Abstract
The presence of lipopolysaccharide (LPS) on the cell surface of Gram-negative bacteria is critical for viability. A conserved β-barrel membrane protein LptD translocates LPS from the periplasm across the outer membrane (OM). In Escherichia coli, this protein contains two disulfide bonds and forms the OM LPS translocon with the lipoprotein LptE. Here we identified seven in vivo states on the oxidative folding pathway of LptD. Proper assembly involved a nonfunctional intermediate containing non-native disulfides. Intermediate formation required the oxidase DsbA, and subsequent maturation to the active form with native disulfides was triggered by LptE. Thus, disulfide bond-dependent protein folding of LptD requires the proper assembly of a two-protein complex in order to promote disulfide bond rearrangement.
A defining feature of Gram-negative organisms is the presence of lipopolysaccharide (LPS) on the cell surface (1). LPS must be properly assembled in the outer leaflet of the outer membrane (OM) to establish a permeability barrier against toxic compounds, including antibiotics (2, 3). In Escherichia coli, a translocon responsible for LPS movement across the OM is composed of two essential OM proteins: an integral β-barrel protein, LptD (for lipopolysaccharide transport protein D) and a lipoprotein, LptE (4–6). The LptD/E complex forms part of the trans-envelope LPS exporter, which contains five other essential Lpt proteins that collectively move LPS from the inner membrane (IM) to the cell surface (7). Assembly of the OM LPS translocon presents a challenging protein-folding problem because LptE resides inside LptD (5, 6) and formation of the correct disulfide bonds in LptD is required for this translocon to function (8). How the cell coordinates assembly of the OM complex with the formation of the rest of the trans-envelope exporter is unknown.
To understand the assembly of the functional OM LPS translocon, we examined the biogenesis of LptD. E. coli LptD contains an N-terminal periplasmic domain (a.a. 25–202) and a C-terminal integral β-barrel domain (a.a. 203–784) (5), which is folded and inserted into the OM by the Bam complex (β-barrel assembly machine) (9–11). LptD has four cysteines, two in the N- terminal domain (Cys31 and Cys173), and two very near the C-terminus (Cys724 and Cys725). In its mature form, LptD contains two long-range non-consecutive disulfide bonds connecting the N- and C-terminal domains, one between the first and third cysteines (Cys31-Cys724; [1–3]), and the other between the second and fourth cysteines (Cys173-Cys725; [2–4]) (Fig. 1A) (8). Either of these two disulfides is sufficient for LptD to function (8).
Fig. 1.
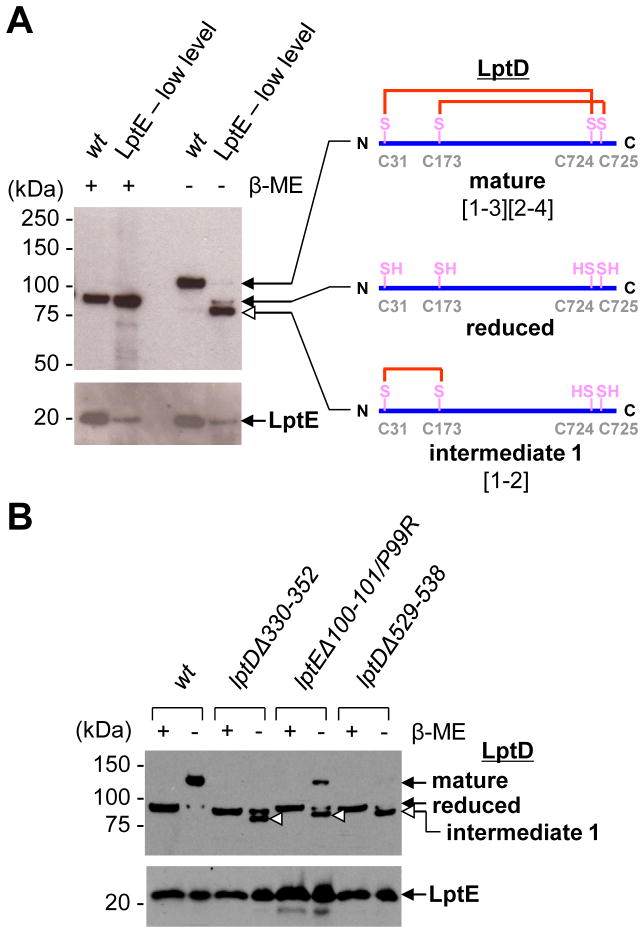
A non-native disulfide-bonded LptD intermediate (1, [1–2]-LptD) accumulates in strains with defective LptD or LptE. (A) α-LptD and α-LptE immunoblot analyses of OM fragments obtained from wild-type (wt) and LptE-limiting strains grown to early-log phase (see Fig. S1). More membranes (3x) from cells expressing limited LptE were loaded. (B) α-LptD and α-LptE immunoblot analyses of OM fragments obtained from wt, lptDΔ330–352, lptEΔ100–101/P99R and lptDΔ529–538 strains. Open arrowheads indicate the position of [1–2]-LptD in each strain. Where indicated, β-mercaptoethanol (β-ME) was used to reduce disulfide bonds.
LptE is required to promote assembly of mature LptD (8). We wondered if an LptD intermediate would accumulate in vivo when LptE is the limiting reagent. OM fragments isolated from wild-type cells contain only mature LptD, which migrates more slowly during SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) than reduced LptD (Fig. 1A) (8, 12–13). In contrast, OM fragments from an LptE-limiting strain, which produced lower levels of LptE (Fig. S1), contained a new LptD species. In the absence of β-mercaptoethanol, this species migrated slightly faster than fully-reduced LptD, suggesting that it was disulfide-bonded (Fig. 1A). To assign the disulfide connectivity of this LptD species (intermediate 1), we examined the SDS-PAGE migration patterns of the six LptD mutant proteins in which two of the cysteines were changed to serine (8). Only the mutant lacking the last two cysteines (LptDCCSS), which can make the Cys31-Cys173 ([1–2]) disulfide bond within the N-terminal domain, exhibited faster mobility than reduced LptD (Fig. S2). Thus, the LptD intermediate 1 that accumulates when LptE is limiting contains the consecutive [1–2] disulfide bond, and LptE is required for the conversion of this intermediate to LptD with its native disulfide bonds. Whether the 3rd and 4th cysteines are ever disulfide-bonded in this intermediate remains unclear, although we have obtained some evidence suggesting that the [3–4] disulfide bond may be formed (Fig. S3).
To examine how defects in either component affect translocon assembly, three mutants, LptDΔ330–352 (LptD4213, 14), LptDΔ529–538 (6) and LptEΔ100–101/P99R (LptE6, 15), were analyzed. Two of these mutations impair interactions between LptD and LptE (6, 15). Although none of these mutations lowered the levels of LptE in the OM, each resulted in substantial accumulation of intermediate 1 ([1–2]-LptD) (Fig. 1B). LptD containing a [1–2] disulfide bond is not functional (8), explaining why these mutants have defective OMs (6, 14–15).
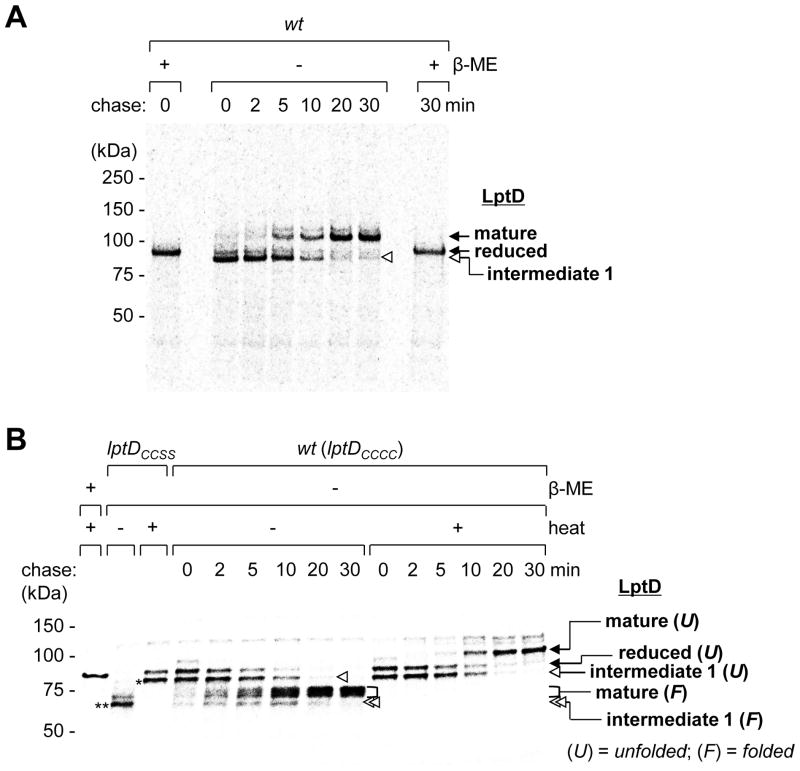
To determine whether intermediate 1 is on the folding pathway of LptD in a wild-type strain, or a dead-end product, we pulse-labeled FLAG3-tagged LptD with [35S]-methionine and monitored its progression to the mature form during a cold-methionine chase. The predominant form at the start of the chase was [1–2]-LptD (intermediate 1), which was slowly converted to mature [1–3][2–4]-LptD (Fig. 2A). The final distribution of LptD species at ~20 min was identical to that at steady state (Fig. S4A and S4B). Thus, a non-native disulfide-bonded species (intermediate 1) is an intermediate along the oxidative folding pathway of LptD in vivo. Furthermore, formation of mature [1–3][2–4]-LptD proceeds via disulfide bond rearrangement.
Fig. 2.
Intermediate 1 is observed along the LptD assembly pathway in vivo. Folding of the LptD β-barrel domain is slow and precedes disulfide bond rearrangement. (A) Wild-type cells expressing LptD-FLAG3 were pulsed with [35S]-methionine and chased with cold methionine. The samples were alkylated with NEM (N-ethylmaleimide), immunoprecipitated with α-FLAG antibody, and analyzed by SDS-PAGE/autoradiography. (B) [35S]-Met pulse-chase of newly-synthesized LptD-FLAG3 where samples were processed using non-denaturing conditions and analyzed by seminative SDS-PAGE to preserve (− heat) or denature (+ heat) the β-barrel. Pulse- labeled LptDCCSS-FLAG3 marks the positions of unfolded (asterisk) and folded (double asterisks) [1–2]-LptD.
We next performed pulse-chase experiments under conditions that preserve a folded β-barrel to determine whether disulfide bond rearrangement occurs before or after β-barrel folding. When samples were not heated prior to SDS-PAGE, mature [1–3][2–4]-LptD migrated as a broad band with faster gel mobility than the heat-denatured form, indicating that the β-barrel was folded (Fig. 2B) (5). The amount of folded [1–3][2–4]-LptD increased with time. A new fast-migrating LptD species, appearing at the start and chasing away by 20 min, could be detected (Fig. 2B). This species was also heat-modifiable and ran at the same position as the major band in a non-heated LptDCCSS sample containing the [1–2] disulfide (Fig. 2B). We assign this LptD species as intermediate 1 containing a folded β-barrel. Because folded intermediate 1 chased to folded mature [1–3][2–4]-LptD, we conclude that the LptD β-barrel folds before disulfide rearrangement occurs. Furthermore, monitoring protease susceptibility supported this notion (Fig. S5). Because this [1–2]-LptD species did not accumulate substantially during the chase, we conclude that folding is slower than rearrangement.
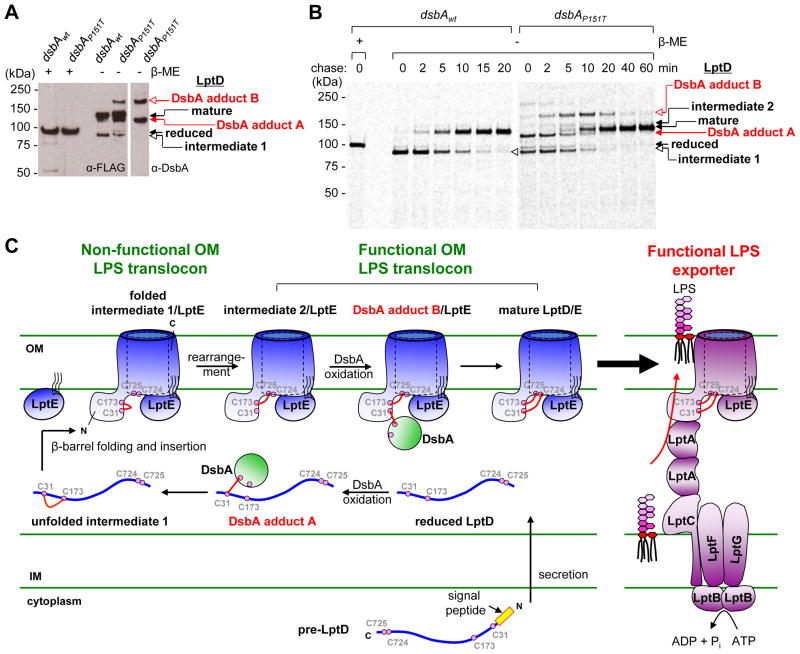
LptD is a substrate of the periplasmic oxidase DsbA (16) and oxidation of LptD is impaired in strains lacking DsbA (8, 12; Fig. S4). We sought to elucidate the role of DsbA in introducing disulfide bonds in LptD by using the DsbAP151T mutant that forms kinetically stable mixed-disulfide intermediates with substrate proteins to see if we could accumulate cross-linked adducts between LptD and DsbA (16). We observed two LptD species that also contained DsbA (Fig. 3A). These new bands were identified as mixed-disulfide intermediates linking DsbA to Cys31 of reduced LptD (DsbA adduct A; [1-DsbA]) and to Cys31 of [2–4]-LptD (DsbA adduct B; [2–4][1-DsbA]) by comparing adducts observed with LptD using Cys-to-Ser mutants (Fig. 3A, 3C and S6). DsbA adduct A was likely to correspond to the first-formed intermediate between LptD and DsbA leading to the formation of the [1–2] disulfide bond (Fig. 3C). When pulse- chase analysis was carried out in the dsbAP151T strain, we detected [35S]-labeled DsbA adduct A at early time points (t = 0–2 min, Fig. 3B), consistent with this interpretation. We also observed the appearance of DsbA adduct B ([2–4][1-DsbA]-LptD) at t = 2 min, which chased by t = 40 min. The fact that this species appeared before (and chased into) [1–3][2–4]-LptD suggests that DsbA adduct B is also along the assembly pathway and that DsbA directly introduces the [1–3] disulfide bond in LptD after an intermediate containing the [2–4] disulfide (intermediate 2, see Fig. S4) is already formed (Fig. 3C). Thus, DsbA makes both the [1–2] and [1–3] disulfide bonds in LptD at different stages of assembly.
Fig. 3.
The oxidative folding pathway of LptD. (A) LptD intermediates containing mixed-disulfide adducts with DsbA can be detected in vivo. α-FLAG immunoprecipitation using wt or dsbAP151T cells containing pET23/42lptD-FLAG3. Samples were analyzed by α-FLAG and α-DsbA immunoblots. (B) [35S]-Met pulse-chase of newly-synthesized LptD-FLAG3 in wt and dsbAP151T backgrounds. (C) Biogenesis of the trans-envelope LPS exporter. The six intermediate LptD species observed experimentally are depicted as they appear after secretion across the IM during oxidative assembly of the OM translocon.
The ability to detect several DsbA adducts with LptD during pulse-chase allowed us to describe the sequence of events leading to the assembly of a functional OM LPS translocon. We could assign seven distinct species along the oxidative folding pathway of LptD in vivo (Fig. 3C). The first non-consecutive disulfide bond was formed between Cys173 and Cys725 ([2–4]) following disulfide bond rearrangement from [1–2]-LptD. Because either of the two non- consecutive disulfide bonds is sufficient for the proper function of LptD (8), the [2–4]-LptD species (intermediate 2) represents the first functional form of LptD. In this regard, it is important to point out that cysteines corresponding to both Cys173 and Cys725 are present in >95% of over one thousand Gram-negative LptD proteins that have non-identical sequences (Fig. S7, 12). In contrast, Cys31 and Cys724 are much less conserved, suggesting that the [2–4] disulfide bond plays a critical structural role in the function of the translocon.
LptD and LptE function together in a single step to insert LPS correctly on the cell surface (4–6). Progression of LptD from an unfolded intermediate, to a pre-assembled nonfunctional complex with LptE containing non-native disulfide bonds (intermediate 1), and then to a rearranged functional state containing native disulfide bonds, provides a mechanism to check that both translocon components have been properly synthesized, targeted, and assembled together in the OM. Mutants in LptD or LptE accumulate intermediate 1 (Fig. 1B), suggesting that assembly of the translocon stalls in a non-functional state if there are defects in either component. Intermediate 1 also accumulates in strains with limited LptE (Fig. 1A), but it can be completely converted to the functional form of LptD if additional LptE is subsequently expressed (Fig. S8). This “rescue” experiment demonstrates that increased LptE levels, which permits formation of the plug-and-barrel, triggers rearrangement of the disulfide bonds. Thus, assembly of the LptD/E complex controls activation of the translocon; only when the complex is properly formed does disulfide bond rearrangement take place efficiently (Fig. 3C). The oxidative assembly of LptD is regulated by disulfide bond rearrangement, triggered by binding to LptE, which, in fact, is not even an oxidoreductase.
Producing a non-functional intermediate 1 prior to activating it via disulfide bond rearrangement also provides a mechanism for quality control to ensure that the trans-envelope LPS exporter is only assembled with a functional OM translocon (Fig. 3C). Proper disulfide bonds in LptD are required for the trans-envelope Lpt bridge to form, via interactions between LptA and LptD (17). The triggering strategy prevents a defective OM translocon from achieving the correct disulfide bond configuration, thereby preventing formation of a non-functional trans-envelope LPS exporter that might release LPS into an inappropriate compartment.
Our ability to visualize and accumulate different oxidation states of LptD allows us to extend to membrane proteins the classic approach of using disulfide bonds to study the folding of soluble proteins in vivo (18, 19). Our studies have established that the rate-determining step in translocon assembly is β-barrel folding. Folding is remarkably slow, taking ~20 minutes, or one-third of the cell cycle (Fig. 2). The porin LamB folds several orders of magnitude faster than LptD (20). LptD folding may be slow because the protein must wrap around LptE, forming a β-barrel that also contains a plug (5, 6). Given the slow rate of LptD folding, it may be possible to design inhibitors against the long-lived non-functional states of the OM LPS translocon, and thus prevent its activation.
Supplementary Material
Acknowledgments
We thank E. Freinkman for pET23/42lptD-FLAG3. J. B. is an American Cancer Society Professor. This work is supported by NIH grants AI081059 (to D.K.), NIGMS 41883 (to J.B.), and in part by JSPS Grant-in-Aid for Scientific Research (C) (21580092) (to H.K.).
Footnotes
References and Notes
- 1.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mühlradt PF, Golecki JR. Asymmetrical distribution and artifactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. Eur J Biochem. 1975;51:343–352. doi: 10.1111/j.1432-1033.1975.tb03934.x. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci USA. 2010;107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freinkman E, Chng SS, Kahne D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci USA. 2011;108:2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chng SS, Gronenberg LS, Kahne D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a trans-envelope complex. Biochemistry. 2010;49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz N, Chng SS, Hiniker A, Kahne D, Silhavy TJ. Non-consecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci USA. 2010;107:12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggert US, et al. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science. 2001;294:361–364. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]
- 10.Wu T, et al. Identification of a multi-component complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 13.Materials and methods are available as supporting material on Science Online.
- 14.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chimalakonda G, et al. Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2011;108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 17.Freinkman E, Okuda S, Ruiz N, Kahne D. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry. 51:4800–4806. doi: 10.1021/bi300592c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadokura H, Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138:1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansens A, van Duijn E, Braakman I. Coordinated nonvectorial folding in a newly synthesized multidomain protein. Science. 2002;298:2401–2403. doi: 10.1126/science.1078376. [DOI] [PubMed] [Google Scholar]
- 20.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. Cold Spring Harbor Laboratory Press; Plainview, NY: 1984. [Google Scholar]
- 22.Kadokura H, Beckwith J. Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 2002;21:2354–2363. doi: 10.1093/emboj/21.10.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.