DOI] [PubMed] [Google Scholar]
- Agrawal AA, Laforsch C, Tollrian R. Transgenerational induction of defences in animals and plants. Nature. 1999;401:60–63. [Google Scholar]
- Agrawal AA, Conner JK, Johnson MTJ, Wallsgrove R. Ecological genetics of an induced plant defense against herbivores: additive genetic variance and cost of phenotypic plasticity. Evolution. 2002;56:2206–2213. doi: 10.1111/j.0014-3820.2002.tb00145.x. [DOI] [PubMed] [Google Scholar]
- ASPB. The Arabidopsis book. Rockville, MD: American Society of Plant Biologists; 2002. Available from http://www.aspb.org/publications/arabidopsis . [Google Scholar]
- Baldwin IT. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proceedings of the National Academy of Sciences, USA. 1998;95:8113–8118. doi: 10.1073/pnas.95.14.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecology Letters. 2008;11:106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Arcuri D, Richards CL, Pigliucci M. Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana. Evolutionary Ecology. 2010;24:541–553. [Google Scholar]
- Cipollini DF, Redman AM. Age-dependent effects of jasmonic acid treatment and wind exposure on foliar oxidase activity and insect resistance in tomato. Journal of Chemical Ecology. 1999;25:271–281. [Google Scholar]
- Coley PD. Effects of plant-growth rate and leaf lifetime on the amount and type of anti-herbivore defense. Oecologia. 1988;74:531–536. doi: 10.1007/BF00380050. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:355–381. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- Dawson W, Fischer M, van Kleunen M. The maximum relative growth rate of common UK plant species is positively associated with their global invasiveness. Global Ecology and Biogeography. 2011;20:299–306. [Google Scholar]
- Delaney TP. Genetic dissection of acquired resistance to disease. Plant Physiology. 1997;113:5–12. doi: 10.1104/pp.113.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gols R, Wagenaar R, Bukovinszky T, et al. Genetic variation in defense chemistry in wild cabbages affects herbivores and their endoparasitoids. Ecology. 2008;89:1616–1626. doi: 10.1890/07-0873.1. [DOI] [PubMed] [Google Scholar]
- Grime JP. Plant strategies, vegetation processes, and ecosystem properties. 2nd edn. New York: John Wiley and Sons; 2001. [Google Scholar]
- Grime JP, Hunt R. Relative growth-rate: its range and adaptive significance in a local flora. Journal of Ecology. 1975;63:393–422. [Google Scholar]
- Handley R, Ekbom B, Agren J. Variation in trichome density and resistance against a specialist insect herbivore in natural populations of Arabidopsis thaliana. Ecological Entomology. 2005;30:284–292. [Google Scholar]
- Herrera CM, Bazaga P. Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet Viola cazorlensis. New Phytologist. 2010;187:867–876. doi: 10.1111/j.1469-8137.2010.03298.x. [DOI] [PubMed] [Google Scholar]
- Herms DA, Mattson WJ. The dilemma of plants – to grow or defend. Quarterly Review of Biology. 1992;67:283–335. [Google Scholar]
- Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Quarterly Review of Biology. 2009;84:131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000530. e1000530. http://dx.doi.org/10.1371/journal.pgen.1000530 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban R, Baldwin IT. Induced responses to herbivory. Chicago: University of Chicago Press; 1997. [Google Scholar]
- Karban R, Agrawal AA, Thaler JS, Adler LS. Induced plant responses and information content about risk of herbivory. Trends in Ecology and Evolution. 2002;14:443–447. doi: 10.1016/s0169-5347(99)01678-x. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Richards EJ. Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Research. 1995;23:130–137. doi: 10.1093/nar/23.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempel A, Schadler M, Chrobock T, Fischer M, van Kleunen M. Tradeoffs associated with constitutive and induced plant resistance against herbivory. Proceedings of the National Academy of Sciences, USA. 2011;108:5685–5689. doi: 10.1073/pnas.1016508108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, et al. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiology. 2001;126:811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Mitchell-Olds T. The glucosinolate–myrosinase system in an ecological and evolutionary context. Current Opinion in Plant Biology. 2005;8:264–271. doi: 10.1016/j.pbi.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occuring genetic variation in Arabidopsis thaliana. Annual Review of Plant Biology. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Linhart YB, Grant MC. Evolutionary significance of local differentiation in plants. Annual Review of Ecology and Systematics. 1996;27:237–277. [Google Scholar]
- Mauricio R, Rausher MD. Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution. 1997;51:1435–1444. doi: 10.1111/j.1558-5646.1997.tb01467.x. [DOI] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Evolution of phenotypic plasticity: where are we going now? Trends in Ecology and Evolution. 2005;20:481–486. doi: 10.1016/j.tree.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Raskin I. Role of salicylic acid in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1992;43:439–463. [Google Scholar]
- Reich PN, Walters MB, Ellsworth DS. Leaf life-span in relation to leaf, plant, and stand characteristic among diverse ecosystems. Ecological Monographs. 1992;62:365–392. [Google Scholar]
- Reinders J, Wulff BBH, Mirouze M, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes and Development. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. Inherited epigenetic variation – revisiting soft inheritance. Nature Reviews Genetics. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Population epigenetics. Current Opinion in Genetics and Development. 2008;18:221–226. doi: 10.1016/j.gde.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Roux F, Colomé Tatché M, Edelist C, et al. Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics. 2011;188:1015–1017. doi: 10.1534/genetics.111.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes TL, Kunkel BN, Richards EJ. Epigenetic variation in Arabidopsis disease resistance. Genes and Development. 2002;16:171–182. doi: 10.1101/gad.952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatra GS, Miranda J, Chinnappa CC, Reid DM. Effect of light quality and 5-azacytidine on genomic methylation and stem elongation in two ecotypes of Stellaria longipes. Plant Physiology. 2000;109:313–321. [Google Scholar]
- Thaler JS, Stout MJ, Karban R, Duffey SS. Exogenous jasmonates simulate insect wounding in tomato plants (Lycopersicon esculentum) in the laboratory and field. Journal of Chemical Ecology. 1996;22:1767–1781. doi: 10.1007/BF02028503. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninck IAMA, et al. Separate jasmonate-dependent and salicylate-dependent defense response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences, USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Traw MB, Bergelson J. Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiology. 2003;133:1367–1375. doi: 10.1104/pp.103.027086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzic M, Lippman Z, et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biology. 2007;5 doi: 10.1371/journal.pbio.0050174. e174. http://dx.doi.org/10.1371/journal.pbio.0050174 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinig C, Stinchcombe JR, Schmitt J. Evolutionary genetics of resistance and tolerance to natural herbivory in Arabidopsis thaliana. Evolution. 2003;57:1270–1280. doi: 10.1111/j.0014-3820.2003.tb00335.x. [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA. Glucosinolate research in the Arabidopsis era. Trends in Plant Science. 2002;7:263–270. doi: 10.1016/s1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]