Abstract
We analyzed the biochemical composition of the magnetosome membrane (MM) in Magnetospirillum gryphiswaldense. Isolated magnetosomes were associated with phospholipids and fatty acids which were similar to phospholipids and fatty acids from other subcellular compartments (i.e., outer and cytoplasmic membranes) but were present in different proportions. The binding characteristics of MM-associated proteins were studied by selective solubilization and limited proteolysis. The MM-associated proteins were further analyzed by various proteomic approaches, including one- and two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Edman and mass spectrometric (electrospray ionization-mass spectrometry-mass spectrometry) sequencing, as well as capillary liquid chromatography-mass spectrometry-mass spectrometry of total tryptic digests of the MM. At least 18 proteins were found to constitute the magnetosome subproteome, and most of these proteins are novel for M. gryphiswaldense. Except for MM22 and Mms16, all bona fide MM proteins (MMPs) were encoded by open reading frames in the mamAB, mamDC, and mms6 clusters in the previously identified putative magnetosome island. Eight of the MMPs display homology to known families, and some of them occur in the MM in multiple homologues. Ten of the MMPs have no known homologues in nonmagnetic organisms and thus represent novel, magnetotactic bacterium-specific protein families. Several MMPs display repetitive or highly acidic sequence patterns, which are known from other biomineralizing systems and thus may have relevance for magnetite formation.
Many prokaryotes build more or less complex subcellular structures, such as intracellular membranes or structures generally described as inclusions. Analysis of many examples of these structures has revealed a remarkable degree of intracellular differentiation and association with distinct subsets of proteins, including in structures such as endospores (27, 58), chlorosomes (13), gas vesicles (34), carboxysomes (14), acidocalcisomes (52), and polyhydroxybutyrate (PHB) granules (30).
Among the most intriguing examples of subcellular structures are magnetosomes, which are formed by magnetotactic bacteria (MTB) (5, 10). Magnetosomes are nanometer-size magnetic particles which are arranged in a bacterial cell in chain-like structures that are thought to serve as a navigational device in bacterial magnetotaxis (7, 17).
The superior crystalline and magnetic characteristics of bacterial magnetosomes make them potentially useful in a number of biotechnological applications, such as in immobilization of bioactive compounds, as contrast agents for magnetic resonance imaging, and in magnetic drug targeting (21, 42, 49). The characteristics of bacterial magnetosomes have recently even been considered for use as biosignatures to identify presumptive Martian magnetofossils (56).
The magnetosome particles consist of crystals of a magnetic iron mineral which are enclosed within membrane vesicles. Although the biomineralization of magnetosomes is poorly understood at the molecular and biochemical levels, it has been generally assumed that the magnetosome membrane (MM) is crucial in the biological control of mineral formation (3, 4, 7, 18). In addition to phospholipids associated with isolated magnetosome particles, Gorby and coworkers in an initial study detected two of the numerous proteins in the MM that have not been found in other cell fractions (18). In other studies, several genes encoding magnetosome-associated proteins were identified by reverse genetics in Magnetospirillum magnetotacticum and Magnetospirillum strain AMB-1 (29, 35, 36, 37). In a recent study the workers identified two low-molecular-mass proteins which were tightly bound to the MM in strain AMB-1 along with other proteins (3). One of these low-molecular-mass proteins (Mms6) had an effect in in vitro magnetite nucleation. All the previous approaches, however, were focused on analysis of individual proteins or a limited number of proteins, and the overall biochemical composition of the MM in different MTB has for the most part remained elusive.
Magnetospirillum gryphiswaldense produces up to 60 cubo-octahedral magnetosomes which are approximately 45 nm in diameter and consist of membrane-bound crystals of magnetite (Fe3O4) (44, 46). Because M. gryphiswaldense is genetically tractable (51) and can be readily grown by microaerobic mass cultivation (22), it has been used as a model for investigation of magnetosome formation in a number of studies (47, 48, 50). Recently, an initial analysis of isolated magnetosomes led to identification of at least 13 MM-specific protein bands (20). Cloning of the genes encoding four of the most abundant MM-associated proteins revealed that these genes are arranged in several operon-like gene clusters which are highly conserved in different MTB. Identified MM proteins (MMPs) were found to exhibit homology to tetratricopeptide repeat proteins (MamA), cation diffusion facilitators (MamB), and HtrA-like serine proteases (MamE) or to exhibit no similarity to known proteins (MamC and MamD). In another study, three operons encoding MMPs were identified as parts of a larger putative magnetosome island. This apparently unstable 35- to 80-kb genomic region was functionally linked to magnetosome biosynthesis in a deletion mutant and seemed to encode many other functions required for biomineralization. The presence of additional MTB-specific open reading frames (ORFs) in this region led to the conclusion that the specific subset of MMPs is more complex, and many of the previously identified mam and mms genes were predicted to encode additional MM-associated proteins (45).
In this study, we examined the biochemical characteristics of the MM by using different approaches and analyzed its protein composition. A number of proteins constituting the MM subproteome were detected, and their genes were identified in a preliminary genome analysis of M. gryphiswaldense. Most of the bona fide MMPs were assigned to ORFs in the mamAB, mamDC, and mms clusters in the putative magnetosome island.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. gryphiswaldense MSR-1 (= DSM 6361), M. magnetotacticum MS-1 (= ATCC 31632), and Magnetospirillum sp. strain AMB-1 (= ATCC 700264) were used in this study. The strains were grown under microaerobic conditions in an oxystat fermentor as described previously (22).
Isolation and biochemical analysis of magnetosomes.
The protocol used for magnetosome isolation was the protocol described previously (20), with minor modifications. Briefly, 10 g (wet weight) of M. gryphiswaldense cells suspended in 50 ml of 50 mM HEPES-4 mM EDTA (pH 7.4) was disrupted by three passes through a French pressure cell (20 000 lb/in2). All buffers used for magnetosome isolation contained 0.1 mM phenylmethylsulfonyl fluoride as a protease inhibitor. Unbroken cells and cell debris were removed from the sample by centrifugation (5 min, 680 × g). The cell extract was passed through a MACS magnetic separation column (Miltenyi Biotec). Columns were placed between two Sm-Co magnets, which generated a magnetic field that magnetized the column wire matrix and produced strong magnetic field gradients near the wires that resulted in trapping of the magnetic particles in the matrix. Bound magnetic particles were rinsed first with 50 ml of 10 mM HEPES-200 mM NaCl (pH 7.4) and then with 100 ml of 10 mM HEPES (pH 7.4). After the column was removed from the magnet, magnetic particles were eluted from the column by flushing it with 10 mM HEPES buffer. Finally, the magnetosome suspension was loaded on top of a sucrose cushion (55% [wt/wt] sucrose in 10 mM HEPES [pH 7.4]) and subjected to ultracentrifugation in a swinging bucket rotor. The magnetic particles were completely pelleted after centrifugation for 12 h at 280,000 × g and 4°C.
For analysis of protein binding characteristics, the isolated magnetosomes of MSR-1 were subjected to two different treatments, as follows.
For selective solubilization, aliquots (50 μg, wet weight) of isolated magnetosomes were resuspended in either 2% (wt/vol) Triton X-100, 2% (wt/vol) Tween 20, 500 mM octylglucoside (zwitterionic), 5% (wt/vol) sodium dodecyl sulfate (SDS) (cationic), 5 M urea, or 2 M NaCl. After they were shaken at room temperature for 2 h, the suspensions were centrifuged for 30 min at 13,000 × g. The pellets were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) after extensive washing.
For limited proteolytic digestion, aliquots (50 μg) of isolated magnetosomes were resuspended in 10 mM potassium phosphate buffer (pH 7.2) and were separately treated with 40 μg of trypsin (20 min), proteinase K (5 min), or pronase (5 min) per ml and incubated at 37°C with slight shaking. Digestion was stopped by addition of either trypsin inhibitor or trichloroacetic acid for proteinase K. Pronase was removed by extensive washing with 10 mM Tris-HCl. After centrifugation for 10 min at 13,000 × g, the pellets were analyzed by SDS-PAGE.
Gel electrophoresis.
Protein concentrations were measured with a BCA-Protein Micro assay kit (Pierce) used according to the manufacturer's instructions. For one-dimensional (1D) SDS-PAGE of magnetosome-associated proteins we used the procedure of Laemmli (26). An amount of magnetosome particles or solubilisate equivalent to 20 μg of protein was mixed with electrophoresis sample buffer containing 2% (wt/wt) SDS and 5% (wt/vol) 2-mercaptoethanol. After the samples were boiling for 5 min, they were centrifuged for 3 min. The supernatants were loaded onto polyacrylamide gels containing various concentrations of polyacrylamide (8 to 16%). Tricine-SDS-PAGE was performed as described by Schägger and von Jagow (43).
Two-dimensional (2D) PAGE was carried out as described previously (9, 15, 28). Approximately 60 μg of magnetosome protein was used for isoelectric focusing. Carrier ampholytes (BDH Laboratory Supplies, Poole, United Kingdom) were used to generate a pH gradient from 3.5 to 10 with a model 175 chamber from Bio-Rad (Munich, Germany). For the second dimension, Tris-SDS-PAGE was used (9 to 16% acrylamide gradient gels; Bio-Rad Protean II). 1D and 2D gels were stained with Coomassie brilliant blue R-250 (Serva) or with silver.
Digitized gels were analyzed with the Image Master 1D software (v.3.0; Amersham-Pharmacia) or with the Melanie II software package (2D gels) (2).
After electrophoresis, the proteins were electroblotted onto a polyvinylidene difluoride membrane (ProBlott; Applied Biosystems) according to the manufacturer's instructions by using a semidry blot apparatus (Hoefer).
N-terminal amino acid sequence analysis.
Proteins (20 to 50 pmol) were sequenced after blotting onto a polyvinylidene difluoride membrane by automated N-terminal Edman degradation with an Applied Biosystems (Foster City, Calif.) pulsed liquid-phase sequencer (model 477A) under standard conditions. Phenylthiohydantoin derivatives of amino acids were identified with an on-line analyzer (model 120A; Applied Biosystems) with a repetitive yield of 92 to 95%.
Total tryptic digest of whole magnetosomes.
An aliquot of a magnetosome suspension equivalent to approximately 40 μg of protein was pelleted with a Dynal magnet (Dynal A.S., Oslo, Norway). The isolated wet magnetosomes were resuspended in 50 μl of degassed 6 M guanidine hydrochloride-0.5 M Tris-HCl-2 mM EDTA (pH 7.5) and reduced for 1 h at 37°C with 2 μl of an aqueous solution containing 2 μmol of dithiothreitol. Alkylation was performed by adding of 20 μl (20 μmol) of a 1 M solution of iodoacetamide in 6 M guanidine hydrochloride for 30 min. The alkylated magnetosomes were pelleted again and washed three times with 200 μl of 0.1 M Tris-HCl (pH 8.0). After resuspension in 50 μl of 0.1 M Tris-HCl (pH 8.0) containing 10% acetonitrile, the magnetosome-associated proteins were digested with 5 μg of trypsin (Promega) overnight. The magnetite moiety was separated with a magnet, and the supernatant was used for mass spectrometric (MS) experiments.
MS analysis.
Samples were identified after chromatographic separation of the peptide mixture on an LC Packings 75-m PepMap C18 column (Dionex, Idstein, Germany) by using a capillary liquid chromatography (CapLC) system delivering a gradient of from 5.7 to 80% acetonitrile and 0.1% formic acid at 200 nl/ml. The eluting peptides were ionized by electrospray ionization by using a Q-TOF hybrid mass spectrometer (Micromass, Manchester, United Kingdom). This instrument, in automated switching mode, selected precursor ions based on intensity for peptide sequencing by collision-induced fragmentation tandem MS. The MS-MS analyses were conducted by using collision energy profiles that were chosen based on the m/z value, the charge state of the parent ion, and the fragment ion masses and intensities, and the results were correlated with the preliminary MSR-1 nucleic acid databases by using the Mascot software (38). The results were validated manually.
Analytical methods.
The iron content of magnetosomes was determined as described previously (20, 22). Lipids were extracted from the magnetosomes and whole cells as previously described (8, 11). Quantitative determination of phospholipids was performed as described by Kates (25). Fatty acids were analyzed as the methyl ester derivatives prepared from 10 mg of dry cell material. Cells were subjected to differential hydrolysis in order to detect ester-linked and non-ester-linked (amide-bound) fatty acids (B. Tindall, unpublished data). Fatty acid methyl esters were analyzed by gas chromatography by using a nonpolar capillary column (0.2 μm by 25 m; film thickness, 0.33 μm; internal column diameter, 0.2 mm; Hewlett-Packard Ultra 2) and flame ionization detection. The conditions used were as follows: injection and detector port temperature, 300°C; carrier gas, hydrogen at an inlet pressure of 60 kPa; split ratio, 50:1; injection volume, 1 μl; and a temperature program in which the temperatures was increased from 130°C (held for 2 min) to 310°C at a rate of 4°C min−1.
The unambiguous position of double bonds and the presence of hydroxy fatty acids were confirmed by gas chromatography-MS by using a Finnigan MAT GCQ as described previously (24). Hydroxy fatty acids were detected by their characteristic fragments at m/z 103 for 3-OH fatty acids and at m/z M+-59 for 2-OH fatty acids. Thin-layer chromatography and detection of polar lipids were carried out as described by Skipsi and Barlday (53).
Heme and glycostaining.
Isolated magnetosomes (approximately 20 μg of protein) were suspended in sample buffer containing 2% SDS and 5% 2-mercaptoehanol and were separated by 1D SDS-PAGE by using 16% polyacrylamide gels as described above; however, boiling of protein samples before electrophoresis was omitted. Heme staining was performed by using 3,3′,5,5′-tetramethylbenzidine as described by Thomas et al. (55) before Coomassie blue staining.
Protein glycosylation was assayed with an ECL glycoprotein detection module (Amersham-Pharmacia) used according to the manufacturer's instructions.
Electron microscopy.
Preparation and negative staining of magnetosomes were performed as described previously (20). Micrographs were taken with a Philips EM301 electron microscope at an acceleration voltage of 80 kV.
Analysis of DNA and protein sequence data.
Genome sequence data from M. gryphiswaldense MSR-1 were used from the whole genome shotgun in progress (45), at the present stage of eightfold sequencing coverage. The assembled shotgun sequences have a constant total contig length approaching 4.6 Mb; hence, they represent nearly the genome size of M. gryphiswaldense MSR-1 as estimated by pulsed-field gel electrophoresis.
Peptide sequences determined in this study were compared to the translated preliminary genome of MSR-1 and to the partial sequence encoded by the magnetosome island of MSR-1 (45) (accession number BX571797) by using the BLAST algorithm (1). Basic analyses of DNA and protein sequences were performed by using by the MacVector 7.0 software package (Oxford Molecular Ltd.). Sequence alignment was carried out by using the ClustalW algorithm (57), which is part of the same software. Protein sequences were compared to the GenBank, EMBL, and SwissProt databases. Motif searches were carried out by using the Prosite program (23). Protein location was analyzed by the PSORT program (31).
Sequences of all bona fide magnetosome genes were finished to satisfy the Bermuda Rules (http://www.ornl.gov/hgmis/research/Bermuda.html#1). Se-quences of proteins that represented putative contaminants (see Table 3) were analyzed by high-throughput genome annotation (40).
TABLE 3.
Summary of features of presumed protein contamination of the MM of M. gryphiswaldense identified by various sequencing methods
| Protein | 1D band no. | 2D spot no. | No. of peptides
|
Best Blast hit (accession no.)a | E value | |
|---|---|---|---|---|---|---|
| 2D PAGE + MS-MS | Total tryptic digestion + MS-MS | |||||
| Outer membrane protein | 7, 8 | 4 | Bracella melitensis (Q44662) | 4e-54 | ||
| ATP synthase alpha | 8, 9 | 6 | Rhodospirillum rubrum (PO5036) | 3e-11 | ||
| ATP synthase beta | 6 | 6 | 6 | 1 | Rhodospirillum rubrum (PO5038) | 0.0 |
| Ribosomal protein S1 | 4 | 4 | Bradyrhizobium japonicum (NP_767380) | 0.0 | ||
| EF-Tu | 7 | 4 | 2 | Masorhizobium loti BAC50667 (NP_102118) | 1e-07 | |
| FusA | 1 | 5 | Rickettsia bellii (AAM90927) | 0.0 | ||
| DnaK | 3 | 6 | Rhodopseudomonas sp. (O05700) | 0.0 | ||
| GroEL | 5 | 10 | Rhizobium meliloti (NP_437546) | 0.0 | ||
| Cytochrome c1 | 1 | Rhodospirillum rubrum (P23135) | 1e-63 | |||
| cb-type cytochrome c oxidase CcoO subunit | 1 | Brucella melitensis (NP_540482) | 1e-90 | |||
| Cytochrome b | 2 | Bradyrhizobium japonicum (NP_769126) | 1e-140 | |||
Excluding hits to other MTB.
Nucleotide sequence accession numbers.
Sequences of proteins that represented putative contaminants have been deposited in the GenBank, EMBL, and DDJB data libraries as draft sequences for the whole genome shotgun under accession number BX640510. The MM22 sequence has been deposited under accession number BX640511.
RESULTS
Isolation and biochemical analysis of magnetosomes from M. gryphiswaldense.
Using the procedure described in Material and Methods section, we obtained 40 mg of purified magnetosomes from 1 g (dry weight) of magnetic cells, which included approximately 99.5% of the total cell-bound iron. The higher magnetosome yields in this study than in a previous study (20) were due to the improved cultivation of bacteria in an oxystat fermentor, which resulted in a vastly increased intracellular magnetosome content (22).
The profile of MM-associated fatty acids is shown in Table 1. A number of common fatty acids were identified both in the whole cells and in the MM fraction. However, the amide-linked fatty acids 3-hydroxyhexadecanoic acid (3 OH 16:0), 3-hydroxyoctadecanoic acid (3OH 18:0), and 2-hydroxydecenoic acid (2OH 18:1), which are typically present in the outer membrane of gram-negative bacteria (41), were not detected in the MM fraction. The results of thin-layer chromatography in this study essentially confirmed the previous gas chromatographic analysis of polar lipids (4). Specifically, phosphatidylethanolamine and phosphatidylglycerol were identified as the most abundant polar lipids both in whole-cell extracts and MM, whereas ornithinamid lipid and an unidentified amino lipid were less abundant in the MM than in the fraction of lipids from whole cells (data not shown).
TABLE 1.
Fatty acid compositions of whole cells and magnetosomes of M. gryphiswaldense MSR-1 as determined by gas chromatography-MS
| Fatty acid | Relative amt (%)
|
|
|---|---|---|
| Whole-cell polar lipids | Magnetosome polar lipids | |
| 16:1ω7c | 21.384 | 21.707 |
| 16:0 | 8.947 | 11.860 |
| NDa | 0.411 | 1.671 |
| 3 OH 16:0 | 2.694 | |
| 18:1ω9c | 0.508 | 2.299 |
| 18:1ω7c | 55.972 | 57.734 |
| 18:1 | 0.882 | 2.724 |
| 2OH 18:1 | 2.636 | |
| 3OH 18:0 | 1.909 | |
| 2OH 19:1/cy 19:0 | 3.083 | 2.005 |
ND, fatty acid could not be identified by gas chromtography-MS due to an overlap with antifoam peaks.
Analysis of MM-associated proteins from M. gryphiswaldense by 1D SDS-PAGE.
To investigate the effect of the preparation protocol on the protein composition of the MM, the conditions used for magnetosome purification were varied; the method used for cell disruption, the buffer composition, the pH, the ionic strength, and other parameter were varied in numerous experiments. In summary, the standard procedure yielded the optimum results in terms of protein yield, reproducibility, and specificity (data not shown). For example, alternative methods used for cell disruption, including ultrasonication and lysozyme treatment, resulted in poor magnetosome yields and poor reproducibility. Both ultracentrifugation and subsequent magnetic separation were found to be crucial, as the omission of purification steps resulted in a complex, highly nonspecific protein pattern.
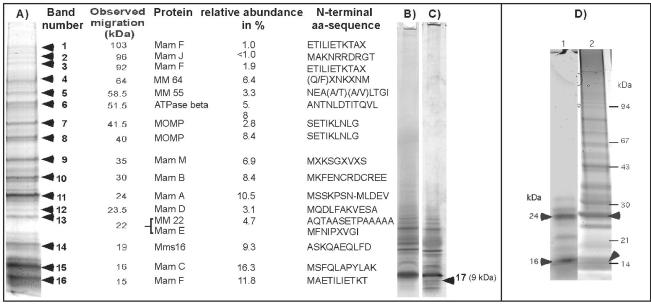
Depending on the electrophoresis conditions, between 15 and 20 major bands were detected by Coomassie blue staining in 1D PAGE gels (Fig. 1A). Several additional low-molecular-mass proteins, including band 17, were identified in Tris-Tricine-SDS gels (Fig. 1B and C) (43). With the exception of bands 4, 5, and 17, which yielded poor results, unambiguous N-terminal amino acid sequences were derived from 14 proteins.
FIG. 1.
Separation of MM-associated proteins by 1D PAGE. (A) Summary of MMPs detected by Coomassie blue staining in 1D SDS-16% PAGE gels. (B and C) Coomassie blue-stained (B) and silver-stained 16% SDS-Tricine gels of MMPs. The heterogeneous band at 9 kDa (band 17) revealed an ambiguous N-terminal amino acid sequence, M(G/Y/F/I)(P/Q/T/K)(L/I/V)(K/A)(M/G/V)(A/T/V/I), which partially contains the N terminus of MamG. (D) Heme staining of MMPs (lane 1), which were subsequently stained with Coomassie blue (lane 2). The arrowheads indicate the positions of heme-positive bands. aa, amino acid.
MM-associated proteins were tested for the presence of covalently bound heme by SDS-PAGE, followed by detection of heme-associated peroxidase activity (55). Two major positive bands at approximately 25 and 16 kDa, as well as several minor bands, were detected (Fig. 1D). Likewise, 1D PAGE-resolved MMPs were tested for glycosylation. However, no bands reacting with the glycostaining kit were detected in the MM.
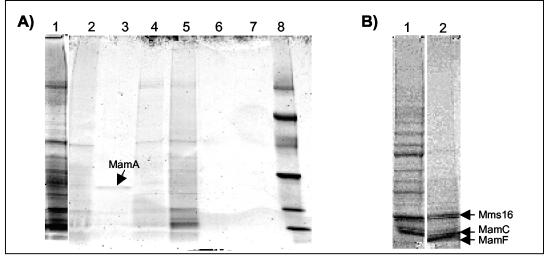
The characteristics of the MM-associated proteins were further analyzed by treating purified magnetosomes with various solubilizing agents (Fig. 2A). Treatment with hot (95°C) 1% SDS readily removed all proteins, whereas solubilization was less effective with cold (room temperature) 5% SDS. Several of the smaller bands (e.g., bands at 16 and 19 kDa) apparently were more resistant to solubilization, indicating that they are relatively tightly bound to the MM. While cationic denaturing detergents like SDS completely solubilized the MM, treatment with nonionic detergents (Triton X-100, Tween 20), as well as with zwitterionic detergents like octylglycosid, was less effective. A 24-kDa protein that was identified as MamA by N-terminal amino acid sequence analysis could be selectively solubilized by Tween 20. The apparently loose binding of this protein to the MM is consistent with its suggested peripheral association (37). Treatment with 5 M NaCl and urea had virtually no effect on the solubilization of MMPs.
FIG. 2.
Binding characteristics of the MM-associated proteins. (A) SDS-PAGE of proteins solubilized from the MM after treatment with various agents. Lane 1, 1% hot SDS (5 min); lane 2, Triton X-100 (2%); lane 3, Tween 20 (2%); lane 4, octylglucoside (500 mM); lane 5, SDS (5%); lane 6, urea (5 M); lane 7, NaCl (2 M); lane 8, low-molecular-weight marker. The arrow indicates the position of a 24-kDa band which was selectively solubilized by Tween 20 and was identified as MamA by its N-terminal amino acid sequence (MSSKPSN). (B) Lane 1, proteins from untreated magnetosomes; lane 2, proteins from magnetosomes after tryptic digestion. The arrows indicate the positions of protein bands resistant to partial tryptic digestion, which were identified as Mms16 (ASKQAEQLFD), MamC (MSFQLAPYLAK), and MamF (MAETILIETKT).
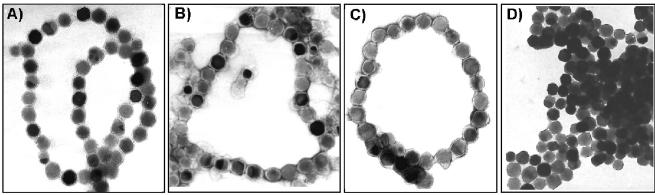
While pronase and proteinase K treatment caused total degradation of MM-associated proteins (data not shown), bands at molecular masses of 19, 16, and 15 kDa remained associated with the MM after limited tryptic digestion (Fig. 2B). These bands were identified as MamC, MamF, and Mms16. The incomplete digestion of these proteins suggests that they are partially protected against proteolytic degradation, perhaps because of their integral membrane localization. Protease treatment of magnetosomes resulted in macroscopic agglomeration of the isolated magnetosome particles, as observed after solubilization of the MM with detergents. This indicates that the presence of proteins is crucial for maintaining the integrity of the MM vesicles. Under these conditions, however, the membrane structure enveloping the crystals apparently was not entirely removed. Even after boiling with 1% SDS, which led to instant agglomeration of particles, traces of organic material remained attached to the crystals and a residual tendency to form chains was observed, although the particles were tightly spaced due to the MM solubilization (Fig. 3).
FIG. 3.
Electron micrographs of isolated magnetosome particles. (A) Untreated magnetosomes; (B) magnetosomes after treatment with Triton X-100; (C) magnetosomes after tryptic digestion; (D) magnetosomes after boiling with 1% SDS.
Comparison of MM-associated proteins from M. magnetotacticum and Magnetospirillum strain AMB-1.
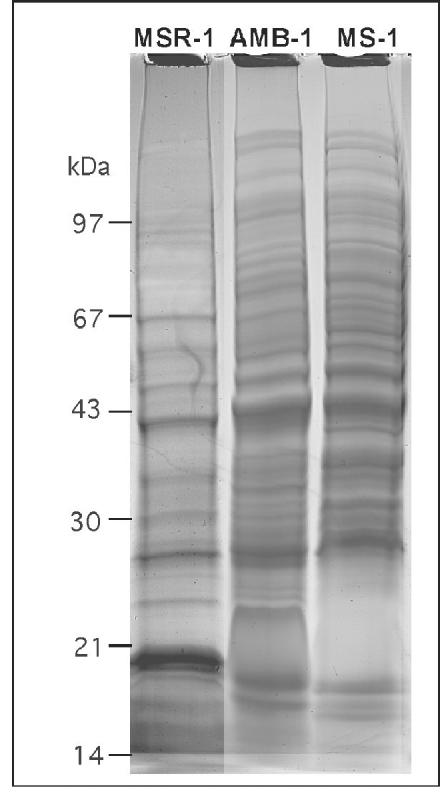
The closely related Magnetospirillum strains produce magnetite crystals which are virtually identical in terms of size, alignment, and crystal morphology. However, preliminary biochemical analysis of the MM by using different protocols with M. magnetotacticum MS-1 and Magnetospirillum strain AMB-1 revealed divergent protein patterns of the MM for strains MS-1, AMB-1, and MSR-1 (3, 18, 35-37). To clarify whether the observed differences were due to differences in the isolation procedure, magnetosome particles were purified from M. magnetotacticum and strain AMB-1 by using the same protocol that was used for M. gryphiswaldense MSR-1. The protein profiles of the solubilized MM from the three strains are shown in Fig. 4. Approximately 20 to 25 bands were identified in magnetosome preparations from MS-1 and AMB-1. While the protein patterns of AMB-1 and MS-1 were similar to each other and shared a number of bands, their composition was clearly distinct from that of MSR-1.
FIG. 4.
SDS-16% PAGE of MMPs from M. gryphiswaldense MSR-1, Magnetospirillum strain AMB-1, and M. magnetotacticum MS-1.
Identification of MM-associated proteins by MS analysis.
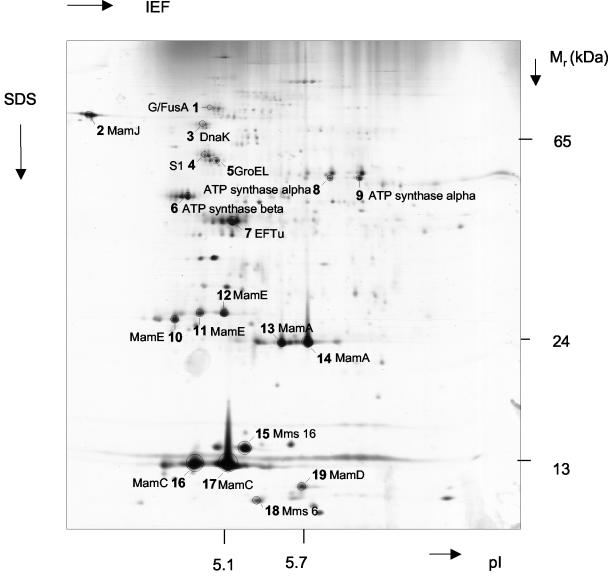
Results of the 2D analysis are shown in Fig. 5. Proteins were excised from 19 major spots on the Coomassie blue-stained gel and subjected to tryptic digestion and subsequent MS analysis. Several horizontal chains of spots with nearly identical molecular weights but different pIs were detected, which may be attributed to mistranslation, artificial chemical modification during protein preparation, or posttranslational modification.
FIG. 5.
Silver-stained 2D PAGE of MMPs from M. gryphiswaldense. Proteins from marked spots were excised from the corresponding Coomassie blue-stained gel and identified by electrospray ionization-MS-MS after tryptic digestion. IEF, isoelectric focusing.
Alternatively, proteins from magnetosome preparations were directly subjected to complete tryptic digestion and subsequent CapLC-MS-MS analysis without prior separation by electrophoresis. Overall, approximately 140 different spectra were identified, which could be assigned to at least 27 different proteins (Tables 2 and 3).
TABLE 2.
Summary of features of bona fide MMPs of M. gryphiswaldense identified by various sequencing methods
| Protein | Edman sequencing ID band no. | 2D spot no. | 2D PAGE + MS-MS
|
Total tryptic diges- tion + MS-MS
|
Length (amino acids) | Predicted molecular mass (kDa) | pI | Best Blast hit (accession no.)a | E value | Putative function | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coverage (%) | No. of peptides | Coverage (%) | No. of peptides | |||||||||
| MamA (Q93DY9) | 11 | 13, 14 | 35 | 7 | 40.6 | 7 | 217 | 24.01 | 5.64 | MM2348 of Methanosarcin (AE013478) | 3e-15 | Tetratricopeptide repeat protein |
| MamB (Q93DY6) | 10 | 13.8 | 4 | 297 | 31.96 | 5.25 | YdfM of Bacillus subtilis (C69781) | 1e-37 | CDF transporter | |||
| MamC (Q93DY1) | 15 | 16, 17 | 38.7 | 4 | 54.8 | 5 | 125 | 12.40 | 4.88 | Unknown | ||
| MamD (Q93DY2) | 12 | 19 | 6.1 | 2 | 14.3 | 3 | 314 | 30.20 | 9.68 | Unknown | ||
| MamE | 13 | 10 | 14.5 | 8 | 6.7 | 7 | 772 | 78.00 | 8.69 | MLL5022 of Rhizobium loti (Q98CS8) | 9e-35 | Serine protease |
| MamF | 1, 3, 16 | 26.8 | 3 | 111 | 12.30 | 9.57 | Unknown | |||||
| MamG | 17 | 84 | 7.70 | 9.28 | Unknown | |||||||
| MamJ | 2 | 2 | 12.7 | 5 | 16.3 | 4 | 466 | 48.51 | 3.80 | Unknown | ||
| MamM | 9 | 12.9 | 3 | 318 | 34.50 | 5.82 | BH 1238 of Bacillus halodurans (F83804) | 1e-33 | CDF transporter | |||
| MamN | 6.4 | 2 | 437 | 46.14 | 6.70 | TM0934 of Tharmatoga maritima (AE001757) | 6e-30 | Inorganic ion transport | ||||
| MamO | 17.5 | 7 | 632 | 65.40 | 6.51 | CC1282 of Canlobacter crescentus (C87408) | 6e-15 | Serine protease | ||||
| MamQ | 10.3 | 2 | 272 | 30.00 | 8.48 | T. maritima (AE001759) | 3e-16 | Unknown | ||||
| MamR | 27.4 | 2 | 72 | 8.10 | 8.48 | Unknown | ||||||
| MamS | 7.7 | 1 | 180 | 18.71 | 7.02 | Unknown | ||||||
| MamT | 5.7 | 1 | 174 | 18.88 | 10.05 | Unknown | Heme binding | |||||
| Mms6 | 18 | 17.5 | 2 | 136 | 14.26 | 9.79 | —b | Iron binding | ||||
| Mms16 | 14 | 15 | 67.6 | 9 | 145 | 16.35 | 5.49 | Apm of Rhodospirillum rubrum (ZP_00014946)c | 6e-28 | Activator for PHB depolymerase | ||
| MM22 | 13 | 196 | 20.00 | 7.14 | Enterococcus faecalis V583 (NP_816281) | 3e-07 | Conserved hypothetical protein | |||||
Identification and analysis of genes encoding MM-associated proteins.
ORFs corresponding to all of the peptide sequences determined were identified in the unfinished genome assembly of MSR-1. The majority of these ORFs could be assigned to genes in the previously identified putative magnetosome island of M. gryphiswaldense (BX571797) (45).
Based on sequence analysis, the MM-associated proteins can be divided into two groups. The first group comprises proteins classified as bona fide MMPs based on (i) their presence in major bands or spots in 1D and 2D gels, (ii) no or only remote similarity to proteins in organisms other than MTB, and (iii) colocalization in operons together with other genes encoding known MMPs. These known MMPs include MamA, MamB, MamC, MamD, MamE, MamF, MamG, MamJ, MamM, MamN, MamO, MamQ, MamR, MamS, MamT, MM22, Mms6, and Mms16 (Table 2). While MamA, MamB, MamC, MamD, and MamE were previously identified in the MM of M. gryphiswaldense (20), the rest of the MMPs are new for this organism and are described below.
The second group comprises proteins with unequivocal homology to ubiquitous, biochemically well-characterized, and very abundant proteins in other (nonmagnetic) organisms (Table 3). They most likely do not represent true MMPs, but their association with the MM is due to nonspecific adsorption during cell disruption.
Characteristics of bona fide MMPs. (i) Mms16.
Spot 15 and band 14 correspond to a protein that exhibits 80% identity to Mms16 of Magnetospirillum strain AMB-1 (SwissProt accession number Q93UW2), which was the most abundant protein associated with the magnetosomes in this strain (35). Because of its observed in vitro GTPase activity, it has been suggested that this protein is involved in the formation of MM vesicles. Genes with similarity to mms16 are present in the genomes of a number of nonmagnetic bacteria. For instance, in Rhodospirillum rubrum, a similar protein (Apd; 70% similarity) seems to be involved in the activation of a depolymerase for hydrolysis of PHB (R. Handrick and D. Jendrossek, unpublished data). This protein also displays weaker sequence similarity to phasine-like, PHB granule-associated proteins from several bacteria.
(ii) Mms6 and MamG.
The apparent molecular mass of the Mms6 protein of M. gryphiswaldense (spot 18) is significantly less than 12.7 kDa, as calculated from the full-length gene, potentially due to proteolytic cleavage of a premature propeptide. It displays homology only to other proteins found exclusively in MTB, especially Mms6 of Magnetospirillum strain AMB-1 (Q83VL7; 67% identity). Recently, Mms6 was described in the latter strain as a tightly bound constituent of the MM, which exhibited in vitro iron binding activity (3).
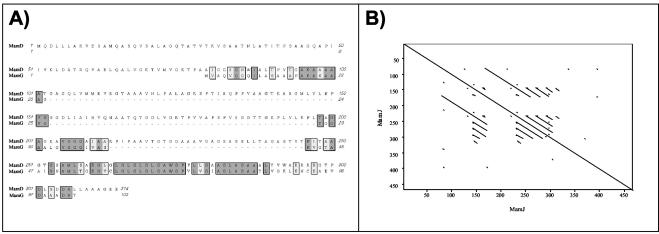
The ambiguous N-terminal amino acid sequence obtained from heterogeneous band 17 partially matches the N terminus of the deduced polypeptide encoded by the mamG gene (45) and thus seems to represent the MamG protein that is similar to Mms5 from AMB-1 (Q83VL6) (3). Interestingly, MamG and MamD exhibited extensive sequence identity with each other (Fig. 6A) and had the hydrophobic sequence motif KGXXLGLGL/MGLGAWGPXXLG in common with Mms6. This motif exhibits an intriguing similarity to LG-rich repetitive sequences found in silk-like (fibroin) proteins (59) and mollusk shell framework proteins (54), as well as elastins and cartilage proteins (12), which are known to have a remarkable tendency for self-aggregation; several of these proteins are involved in other biomineralization processes.
FIG. 6.
Selected sequence characteristics of several MMPs. (A) Sequence alignment of MamG and MamD of M. gryphiswaldense. (B) Dot blot analysis of MamJ by Pustell protein matrix analysis (39) (pam 250 matrix).
(iii) MamJ.
MamJ was identified by 1D and 2D PAGE, as well as in the MMP digest. The apparent mass (approximately 96 kDa) in 1D PAGE suggests that this band may represent an undissociated homodimer of MamJ. The MamJ protein exhibits extensive self-similarity and is particularly rich in acidic residues (14% E, 4.75% D), which are organized in several conspicuous repetitive motifs (Fig. 6B).
(iv) MamM.
Band 9 and three peptide fragments from the MM tryptic protein digest correspond to a protein encoded by the mamM gene of the mamAB cluster in M. gryphiswaldense (45). The MamM protein exhibits sequence similarity (47%) to the previously identified MamB protein of MSR-1 and represents another MMP with significant homology to cation diffusion facilitator (CDF) transporters (20).
(v) MamF.
The considerable difference between the calculated masses and the molecular masses of the two large bands (92 and 103 kDa) in 1D PAGE gels is consistent with the occurrence of undissociated oligomers (hepta- or hexamers) of the MamF protein, while the 15-kDa band seems to correspond to the monomeric protein. Altogether, the three bands (bands 1, 3, and 16) represent the second-most-abundant protein (14.8%) in the MM after MamC (16.3%). Hydropathy plots of MamF predict that it is a highly hydrophobic protein with two transmembrane helices. An integral membrane location would be also consistent with the observed resistance of this protein to limited tryptic digestion. BLAST searches failed to identify homologues of MamF except for hypothetical proteins in magnetotactic strains MS-1 and MC-1.
(vi) MM22.
An ORF encoding the N-terminal sequence of band 13 was identified, and this sequence is preceded by five additional amino acid residues, including the initial methionine. The MM22 protein is likely to be associated with the membrane and exhibits similarity with conserved hypothetical proteins from various bacteria. No homologues were identified in other MTB.
(vii) MamN, MamO, MamQ, MamR, MamS, and MamT.
The MamN, MamO, MamQ, MamR, MamS, and MamT proteins were identified only in tryptic digests of the total MM, while no corresponding bands or spots were detected in 1D and 2D PAGE gels. With the exception of MamN and MamO, there is no similarity or only a low level of similarity with well-characterized proteins in organisms other than MTB. Both MamO and the previously identified MamE protein exhibit sequence similarity to HtrA-like serine proteases, although MamO apparently does not contain a highly conserved PDZ domain and the two proteins exhibit only relatively weak (31%) sequence similarity to each other. MamN exhibits some similarity to several transport proteins. MamT contains two putative cytochrome c heme binding sites (Cys-X-X-Cys-His), which are conserved in the predicted MamT proteins from MS-1 and MC-1 (data not shown). As the deduced mass of MamT (19 kDa) differs from the masses of the MM heme-positive bands in 1D SDS-PAGE gels (Fig. 1D), it is not clear that MamT corresponds to one of these bands.
DISCUSSION
Whereas the lipid and fatty acid patterns were similar for the MM and other subcellular compartments and mainly differed in the relative proportions, the subproteome of the MM is complex and distinct, as revealed by different approaches.
While 1D analysis has limited resolving power, it has several advantages over 2D analysis, such as the ability to separate highly hydrophobic membrane proteins and a tolerance for higher protein loads. Accordingly, 11 MMPs were identified by 1D analysis, compared with the 7 proteins identified by 2D analysis, while one protein (Mms6) found in the 2D analysis was absent from 1D patterns. The highest number of MMPs was identified in unresolved, tryptic digests of the entire MM by CapLC-MS-MS, and these proteins included all of the proteins found by 1D and 2D analyses except Mms6. For some proteins, total sequence coverage of more than 50% was obtained. Due to its high sensitivity, MS analysis also detected a high proportion of putative contaminants. Other advantages of liquid chromatography-MS-MS methods are that they can handle extremely complex peptide mixtures and the sample remains in solution throughout preparation and subsequent analysis. Hence, losses associated with poor recovery from gels are eliminated. In conclusion, the combination of the different approaches is likely to provide the maximum amount of information.
Although the procedure for magnetosome isolation was quite stringent, we found a number of proteins associated with preparations of the MM which apparently are not bona fide MMPs. Most of these proteins were only minor constituents of the MM and escaped detection by Coomassie blue staining. They mostly included very abundant and ubiquitous cellular constituents, such as outer membrane proteins, ATPase subunits, ribosomal proteins, and respiratory chain constituents. Thus, they are likely to represent contamination originating from other subcellular compartments, which became adsorbed to the particles during cell disruption. Cross-contamination by tightly bound proteins from other cell fractions is a common observation that has also been reported for isolation of other complex intracellular structures, such as PHB granules and endospores (27, 30, 58). However, the classification of putative contaminants is somewhat arbitrary, and we cannot entirely exclude the possibility that some of these proteins are associated with the MM in vivo. For example, it could be envisioned that proton-translocating ATPase enzymes, which were among the putative contaminants, might play a role in the energetization of the MM. On the other hand, we classified Mms16 of M. gryphiswaldense MSR-1 as a bona fide MMP based on its high level of identity to the AMB-1 protein, which was reported to be essential in magnetosome formation (35). However, because of the striking homology to proteins involved in PHB metabolism, the specific role of this protein in biomineralization requires further clarification.
Of the 18 bona fide MMPs that were identified in this study, 13 are novel for MSR-1, and 11 have not been identified in other MTB previously. With the exception of MM22, all MMPs have highly similar homologues in other MTB. Mms16 appears to be restricted to magnetospirilla and is absent from strain MC-1. MamC, MamD, MamF, MamG, MamJ, MamR, MamS, MamT, and Mms6 exhibit no homology to known proteins in nonmagnetic organisms and therefore seem to represent MTB-specific protein families. Other MMPs (MamA and MamQ) have homologues with diverse or unknown functions in nonmagnetic organisms. The other MMPs can be assigned to the HtrA-like serine protease family (MamE and MamO) and to the ion-transporting protein group (MamN, MamB, and MamM). Identification of MamM as a second MM-specific CDF transporter in addition to MamB lends further support to the hypothesis that these proteins belonging to the CDF3 subfamily of putative iron transporters (33) are involved in the magnetosome-directed transport of iron. Likewise, the MamO sequence predicts that there is an additional MM-bound serine protease in addition to MamE and corroborates the speculated function of HtrA-like proteins in magnetosome formation (e.g., in the processing and maturation of MMPs during MM assembly). At least one MMP (MamT) was found to contain conserved heme binding sites. The identification of redox proteins may extend the range of putative biochemical functions associated with the MM to redox cycling of iron during the formation of the iron oxide magnetite (Fe3O4).
One noticeable feature common to several MMPs is the presence of repetitive motifs. Examples are found in MamD, Mms6, and MamG, all of which have a hydrophobic LG-rich motif known from other biomineralizing proteins. Another sequence pattern with potential relevance for magnetite biomineralization is found in the acidic protein MamJ; in this protein 18.7% of the acidic amino acid residues are glutamate and aspartate that occur in multiplets. A number of additional conspicuous genes predicting proteins with highly repetitive and/or acidic sequence motifs are found in the genome assemblies of MSR-1 and other MTB (data not shown). Clusters of acidic groups are commonly found in proteins and polysaccharides involved in other biomineralizing systems, such as those in mollusk shells, coccolithophorids, and other organisms (6, 19). Acidic groups have a strong affinity for metal ions and are often involved in the initiation of crystal nucleation by binding of metal ligands. It is therefore tempting to speculate that MamJ and other acidic or repetitive proteins are directly involved in the control of the biomineralization process by providing iron binding activity.
In an effort to clarify the existence of multiple bands for individual gene products, we considered the possibility of posttranslational modification by glycosylation. However, we failed to identify reacting bands in the MM by glycostaining. This finding is of particular interest, as glycoproteins are common constituents of other biomineralization systems (19).
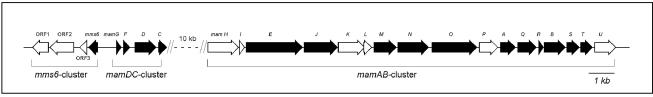
With the exception of Mms16 and MM22, all bona fide MMPs could be assigned to previously identified genes in MSR-1, which are organized in the mms6, mamDC, and mamAB gene clusters located in the putative magnetosome island (20, 45). A scheme for the assignment of MMP-encoding genes and their organization is shown in Fig. 7. Of the 25 identified genes in these three operon-like clusters, 16 now can be assigned to proteins associated with the MM, thereby confirming previous speculation that these clusters encode other magnetosome proteins (20, 45). Nevertheless, we were unable to assign 9 of the 25 genes from this region to identified MMPs. Despite the high sensitivity of our method, some of the proteins might have escaped detection, might have resisted efficient solubilization, or might have been lost during purification due to their loose association with the MM. Other proteins might play different roles related to magnetosome biomineralization, such as roles in regulation or intracellular differentiation, which are not necessarily associated with localization at the MM. Further experiments, such as in situ localization studies, are required to analyze the expression of these gene products.
FIG. 7.
Molecular organization of a region from the putative magnetosome island of M. gryphiswaldense comprising the entire mms6, mamDC, and mamAB gene clusters (45). The solid arrows indicate genes encoding MMPs, which were identified in this study.
Although their genes are present in the M. gryphiswaldense MSR-1 genome, we also failed to detect homologues of MpsA and MagA, which were previously found to be associated with isolated magnetosomes in strain AMB-1 (32, 36). Analysis of the MMP patterns for other magnetospirilla revealed a remarkable dissimilarity between MSR-1 and the two other strains analyzed. This was somewhat surprising, because a close genetic similarity between MSR-1 and MS-1 can be inferred from available genome data. Moreover, several MMPs (MamA, MamD, MamC, Mms6, and Mms16) were previously reported to be present in strain AMB-1 (3, 35, 36).
With the emerging picture of the astonishingly complex protein composition of the MM, the following questions arise: How is such a macromolecular structure assembled, and how are the MMPs targeted specifically to the MM? Several bands and spots (e.g., MamE and Mms6) had an apparent mass that was substantially lower than that predicted on the basis of their genes, which might have been due to posttranslational cleavage at presumptive signal sequences during maturation and MM insertion. However, in a preliminary analysis we failed to detect any obvious sequence motifs or sorting signals universal to all MMPs. While PSORT analysis predicted an inner membrane localization for most of the MMPs, for others (e.g., MamA, MamR, and Mms16) a cytoplasmic localization was presumed. Consistent with this, most MMPs have the characteristics of typical membrane proteins based on hydropathy plots and structural predictions, whereas other MMPs are hydrophilic. This means that the binding to the MM cannot be exclusively by hydrophobic interactions but for some proteins may involve other types of interactions, such as protein-protein interactions or direct interactions with the mineral surface of magnetite crystals. Alternatively, it might be speculated that assembly of the MM is mediated by scaffolding proteins, which are often required for proper assembly of macromolecular complexes, such as virus capsids and other complex subcellular structures (16). Involvement of similar functions might be envisaged during MM assembly.
In conclusion, our study was the most comprehensive biochemical analysis of the MM in an MTB so far. Identification of the magnetosome subproteome provided candidates for further biochemical and genetic analysis to elucidate their specific functions in magnetosome assembly and magnetite biomineralization. In addition, our findings have relevance for future uses of bacterial magnetosomes in biotechnological applications.
Acknowledgments
This study was supported by DFG grant SPP 1104 and by the BMBF Biofuture Program.
We thank Udo Heyen (Max-Planck-Institut, Bremen, Germany) for help with fermentor cultivation and Jana Richter (Max-Delbrück-Centrum, Berlin, Germany) and Katja Schmidt (Max-Planck-Institut, Bremen, Germany) for excellent technical assistance. We are grateful to Brian Tindall (Deutsche Sammlung von Microorganismen und Zellkulturen, Braunschweig, Germany) for his help with fatty acid and lipid analysis and to Mohamed Madkour and Frank Mayer (Universität Göttingen) for help with electron microscopy. Cathrin Wawer (Max-Planck-Institut, Bremen, Germany) is gratefully acknowledged for critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appel, R. D., P. M. Palagi, D. Walther, J. R. Vargas, J. C. Sanchez, F. Ravier, C. Pasquali, and D. F. Hochstrasser. 1997. Melanie II-A third-generation software package for analysis of two-dimensional electrophoresis images. I. Features and user interface. Electrophoresis 18:2724-2734. [DOI] [PubMed] [Google Scholar]
- 3.Arakaki, A., J. Webb, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278:8745-8750. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerlein, E. 2000. Single magnetic crystals of magnetite (Fe3O4) synthesized in intracytoplasmic vesicles of Magnetospirillum gryphiswaldense, p. 61-80. In E. Baeuerlein (ed.), Biomineralization. Wiley-VCH, Weinheim, Germany.
- 5.Balkwill, D., D. Maratea, and R. Blakemore. 1980. Ultrastructure of a magnetotactic spirillum. J. Bacteriol. 141:1399-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäuerlein, E. 2003. Biomineralization of unicellular organisms: an unusual membrane biochemistry for the production of inorganic nano- and microstructures. Angew. Chem. Int. Ed. Engl. 42:614-641. [DOI] [PubMed] [Google Scholar]
- 7.Bazylinski, D. 1995. Structure and function of the bacterial magnetosome. ASM News 61:337-343. [Google Scholar]
- 8.Benning, C., J. T. Beatty, R. C. Prince, and C. R. Somerville. 1993. The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc. Natl. Acad. Sci. 90:1561-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjellqvist, B., C. Pasquali, F. Ravier, J. C. Sanchez, and D. Hochstrasser. 1993. A nonlinear wide-range immobilized pH gradient for two-dimensional electrophoresis and its definition in a relevant pH scale. Electrophoresis 14:1357-1365. [DOI] [PubMed] [Google Scholar]
- 10.Blakemore, R. 1975. Magnetotactic bacteria. Science 190:377-379. [DOI] [PubMed] [Google Scholar]
- 11.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 12.Bochicchio, B., A. Pepe, and A. M. Tamburro. 2001. On (GGLGY) synthetic repeating sequences of lamprin and analogous sequences. Matrix Biol. 20:243-250. [DOI] [PubMed] [Google Scholar]
- 13.Bryant, D. A., E. V. Vassilieva, N. U. Frigaard, and H. Li. 2002. Selective protein extraction from Chlorobium tepidum chlorosomes using detergents. Evidence that CsmA forms multimers and binds bacteriochlorophyll a. Biochemistry 41:14403-14411. [DOI] [PubMed] [Google Scholar]
- 14.Cannon, G. C., C. E. Bradburne, H. C. Aldrich, S. H. Baker, S. Heinhorst, and J. M. Shively. 2001. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl. Environ. Microbiol. 67:5351-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diederichs, J. E., D. Groth, and R. Reszka. 1998. Plasma protein adsorption patterns onto cationic liposomes and lipoplexes—influence of cationic lipid head group. J. Liposome Res. 8:52-53. [Google Scholar]
- 16.Dokland, T. 1999. Scaffolding proteins and their role in viral assembly. Cell. Mol. Life Sci. 56:580-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel, R. B., D. A. Bazylinski, M. S. Johnson, and B. L. Taylor. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorby, Y. A., T. J. Beveridge, and R. P. Blakemore. 1988. Characterization of the bacterial magnetosome membrane. J. Bacteriol. 170:834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotliv, B. A., L. Addadi, and S. Weiner. 2003. Mollusk shell acidic proteins: in search of individual functions. Chem. Biochem. 4:522-529. [DOI] [PubMed] [Google Scholar]
- 20.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herborn, C. U., N. Papanikolaou, R. Reszka, K. Grünberg, D. Schüler, and J. F. Debatin. 2003. Magnetosomes as biological model for iron binding: relaxivity determination with MRI. Rofo Fortschr. Geb. Rontgenstr. Neuen. Bildgeb. Verfahr. 175:830-834. [DOI] [PubMed] [Google Scholar]
- 22.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hougardy, A., B. J. Tindall, and J. H. Klemme. 2000. Rhodopseudomonas rhenobacensis sp. nov., a new nitrate-reducing purple non-sulfur bacterium. Int. J. Syst. Evol. Microbiol. 50:985-992. [DOI] [PubMed] [Google Scholar]
- 25.Kates, M. 1972. Laboratory techniques in biochemistry and molecular biology, vol. 3. American Elsevier Publishing Co., Inc., New York, N.Y.
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lai, E. M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lück, M., B. R. Paulke, W. Schröder, T. Blunk, and R. H. Müller. 1998. Analysis of plasma protein adsorption on polymeric nanoparticles with different surface characteristics. J. Biomed. Mater. Res. 39:478-485. [DOI] [PubMed] [Google Scholar]
- 29.Matsunaga, T., N. Tsujimura, H. Okamura, and H. Takeyama. 2000. Cloning and characterization of a gene, mpsA, encoding a protein associated with intracellular magnetic particles from Magnetospirillum sp. strain AMB-1. Biochem. Biophys. Res. Commun. 268:932-937. [DOI] [PubMed] [Google Scholar]
- 30.McCool, G. J., and M. C. Cannon. 1999. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J. Bacteriol. 181:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura, C., T. Kikuchi, J. G. Burgess, and T. Matsunaga. 1995. Iron-regulated expression and membrane localization of the magA protein in Magnetospirillum sp. strain AMB-1. J. Biochem. 118:23-27. [DOI] [PubMed] [Google Scholar]
- 33.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 34.Offner, S., U. Ziese, G. Wanner, D. Typke, and F. Pfeifer. 1998. Structural characteristics of halobacterial gas vesicles. Microbiology 144:1331-1342. [DOI] [PubMed] [Google Scholar]
- 35.Okamura, Y., H. Takeyama, and T. Matsunaga. 2001. A magnetosome-specific GTPase from the magnetic bacterium Magnetospirillum magneticum AMB-1. J. Biol. Chem. 276:48183-48188. [DOI] [PubMed] [Google Scholar]
- 36.Okamura, Y., H. Takeyama, and T. Matsunaga. 2000. Two-dimensional analysis of proteins specific to the bacterial magnetic particle membrane from Magnetospirillum sp. AMB-1. Appl. Biochem. Biotechnol. 84-86:441-446. [DOI] [PubMed] [Google Scholar]
- 37.Okuda, Y., K. Denda, and Y. Fukumori. 1996. Cloning and sequencing of a gene encoding a new member of the tetratricopeptide protein family from magnetosomes of Magnetospirillum magnetotacticum. Gene 171:99-102. [DOI] [PubMed] [Google Scholar]
- 38.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 39.Pustell, J., and F. C. Kafatos. 1984. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 12:643-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabus, R., M. Kube, A. Beck, F. Widdel, and R. Reinhardt. 2002. Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 178:506-516. [DOI] [PubMed] [Google Scholar]
- 41.Ratledge, C., and S. G. Wilkinson. 1988. Microbial lipids, vol. 1. Academic Press, London, United Kingdom.
- 42.Reszka, R. 2000. Applications for magnetosomes in medical research, p. 81-92. In E. Baeuerlein (ed.), Biomineralization. Wiley-VCH, Weinheim, Germany.
- 43.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 44.Schleifer, K., D. Schüler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig, and M. Köhler. 1991. The genus Magnetospirillum gen. nov., description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst. Appl. Microbiol. 14:379-385. [Google Scholar]
- 45.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schüler, D. 2002. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int. Microbiol. 5:209-214. [DOI] [PubMed] [Google Scholar]
- 47.Schüler, D., and E. Baeuerlein. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180:159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schüler, D., and E. Baeuerlein. 1996. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 166:301-307. [DOI] [PubMed] [Google Scholar]
- 49.Schüler, D., and R. B. Frankel. 1999. Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl. Microbiol. Biotechnol. 52:464-473. [DOI] [PubMed] [Google Scholar]
- 50.Schüler, D., R. Uhl, and E. Baeuerlein. 1995. A simple light-scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132:139-145. [Google Scholar]
- 51.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 52.Seufferheld, M., M. C. Vieira, F. A. Ruiz, C. O. Rodrigues, S. N. Moreno, and R. Docampo. 2003. Identification of organelles in bacteria similar to acidocalcisomes of unicellular eukaryotes. J. Biol. Chem. 278:29971-29978. [DOI] [PubMed] [Google Scholar]
- 53.Skipsi, V. P., and M. Barlday. 1969. Detection of lipids on thin-layer chromatograms. Methods Enzymol. Lipids 14:541-548. [Google Scholar]
- 54.Sudo, S., T. Fujikawa, T. Nagakura, T. Ohkubo, K. Sakaguchi, M. Tanaka, K. Nakashima, and T. Takahashi. 1997. Structures of mollusc shell framework proteins. Nature 387:563-564. [DOI] [PubMed] [Google Scholar]
- 55.Thomas, P. E., D. Ryan, and W. Levi. 1976. An improved staining procedure for the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 56.Thomas-Keprta, K. L., S. J. Clemett, D. A. Bazylinski, J. L. Kirschvink, D. S. McKay, S. J. Wentworth, H. Vali, E. K. Gibson, Jr., and C. S. Romanek. 2002. Magnetofossils from ancient Mars: a robust biosignature in the martian meteorite ALH84001. Appl. Environ. Microbiol. 68:3663-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Todd, S. J., A. J. Moir, M. J. Johnson, and A. Moir. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J. Bacteriol. 185:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zurovec, M., and F. Sehnal. 2002. Unique molecular architecture of silk fibroin in the waxmoth, Galleria mellonella. J. Biol. Chem. 277:22639-22647. [DOI] [PubMed] [Google Scholar]