Abstract
Many lineage-specific genes are poised and silenced in stem cells. Upon differentiation, genes that are related to self-renewal and alternative lineages are stably silenced. CpG methylation at proximal promoters and PRC2-mediated H3K27me3 play a role in silencing genes temporarily or permanently, with or without coexistence of active epigenetic marks, respectively. Interestingly, DNA methylation on neuronal genes that is distal to transcription start site enable transcription activation owing to its ability to repel PRC2-mediated inhibition. In addition, DNA demethylase Tet proteins play a role in regulation of changes in DNA methylation and related H3K27me3 during differentiation. Collectively, a complex epigenetic network formed by H3K4me3, histone acetylation/deacetylation, H3K27me3 and DNA methylation/demethylation act together to regulate stem cell self-renewal and differentiation.
Introduction
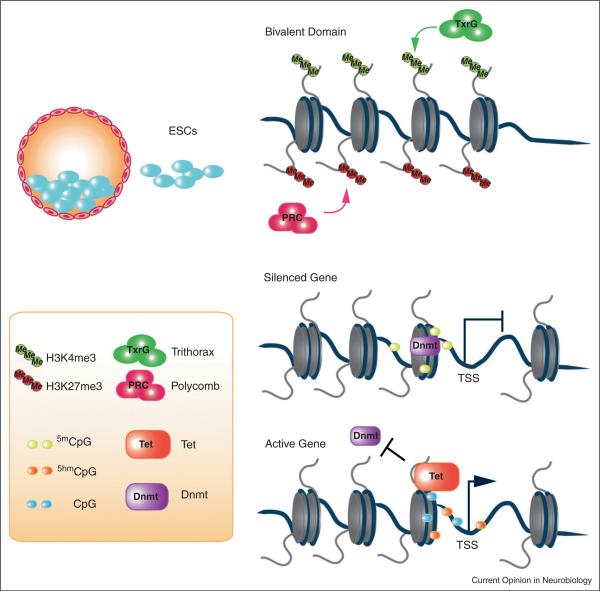
The process of differentiation involves intricate relationships between programs that activate and inhibit gene expression. Through a long cascade of events from embryonic stem cells (ESCs) all the way to differentiated neural cells, significant amount of gene silencing and activation takes place to regulate the potential of each cell. The silencing of developmental genes is achieved by two different mechanisms. Initially, genes are silenced via a temporary process that creates bivalent domains that are poised for activation. Subsequently, a more permanent mechanism ensures long-term silencing of genes during development. Collectively, these mechanisms ensure proper development of the central nervous system (CNS). The poised nature of early gene silencing events prepares a cell for imminent gene activation and differentiation, while preventing precocious differentiation. In ESCs, repression of key regulatory genes, as well as maintenance of ESCs, are regulated by a class of histone modification enzymes, plycomb group (PcG) proteins. Through the action of PcG proteins, differentiation of ESCs is inhibited with no effects on their self-renewing capacity. For instance, factors that are important for differentiation of ESCs are trimethylated at histone H3 lysine 27 (H3K27me3) by polycomb repressive complexes (PRCs) and elimination of PcG results in derepression of their target genes and differentiation of ESCs [1–3]. Despite the fact that most PcG target sites show the H3K27me3 inhibitory epigenetic mark, they also carry marks associated with gene activation, such as H3K4 trimethylation (H3K4me3) [4,5••]. The combination of inhibition and activation marks sets up a `biva-lent' chromatin mark, and collectively, they maintain genes in a poised state for activation (Figure 1). Such bivalent domains are resolved during differentiation, and as a result, most genes end up with either of the two opposing histone marks that correspond to their expression states. Here, we discuss the involvement of epigenetic regulation during neural development.
Figure 1.
In ESCs, the key transcription regulators are bivalently marked with both active and repressive histone markers, H3K4me3 and H3K27me3, respectively. Coexistence of these two histone markers is critical to maintain a poised state within ESCs. Upon differentiation, one of the histone markers would eventually be enhanced through interacting with either PcG or TxrG complex. DNA methylation at the promoter region, induced by de novo Dnmts, is associated with gene silencing. The oxidation of 5mC to 5hmC by Tet proteins relieves this repression and also blocks the binding of Dnmts to the promoter site.
DNA methylation and neural development
DNA methylation plays important roles in genomic imprinting, X-chromosome inactivation, regulation of gene expression and maintenance of epigenetic memory. Two catalytically active de novo DNA methyltransferases, Dnmt3a and Dnmt3b, establish DNA methylation patterns by adding methyl groups onto unmethylated DNA. Dnmt3l, a close homolog of Dnmt3a and Dnm3b, lacks the catalytic domain but interacts with unmethylated H3K4. This interaction recruits and activates methylation activity of Dnmt3a/b [6]. When cells divide, another DNA methyltransferase, Dnmt1, maintains the pattern of DNA methylation by converting the hemimethylated DNA synthesized during replication to fully methylated state [7–10]. Deletion of Dnmt1 or Dnmt3b results in embryonic lethality, whereas Dnmt3a knockout animal die around postnatal day 30 (P30), collectively indicating the essential role of DNA methyltransferases during development [11•,12]. In the classical view of DNA methylation, gene silencing is achieved through inhibiting transcription factor binding to DNA by methylation at the promoter CpG sites and recruitment of methyl-CpG binding proteins (MBDs), which further recruit HDAC repressor complexes, collectively resulting in a repressive state of the chromatin. Even though, most CpG islands overlap with proximal promoters and mediate gene silencing, DNA methylation in intergenic regions and gene bodies is widespread, indicating the importance of distal-promoter methylation and its role in tissue-specific gene expression [13–16]. During early embryogenesis, DNA methylation pattern is established gradually upon fertilization and development of the zygote. At first, methylation patterns of both paternal and maternal genomes are largely removed, owing to Dnmt1 being excluded from the nucleus [17]. Subsequently, de novo DNA methyltransferases begin to re-establish DNA methylation patterns during implantation and subsequent germ layer and cell type differentiation [11•]. The genome-wide eradication of DNA methylation in pre-implantation embryos, followed by re-methylation to establish DNA methylation patterns, is an important process for setting up the pluripotency in early embryonic stem cells [18,19•]. In continuously self-renewing stem cells, genes that regulate differentiation need to be repressed in a stable manner, and this repression needs to be heritable once cells undergo division. Therefore, DNA methylation as well as histone modifications, work in combination to regulate stem cell self-renewal, differentiation, as well as reprogramming, including de-differentiation and trans-differentiation.
DNA methylation and histone modifications play important roles in stem cell differentiation along the neural lineage. During neural development, neurogenesis always precedes gliogenesis, and this correlates with tight spatiotemporal changes in the epigenetic landscape within the genome. Specifically, numerous neuronal genes, in addition to being inhibited by PRC2, are also inhibited by the action of REST (RE1 silencing transcription factor) in undifferentiated stem cells and in nonneuronal cells in the developing embryo. REST exerts its effects through RE1 binding site on promoters of target genes, and by recruiting histone modifiers and chromatin-binding proteins [20]. Highly expressed REST in ESCs and NPCs preferably binds to neuronal genes to prevent premature neuronal differentiation by maintaining neuronal lineage genes in a poised state. The dissociation of REST from its target genes is also concomitant with REST repression as NPCs differentiate into neurons [21]. However, the action of REST on NPCs and non-neuroectodermal lineages differ fundamentally. In ESCs and NPCs, REST recruits corepressor complex that consists of CoREST, HDAC, mSin3A and MeCP2 to the RE1 site. The HDAC within the complex plays a crucial role of maintaining the reversible repression of neuronal genes in ESCs and NPCs. Although the REST complex found in non-neuroectodermal cells is composed of similar core regulators, the chromatin state of neuronal genes is strikingly different compared to that of ESCs and NPCs. The REST complex in non-neuroectodermal cells has a higher association with H3K9 methyltransferase (G9A), SUV39H1 and the H3K4 demethylase, JARID1C. This REST-associated complex found in non-neuroectodermal cells induces permanent silencing of neuronal genes via H3K9 methylation, DNA methylation and H3K4 demethylation [20–22]. On the contrary, the inactive – but, poised – neuronal promoters in stem cells are not associated with DNA methylation or histone H3K9 methylation. The transcription of neuronal genes takes place once NPCs commit to a neuronal lineage and the expression of REST is turned off, followed by the removal of the REST repressor complex from neuronal gene promoters.
DNA demethylation and development
DNA methylation has been considered a major epigenetic mechanism for stable, long term gene silencing in somatic cells, and it sets up a pluripotent state at the early stages of development [18,19•,23]. Even though, active demethylation of the genome in the zygote has been documented for more than a decade, it was demonstrated only recently that such genome-wide dynamic changes in DNA methylation was carried out by a group of enzymes that efficiently modify methylation patterns during development. Ten-eleven translocation (Tet) proteins convert 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) through oxidation reactions [24,25,26••,27]. Even though, 5hmC was detected in mouse ESCs at high levels, once ESCs initiate differentiation, 5hmC levels decrease significantly [24,28]. Upon completion of the differentiation process, 5hmC levels increase in terminally differentiated cells [29]. Collectively, these findings indicate a significant role for the involvement of 5hmC in the dynamic regulation of DNA methylation during development. The Tet group of proteins in mammals have 3 members: Tet1, Tet2 and Tet3 [30,31]. Tet1 was identified as a 5mC dioxygenase that catalyzes the conversion of 5mC to 5hmC [24], and genome-wide analysis of Tet1 and 5hmC distribution in ESCs revealed the function of Tet proteins in regulation of dynamic changes during the differentiation of pluripotent stem cells [32,33•]. In ESCs, 5hmC and Tet1 are enriched at bivalently marked genes. Surprisingly, deposition of 5hmC by Tet1 is required for PRC2 binding to these targets. It is possible that the maintenance of hypomethylation induced by Tet1 in ESCs allows PRC2 binding. Another scenario suggests that the 5hmC mark might involve RNA polymerase II pausing at the promoter sites of poised genes. The discovery of Tets indicates a potentially novel mechanism for the regulation of DNA methylation during development and the establishment and maintenance of the pluripotent epigenetic state during early embryogenesis.
Regulation of gene expression by non-promoter DNA methylation
Approximately 70% of the human gene promoters are associated with CpG islands. Despite the methylated status of the most scattered CpG sites in the mammalian genome, CpG islands generally remain unmethylated with the exception of CpG islands in intragenic regions [34], collectively setting up a bimodal pattern. As discussed above, the binding of Dnmt3a and Dnmt3b to the chromatin may play a role in the establishment of this pattern [35], although the molecular mechanism underlying this phenomenon still remains elusive. On the contrary, binding of transcription factors may regulate targeting of Dnmt3a and Dnmt3b to promoter regions [36,37••]. Even though, many studies provided clues regarding the function of Dnmts, the extrapolation of locus-specific findings to other regions of the genome required more comprehensive genome-wide analyses. With the recent advances in sequencing-based technologies, generation of high-resolution genome-wide Dnmt occupancy, as well as assessment of DNA methylation state, has been performed using pluripotent stem cells and their differentiated progeny. In combination with the recent discovery of Tet group of proteins that act as active DNA demethylases, DNA methylation is now seen as a dynamic regulatory mechanism rather than only permanently silencing genes. In addition, accumulating evidence has revealed the distribution and function of DNA methylation and Dnmts at the proximal promoter and distal promoter regions, further demonstrating the dynamic nature of DNA methylation in gene regulation.
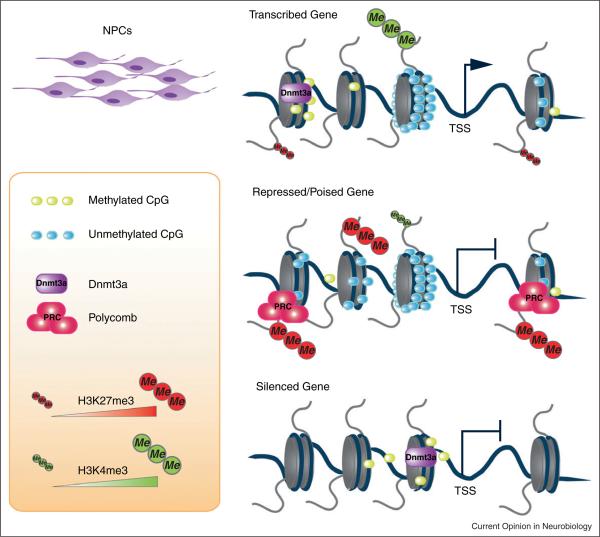
The context-dependent nature of gene regulation by DNA methylation was recently demonstrated through genome-wide DNA methylation analyses in stem cells and their differentiated progeny. For instance, actively transcribed genes were shown to carry high levels of gene body methylation [13,14,38]. In addition, the classic inhibitory view of DNA methylation was challenged and expanded with the finding that DNA methylation may play a role in both gene inhibition and activation using genome-wide mapping of de novo Dnmts [39•• ,40,41]. By methylating proximal promoters in self-renewing postnatal neural stem cells (NSCs), Dnmt3a mediates gene repression. More interestingly, through non-proximal promoter methylation Dnmt3a allows for transcription activation of neurogenic genes in NSCs by antagonizing Plycomb complex-mediated repression [39•• ]. Specifically, in postnatal NSCs, using genome-wide Dnmt3a occupancy analysis, Dnmt3a was shown to be excluded from areas with high CpG content and H3K4me3 marks; however, Dnmt3a was found to be frequently bound to areas with low CpG content promoters or genomic regions flanking H3K4me3 marks [39••] (Figure 2). This phenomenon does not seem to be limited to adult NSCs and may play critical roles during development, since the correlation of non-promoter DNA methylation and transcription of Dnmt3a target genes was also shown when neural differentiation is initiated from mouse embryonic stem cells. In addition, analysis of genome-wide DNA methylation status in human colon cancers also demonstrated that localization of DNA methylation to regions flanking CpG islands (also referred to as `shore regions') instead of CpG-reach promoter regions [16]. The possibility of tissue-specific gene regulation by distal-promoter DNA methylation was supported by the demonstration that CpG-shore DNA methylation played important roles for the lineage specification of hematopoietic stem cells [42]. Human ESCs show extensive non-CpG island DNA methylation, especially in gene bodies of actively transcribed genes, though the function of this methylation has not been fully understood. In addition to non-CpG island methylation, many large domains of the genome show differential methylation patterns [14]. Genes that are part of the partially methylated domains generally show lower levels of expression activity and higher levels of histone marks that are associated with repression, such as H3K27me3 [43]. The hypothesis that non-promoter DNA methylation may facilitate transcription of genes in partially methylated domains was reported in NSCs of the adult brain, in which Dnmt3a-dependent non-proximal-promoter DNA methylation was shown to antagonize PRC2 driven H3K27me3 levels and to ultimately relieve inhibition of a number of neurogenic genes [39]. In this context, non-proximal promoter DNA methylation established by Dnmt3a may represent an additional layer of epigenetic regulation of Polycomb repression in somatic cells [44].
Figure 2.
In postnatal NPCs, DNA methylation, mediated through Dnmt3a, can either repress or activate gene expression based on the context of the chromatin. Dnmt3a induced distal promoter methylation usually coincides at the upstream of actively transcribed genes with CpG island containing promoters. Instead, these CpG island containing promoters are repressed or maintained in a poised state through PRC2-mediated silencing and often associated with high levels of H3K27me3. Genes with low CpG density proximal promoters can be silenced through Dnmt3a-mediated promoter methylation.
Conclusions
During stem cell self-renewal and differentiation, there are two forms of gene silencing events. In undifferentiated stem cells, lineage-specific genes are poised and silenced, while during stem cell differentiation, genes related to stem cell self-renewal and genes related to alternative cell lineage are stably silenced. Both CpG methylation at proximal promoters and PRC2-mediated H3K27me3 are heavily used, taking turns to silence genes either temporarily or permanently. In ESCs, H3K27me3 appears to be the major gene silencing mechanism, while poised state is established by addition of the H3K4me3 mark. DNA methylation is more involved in differentiation. Methylation within proximal promoters can be involved in permanent gene silencing of pluripotent genes, probably in combination with H3K9Me2/3, and in temporal inhibition of differentiation genes, such as glial specific genes during neurogenesis. Interestingly, DNA methylation on neuronal genes that are at distal regions with regards to transcription start site has been shown to enable transcription activation owing to its ability to repel PRC2-mediated inhibition. Consistent with the antagonizing effect between CpG methylation and H3K27me3 modification, it is found that DNA demethylase, Tet proteins allow for PRC2 binding. Upon terminal glial differentiation, the proximal promoter CpG methylation is erased in response to glial differentiation signals, while during neuronal terminal differentiation, CpG methylation globally might also drop, reinstating PRC2-mediated silencing of alternative lineage genes as well as early differentiation genes. Therefore, rather complex epigenetic networks formed by H3K4me3, Histone acetylation/deacetylation, H3K27me3 and DNA methylation/demethylation act coherently to regulate stem cell self-renewal and differentiation.
Acknowledgements
We apologize to colleagues whose work could not be cited owing to space limitations. Y.E.S. is supported by grants from The National Basic Research Program (973 Program, No. 2011CB966200 and 2011CB965100), NIH (P01 GM081621-01A1, 1R01MH082068-01A2) and CIRM (RB3-02129).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 2.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, Koseki H. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 4.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 5 ••.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]; The authors reported the first observation of co-occupancy of both `active' and `repressive' histone modification at the promoter sites of poised genes and defined such regions as `bivalent' domains.
- 6.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 7.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 8.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 9.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 10.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 11 •.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]; By creating Dnmt3a and Dnmt3b deficient mice, the authors showed that both Dnmt3a and Dnmt3b are de novo DNA methyltransferases and are essential for embryonic development.
- 12.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 14.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 18.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 19 •.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]; This paper was the first observation of genes that are highly methylated in sperm that underwent active demethylation in the zygote only hours after fertilization, before the first round of DNA replication commences.
- 20.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorgensen HF, Terry A, Beretta C, Pereira CF, Leleu M, Chen ZF, Kelly C, Merkenschlager M, Fisher AG. REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development. 2009;136:715–721. doi: 10.1242/dev.028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26 ••.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that Tet proteins are capable of demethylating 5mC by converting 5mC to 5hmC. Knocking down Tet1 in pre-implantation embryos demonstrated its role in ES cell maintenance and inner cell mass cell specification.
- 27.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loenarz C, Schofield CJ. Oxygenase catalyzed 5-methylcytosine hydroxylation. Chem Biol. 2009;16:580–583. doi: 10.1016/j.chembiol.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33 •.Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that Tet1 not only modulates DNA methylation levels at CpG-rich promoters, but also promotes transcription of pluripotency factors and participates in the repression of Polycomb targets.
- 34.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 37 ••.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]; By comparing methylomes between mouse ESCs and NPCs, this study identified low-methylated regions (LMRs) that are distal to promoters with little overlap with CpG islands, and further demonstrated the dynamic nature of LMRs during differentiation and the interaction between DNA-binding factors and local DNA methylation.
- 38.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39 ••.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using genome-wide analyses, this study demonstrated that Dnmt3a can mediate repression in postnatal NSCs by methylating proximal promoters, and it also promotes transcription of neurogenic genes by antagonizing Polycomb through distal promoter methylation.
- 40.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trowbridge JJ, Orkin SH. Dnmt3a silences hematopoietic stem cell self-renewal. Nat Genet. 2012;44:13–14. doi: 10.1038/ng.1043. [DOI] [PubMed] [Google Scholar]
- 42.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]