Abstract
Campylobacter jejuni is a major cause of diarrheal disease and food-borne gastroenteritis. The main reservoir of C. jejuni in poultry is the cecum, with an estimated content of 6 to 8 log10 CFU/g. If a flock is infected with C. jejuni, the majority of the birds in that flock will harbor the bacterium. Diagnostics at the flock level could thus be an important control point. The aim of the work presented here was to develop a complete quantitative PCR-based detection assay for C. jejuni obtained directly from cecal contents and fecal samples. We applied an approach in which the same paramagnetic beads were used both for cell isolation and for DNA purification. This integrated approach enabled both fully automated and quantitative sample preparation and a DNA extraction method. We developed a complete quantitative diagnostic assay through the combination of the sample preparation approach and real-time 5′-nuclease PCR. The assay was evaluated both by spiking the samples with C. jejuni and through the detection of C. jejuni in naturally colonized chickens. Detection limits between 2 and 25 CFU per PCR and a quantitative range of >4 log10 were obtained for spiked fecal and cecal samples. Thirty-one different poultry flocks were screened for naturally colonized chickens. A total of 262 (204 fecal and 58 cecal) samples were analyzed. Nineteen of the flocks were Campylobacter positive, whereas 12 were negative. Two of the flocks contained Campylobacter species other than C. jejuni. There was a large difference in the C. jejuni content, ranging from 4 to 8 log10 CFU/g of fecal or cecal material, for the different flocks tested. Some issues that have not yet promoted much attention are the prequantitative differences in the ability of C. jejuni to colonize poultry and the importance of these differences for causing human disease through food contamination. Understanding the colonization kinetics in poultry is therefore of great importance for controlling human infections by this bacterium.
Diarrheal disease and food-borne gastroenteritis are frequently caused by Campylobacter jejuni (10, 11, 25). C. jejuni is a zoonotic microorganism and can be isolated from poultry, cattle, pigs, pets, and wild animals, including birds. This microorganism represents a severe problem in poultry production. Up to 80% of the broiler flocks in several Western countries are infected (17). The distribution, however, is not uniform among different countries; for instance, the incidences are lower in several countries in the northern part of Europe. For example, the reported case for Norway for 1997 was that 6% of flocks were infected (27).
Detection of this important pathogen is difficult due to its special growth requirements, low infectious doses (17), and potential for entering a viable but not culturable state (3). The traditional diagnostic methods are both time-consuming and laborious, requiring prolonged incubations and selective enrichment to reduce the growth of background flora and to promote the growth of C. jejuni (15). Furthermore, the information obtained by traditional enrichment-based diagnostics is qualitative, while quantitative information is often required for control measurements (12).
C. jejuni does not normally multiply outside of the host. Still, it has the ability to survive extended periods in the environment (7). The main reservoir of C. jejuni in poultry is the cecum, with an estimated content of 6 to 8 log10 CFU/g (1). If a flock is infected with C. jejuni, then the majority of the birds in that flock will be bacterial carriers (1, 2, 24, 28). Detection at the flock level could thus be used for monitoring-based control of C. jejuni (4). Such a program has been initiated in Norway, where all the Norwegian flocks were tested for Campylobacter (The Norwegian Veterinary Institute, Oslo, Norway). However, a major challenge is that the traditional enrichment-based detection method takes 2 to 4 days from sampling to result (15).
Nucleic acid-based methods, in particular PCR methods, are promising tools for the rapid and direct detection of C. jejuni in animals used for food production. This is due to both the specificity and the sensitivity of the methods. Several qualitative PCR-based approaches have already been developed for the detection of C. jejuni (5, 6, 8, 18). Recently, quantitative PCR assays for C. jejuni in spiked foods (29), naturally contaminated foods after enrichment (23), and water (13) have also been developed. To our knowledge, no studies have yet utilized the true potential of real-time PCR for the direct quantification of C. jejuni in naturally contaminated material.
An important issue that is not yet addressed with quantitative DNA techniques is the ability of C. jejuni to colonize poultry. Quantitative information is important since the amount of Campylobacter found in poultry products is often correlated with the amount of Campylobacter present in the intestines of the birds. Furthermore, quantifications are important for understanding the colonization kinetics in poultry. This information is crucial in the control of Campylobacter (12).
The aim of the work presented here was to develop and evaluate a PCR-based assay for the rapid detection and quantification of C. jejuni directly from cecal and fecal samples. The challenges in developing such PCR tests are the semisolid nature of the test materials and the fact that these samples may contain very high levels of other bacteria. C. jejuni detection and quantification were done by using the same paramagnetic beads for cell concentration and DNA purification (Fig. 1) (14, 22). This integrated approach enabled a fully automated, rapid, and quantitative sample preparation and DNA extraction method. The integrated sample preparation approach was combined with both traditional end-point and real-time quantitative PCR detection. Furthermore, the integrated cell concentration and DNA purification approach was compared to other commonly used sample preparation methods. Results for direct qualitative and quantitative detection of C. jejuni in both spiked and naturally colonized cecal and fecal samples are presented.
FIG. 1.
Schematic representation of the integrated cell concentration and DNA purification approach. (A) Bacteria bound to the beads. (B) Cells are disrupted, and DNA binds to the beads. (C) Situation after washing. The cell debris is removed, and pure DNA is associated with the beads. Finally, the DNA is detected either by qualitative gel electrophoresis (D) or by real-time quantitative detection (E). A dilution series of C. jejuni from 101 to 105 CFU/ml of bacterial binding buffer (Genpoint AS) is shown for both detection approaches. Cecal content samples were used for this example.
MATERIALS AND METHODS
Test material.
The test material was obtained from chicken (Ross cultivar) flocks in the eastern part of Norway (Prior AS, Oslo, Norway). The chickens were fed with standard feed (Felleskjøopet AS, Oslo, Norway). Unless otherwise stated, <2-h-old feces (without urate crystals) were picked from the transport trays of 30- to 35-day-old chickens at the time of slaughter. The ceca were dissected from the intestinal contents after evisceration. Fecal and cecal samples for use in spiking experiments were stored on ice and were immediately frozen at −80°C upon arrival at the laboratory. Samples collected for the detection of natural infections were individually packed in plastic bags and sent for immediate processing at the test laboratory.
The C. jejuni strain LCD 8617 was used for the main experimental series. The C. jejuni strains ATCC 43442, ATCC 43436, ATCC 43438, and ATCC 43439 were used for investigation of the strain dependence of the assay used in this work. The C. jejuni strains were grown for 24 to 48 h in nutrient broth no. 2 (CM67; Oxoid, Basingstoke, England) at 37°C in a microaerophilic environment.
Culture-based detection.
A sterile Q-Tips cotton swab (Cheseborough-Ponds Inc.) was twisted in each fecal and cecal sample (four to eight and two samples, respectively, per flock, from individual birds) and submerged in a 3.5-ml screw-lid tube (Sarstedt, Nümbrecht, Germany) containing 3 ml of Bolton selective enrichment broth (CM983, SR183E, and SR48; Oxoid). The lids were tightly screwed and the tubes were incubated at 37°C for 24 h. Samples were then spread on Preston agar (CM689, SR204, SR84, and SR48; Oxoid) and incubated microaerophilically at 37°C for 48 h. Finally, at least five samples from all types of colonies, including those not suspected as Campylobacter, were resuspended in 100 μl of distilled H2O, boiled for 5 min, and investigated by PCR (conditions and primers are described below).
Sample preparation for direct PCR-based detection and quantification.
For spiking experiments, 0.5 g of a fecal sample was vortexed in 2 ml of medium (CM 67; Oxoid). The solid materials were subsequently sedimented for 15 min, and the liquid phase was used for the subsequent applications. The semisolid cecal content was separated from the intestinal canal before resuspension in medium (CM 67; Oxoid). For the experiment series to detect Campylobacter in naturally infected samples, a cotton swab was twisted in each fecal and cecal sample (four to eight and two samples, respectively, per flock, from individual birds) and submerged in a 3.5-ml screw-lid tube (Sarstedt, Nümbrecht, Germany) containing 3 ml of Bolton selective enrichment broth (CM983+, SR183, and SR 48; Oxoid). The tubes were inverted three to four times just prior to further use.
The Bugs'n Beads kit (Genpoint, Oslo, Norway) was used for sample preparation and genomic DNA isolation. Aliquots of 20 μl (10 mg/ml) of bacterial binding beads and 800 μl of binding and washing buffer were prepared. The test material (100 μl) was subsequently added, mixed gently by pipetting, and incubated at room temperature for 5 min. A magnetic separator (ABgene, Epsom, United Kingdom) was then used to separate the beads, with bound bacteria, from the solution. After removal of the supernatant, 50 μl of lysis buffer was added. The homogenized sample was then incubated at 80°C for 5 min before the addition of 150 μl of cold ethanol. Following this DNA precipitation step and bead attraction to a magnet, the supernatant was discarded and the DNA-bead complex was washed twice in 1 ml of 70% ethanol. The beads were finally resuspended in 30 μl of water and heated to 80°C for 5 min to remove residual ethanol. The whole process was done both manually and through the application of an automated process.
The process was automated by using a modified Tecan MiniPrep 75 robot (Tecan, San Jose, Calif.). Basically, the same steps were performed as in the manual process, with slight alterations of volumes and incubation times. The volume of the lysis buffer was increased to 100 μl, and the amount of ethanol used was increased to 210 μl. Finally, the DNA was resuspended in 60 μl of water.
A Dynabeads DNA DIRECT system I (Dynal, Oslo, Norway), Prepman sample preparation reagent for gram-negative food pathogens (Applied Biosystems, Foster City, Calif.), and DNeasy tissue kit (Qiagen, Hilden, Germany) were compared to the Bugs'n Beads system by using spiked cecal contents as samples. The starting material for all four kits (made as a master mix for optimal comparison) was a dilution series of C. jejuni spiked in 100-μl aliquots of prepared sample (described previously) corresponding to 10 mg of cecum. Pretreatments of the samples were done where required. For DNA DIRECT, the samples were suspended in 30 μl of buffer (500 mM Tris, 16 mM EDTA, 10 mM NaCl, pH 9.0) and centrifuged at 10,000 × g for 2 min. Samples tested with the Prepman and DNeasy tissue kits were centrifuged at 13,000 × g for 2 min prior to DNA purification.
Facilitators.
The effects of compounds that increase both the sensitivity and the amount of test material tolerated in a PCR were investigated. Bovine serum albumin (BSA) (Sigma, St. Louis, Mo.) was tested at various purities and with various modifications, in addition to polyethylene glycol (PEG) with a molecular weight of 1,500 (Sigma). The additive betaine was also tested. This compound has several proposed features, one of which is the facilitation of PCR with contaminated DNA (30).
Qualitative PCR amplification.
The primer sets U1112 and L1918 (Genpoint) and the amplification primers described by Nogva et al. (13), hereby denoted AB-F (5′CTG AAT TTG ATA CCT TAA GTG CAG C3′) and AB-R (5′CTG AAT TTG ATA CCT TAA GTG CAG C3′), were used for the qualitative amplification of C. jejuni. For U1112 and L1918, the following amplification conditions were used: 94°C for 4 min and then 37 to 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 1 min. An extension period of 7 min at 72°C was included after the reactions. For the primer pair AB-F and AB-R, the cycling conditions were as follows: 40 cycles of 95°C for 15 s and 60°C for 1 min, with denaturation and extension times as previously described. The 50-μl reaction mixtures contained 1× Dynazyme DNA polymerase II reaction buffer, 10 pmol of each primer, a 200 μM concentration of each deoxynucleoside triphosphate, and 0.5 to 1 μl of Dynazyme DNA polymerase II (2 U/μl) (Finnzymes Oy, Espoo, Finland). The reactions contained 0.2% BSA, unless otherwise stated.
5′-Nuclease PCR quantification.
Real-time quantitative PCR amplification was conducted as described by Nogva et al. (13). The 50-μl reaction mixtures contained 1× TaqMan buffer, 5 mM MgCl2, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 400 μM dUTP, 0.02 μM C. jejuni-specific probe (5′TCT CCT TGC TCA TCT TTA GGA TAA ATT CTT TCA CA3′), 0.3 μM (each) C. jejuni-specific primers AB-F and AB-R, 1 U of AmpErase urasil N-glycosylase, and 2.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems). Unless otherwise stated, the reactions contained 0.2% BSA.
The amplification conditions used was as follows: 40 cycles of 95°C for 20 s and 60°C for 1 min. The reactions were performed in an ABI Prism 7700 sequence detection system (Applied Biosystems). Carboxyfluorescein was used as a reporter dye, and 6-carboxy-N,N,N′,N′-tetramethylrhodamine was used as a quencher. A threshold signal was chosen above which the signal could be detected. This gave the threshold cycle (CT), which was the first cycle for which a signal could be detected. This value was then plotted against log input CFU, which gave a standard curve for the determination of amplification efficiency. Standard curves were commonly made from 6-log10 dilution series by linear regression analyses (Microsoft Excel 2000; Microsoft Corp., Redmond, Wash.). The amplification efficiencies were then calculated by using the following formula: 10/2X, where X = the slope of the regression curve. The relationship between the 5′-nuclease PCR signal and CFU was calculated with the following formula: log10 CFU = (39.3 − CT)/3.5, where 39.3 is the theoretical CT value for 1 CFU, CT is the observed CT value, and 3.5 is the mean of the slopes of the regression curves obtained for spiked cecal and fecal samples.
Correlation analyses.
The Pearson correlation coefficient was used to measure the linear relationship between plate counts and 5-nuclease PCR data (Minitab, release 13.30; Minitab Inc., State College, Pa.). The correlation coefficient assumes a value between −1 and +1. If one variable tends to increase as the other decreases, then the correlation coefficient is negative. Conversely, if the two variables tend to increase together, then the correlation coefficient is positive. The correlation coefficient was calculated with the following formula for the two variables x and y: r = Σ(x − mx)(y − my)/(n − 1)sxsy, where mx and sx are the sample mean and the standard deviation for the first variable and my and sy are the sample mean and standard deviation for the second variable.
PCA.
The performances of the different sample preparation approaches tested were evaluated by principal component analyses (PCA) (The Uncrambler; Camo Inc., Corvallis, Oreg.). For PCA, each principal component (PC) is defined as the linear combination of the parameters tested that account for as much as possible of the covariance remaining after previous PCs by using the sum of squares (9). The data were analyzed by using full cross validation with centered data. The independent variables were weighted according to their standard deviations.
RESULTS
Evaluation of C. jejuni strain differences.
Five C. jejuni strains were chosen for testing of the strain variation of the combined cell concentration and DNA purification method. We used ΔCT values in our comparisons since it is very difficult to accurately standardize the amount of input material (Fig. 2). Amplification efficiencies in the range of 0.8 to 1 and S2 values between 99.1 and 99.5% were obtained from linear regression curves for the strains tested. The amplification efficiency determined from a regression curve considering all the strains simultaneously was 0.9, with an S2 value of 99.1%. The Pearson correlation coefficient between the ΔCT values and CFU for the different strains analyzed was 0.997, indicating a low strain-to-strain variation for the sample preparation approach applied.
FIG. 2.
Evaluation of the cell concentration and DNA purification assay with five different C. jejuni strains. The bacteria were spiked directly in bacterial binding and washing buffer (Genpoint), with subsequent analysis by 5′-nuclease PCR. A 10-fold dilution series, from approximately 102 to 106 CFU/ml, is shown. The ΔCT value were obtained by subtracting the CT value for each dilution from the CT value obtained for the log signal for approximately 6 log10 CFU/ml. The following C. jejuni strains were analyzed: •, LCD 8617; ○, ATCC 43438; ×, ATCC 43436; *, ATCC 43439; and +, ATCC 43442. A linear regression curve is shown.
Spiked fecal samples.
C. jejuni cells were spiked in 25-mg fecal samples before isolation. There were log linear correlations between a 10-fold dilution series of the spiked cells over a 4-log10 range and the CT values obtained with real-time quantitative PCR. An amplification efficiency of 1.1 and an S2 value of 99.1% were determined from the linear regression curves. The detection limit obtained was between 2.5 and 25 CFU per PCR.
The presence of PCR inhibitors in the samples was tested by the amount tolerated in a reaction before the enzymatic reaction in an ordinary PCR was inhibited. The effects of compounds that facilitate the amplification reaction were also investigated (Table 1). The reactions were inhibited when DNA from >2.5 mg of fecal material was used per 50-μl PCR, while 11 mg of fecal samples could be used without detectable inhibition when the PCR facilitator BSA was added to the reaction. The addition of BSA also increased the sensitivity of the assay. There were no detectable differences in the facilitator effect between different purities of BSA or BSA from different manufacturers (results not shown). The detection limit without BSA was approximately 1 log10 higher for samples containing fecal material than for pure cultures. However, the addition of BSA gave a detection limit approximately similar to that for pure cultures. The addition of PEG had a slight effect on PCR inhibition in the samples. There was an approximately 1 log10 lower detection limit compared to that for samples without added PEG. The combined addition of BSA and PEG had an effect on both the detection limit and inhibition for the samples. More than 10 mg of fecal sample could be used per reaction, and the detection limit was approximately 1 log10 lower than for pure cultures without facilitators.
TABLE 1.
Amount of fecal or cecal content tolerated in a 50-μl reaction without detectable PCR inhibition
| Matrix | Purifi- cationa | Amt of matrix (mg) in presence of facilitator
|
|||
|---|---|---|---|---|---|
| No facilitator | 0.4% BSA | 4% PEG | 0.4% BSA and 4% PEG | ||
| Feces | Purified | 2.5 | 11 | 5 | 10 |
| Cecum | Purified | 0.1 | 1 | <0.2 | 1 |
| Feces | None | <0.5 | 0.5 | Not tested | Not tested |
| Cecum | None | <0.1 | <0.1 | Not tested | Not tested |
Purified material was purified with the Bugs'n Beads kit from Genpoint.
Spiked cecal content samples.
C. jejuni cells were spiked in 12-mg cecal samples before sample preparation. There were log linear correlations between a 10-fold dilution series of the spiked cells over a 4-log10 range and the CT values obtained with real-time quantitative PCR. An amplification efficiency of 0.9 and an S2 value of 99.6% were obtained. The detection limit was between 2 and 20 CFU.
Samples were tested for both the presence of PCR inhibition and the effect of facilitators (Fig. 3; Table 1) as for the fecal samples, as described above. The cecal samples contained relatively high amounts of PCR inhibitory compounds. DNA from samples containing >0.1 mg of cecal content per 50-μl PCR inhibited the amplification reaction. The addition of BSA overcame much of the inhibition in the samples, and DNA purified from 1 mg of cecal content could be used per 50-μl PCR (Table 1). The addition of PEG had no effect on the PCR inhibition or the detection limit, and there was no combined effect of BSA and PEG (Table 1). The effect of betaine was also tested. There was no detectable effect of betaine on the PCR inhibition or the detection limit for the samples tested here (results not shown).
FIG. 3.
Effect of BSA as a PCR facilitator on cecal content samples. DNAs (2.5%) isolated from 12 mg of cecal samples were tested in 50-μl PCRs with the addition of different concentrations of BSA. Dilution series from 103 to 10 CFU/ml were used for this analysis. The products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining and UV transillumination. mw, molecular weight marker; nt, nucleotides; neg, negative control.
Comparison of different DNA sample preparation methods.
The combined cell concentration and DNA purification approach was compared to standard methods for sample preparation and DNA purification. Samples containing cecal content (12 mg per isolation) were used for these comparisons. To our knowledge, no other integrated system for cell concentration and DNA purification exists. We chose a column-based method (DNeasy; Qiagen), a paramagnetic bead-based method (DNA DIRECT; Dynal), and a sample preparation method based on the selective removal of inhibitory components (Prepman; Applied Biosystems).
The Bugs'n Beads approach gave the same detection limit as the DNeasy and Prepman methods, while the DNA DIRECT method gave a 1-log10 higher detection limit (Table 2). There were more primer dimer formations by the DNeasy, Prepman, and DNA DIRECT methods than by the Bugs'n Beads approach. The S2 values and amplification efficiencies were approximately similar with the DNeasy, Prepman, and Bugs'n Beads kits, whereas with the DNA DIRECT kit, the amplification efficiency and S2 value were lower (Table 2).
TABLE 2.
Comparison of sample preparation methods
| Method | Amplification efficiency | Square regression coefficient (S2) (%) | Detection limita | Performance time | Centrifugation | Primer dimersb |
|---|---|---|---|---|---|---|
| Bugs'n Beads | 0.9 | 99.6 | >2-<20 | 30 min | No | No |
| DNeasy | 0.9 | 99.9 | >2-<20 | 4 h | Yes | Yes |
| Prepman | 1.0 | 99.8 | >2-<20 | 30 min | Yes | Yes |
| DNA DIRECT | 0.7 | 99.2 | >20-<200 | 45 min | Yes | Yes |
The detection limit is between the sample with the lowest CFU giving a signal and the next sample. The values are given relative to the CFU in the original sample.
The level of primer dimer formation was determined by qualitative PCR.
PCA (Fig. 4) showed that the DNA DIRECT method is separate from the other methods tested in the first PC (PC1). PC1 is mainly spanned by the detection limit and amplification efficiency. The DNA DIRECT system has a lower amplification efficiency and a higher detection limit than the other methods. The DNeasy, Prepman, and Bugs'n Beads methods are separated in PC2. The important parameters in PC2 are centrifugation, primer dimer formation, and time of analysis. Bugs'n Beads is the method with the lowest amount of primer dimer formation, and it does not require centrifugation. The distinction between DNeasy and Prepman is mainly the time required for the DNeasy approach.
FIG. 4.
PCA of the comparison of methods. The data presented in Table 2 were subjected to PCA (see Materials and Methods for details). (A) Score diagram for the methods tested. (B) Loading for the different parameters tested. The two PCs (PC1 and -2) explain 50 and 38% of the variance in the data, respectively. AMPEFF, amplification efficiency; REGRESSCO, squared regression coefficient; DETECTLIM, detection limit; time, performance time; CENTRIFUG, centrifugation; PRIMDIMER, primer dimer; BUGSB, Bugs'n Beads; DNADIR, DNA DIRECT.
Qualitative comparisons of enrichment and direct DNA-based testing of naturally colonized poultry.
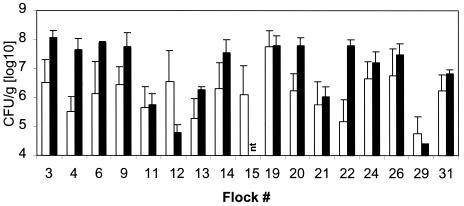
All of the approaches applied gave the same results for 28 of the 31 flocks tested (Table 3). Enrichment and direct qualitative testing gave the same positive and negative determinations for all of the flocks that were analyzed by both approaches. The 5′-nuclease PCR was negative for flocks 8 and 17, while the qualitative PCR analyses were positive. These flocks, however, were probably not colonized with C. jejuni, but with another Campylobacter species (the qualitative PCR assay detects Campylobacter spp., while the 5′-nuclease PCR system detects C. jejuni). For flock 29, 3 of 15 fecal samples and 1 of 3 cecal samples tested positive by quantitative detection, while all of the samples were positive in the qualitative screening (Table 2). The concentration of C. jejuni within this flock, however, was probably on the border of the detection limit for the 5′-nuclease assay, which is approximately 4 log10 CFU/g of fecal or cecal material (Fig. 5).
TABLE 3.
Camplylobacter detection in poultry flocks
| Flock no. | Direct quantitative detectiona,b
|
Direct qualitative detectiona,c
|
Enrichment detectiona,c
|
Presence of Campylo- bacterd | |||
|---|---|---|---|---|---|---|---|
| Feces | Cecum | Feces | Cecum | Feces | Cecum | ||
| 1 | − | − | − | − | − | − | No |
| 2 | − | − | − | − | − | − | No |
| 3 | +++ | +++ | +++ | +++ | +++ | +++ | Yes |
| 4 | +++ | +++ | +++ | +++ | +++ | +++ | Yes |
| 5 | − | − | − | − | − | − | No |
| 6 | ++ | +++ | ++ | +++ | +++ | +++ | Yes |
| 7 | − | − | − | − | − | − | No |
| 8 | − | − | ++ | +++ | + | +++ | Yese |
| 9 | ++ | +++ | ++ | +++ | +++ | +++ | Yes |
| 10 | − | − | − | − | − | − | No |
| 11 | +++ | +++ | +++ | +++ | ++ | +++ | Yes |
| 12 | +++ | +++ | +++ | +++ | +++ | +++ | Yes |
| 13 | +++ | +++ | +++ | +++ | ++ | +++ | Yes |
| 14 | +++ | +++ | +++ | +++ | +++ | +++ | Yes |
| 15 | +++ | NT | +++ | NT | +++ | NT | Yes |
| 16 | − | − | − | − | NT | NT | No |
| 17 | − | − | +++ | +++ | NT | NT | Yese |
| 18 | − | − | − | − | NT | NT | No |
| 19 | +++ | +++ | +++ | +++ | NT | NT | Yes |
| 20 | +++ | +++ | +++ | +++ | NT | NT | Yes |
| 21 | +++ | +++ | +++ | +++ | NT | NT | Yes |
| 22 | ++ | +++ | +++ | +++ | NT | NT | Yes |
| 23 | − | − | − | − | NT | NT | No |
| 24 | +++ | +++ | +++ | +++ | NT | NT | Yes |
| 25 | − | − | − | − | NT | NT | No |
| 26 | +++ | +++ | +++ | +++ | NT | NT | Yes |
| 27 | − | − | − | − | NT | NT | No |
| 28 | − | − | − | − | NT | NT | No |
| 29 | + | + | +++ | +++ | NT | NT | Yes |
| 30 | − | − | NT | NT | NT | NT | No |
| 31 | +++ | +++ | NT | NT | NT | NT | Yes |
Percentage of samples containing Campylobacter, as follows: −, 0; +, ≤50; ++, 50 to 100; +++, 100; NT, not tested.
Detection of C. jejuni.
Detection of Campylobacter spp.
The flock was defined as Campylobacter positive if at least one of the samples gave a positive Campylobacter signal.
Flock contains Campylobacter species other than C. jejuni.
FIG. 5.
Quantification of C. jejuni in poultry flocks. The amount of C. jejuni was determined by real-time quantitative PCR. White bars represent fecal samples, while black bars represent cecal samples. Error bars represent standard deviations for all of the positive samples within a flock. The detection limit of the assay was 4 log10 CFU/g. nt, not tested.
Quantification of C. jejuni in naturally colonized poultry.
The amounts of C. jejuni DNA in both fecal and cecal samples were determined for the C. jejuni-positive flocks (Fig. 5). Fresh fecal samples were collected in transport cages, while cecal samples were collected immediately after slaughter. Each sample was analyzed in duplicate. Generally, the C. jejuni content was higher in the cecal samples than in the fecal samples. Also, the standard deviations were higher for the fecal samples than for the cecal samples. Most of the cecal samples gave DNA signals corresponding to 7.5 to 8.5 log10 CFU/g of material tested. For the fecal samples, these values were between 5.5 and 6.5, although for one flock, 8 log10 CFU/g was detected. The range for the cecal samples was from 8.1 for flock 3 to 4.4 for flock 29, whereas for the fecal samples a log10 value of 8.0 was obtained for flock 19 and of 4.7 was obtained for flock 29. Interestingly, flock 12 contained relatively high amounts of C. jeuni in the fecal samples, while the levels were relatively low in the cecal samples. This flock was probably recently colonized with Campylobacter, since an independent testing conducted 8 days before slaughter was negative (results not shown).
We also investigated the effect of fecal sample quality with samples collected at the farms. A total of 235 samples from 85 flocks were analyzed by 5′-nuclease PCR. The sample quality was determined visually with respect to the content of urate crystals and the water content. The empirical data indicated that sample quality is very important for the reliability of the PCR. Samples with high amounts of urate crystals and dry samples gave consistently lower amounts of Campylobacter than wet samples without urate crystals. We also tested both the urate-containing part and the wet part from the same fecal samples for Campylobacter-positive birds. The Campylobacter content was >10-fold higher in the wet parts than in the urate-containing parts. No cecal droppings were identified in this screening. This is probably due to the rapid adsorption of the cecal droppings by the sawdust on the floors of broiler houses.
Automation.
There were no technical difficulties with fecal or cecal samples in the automated sample preparation approach, using the modified protocol presented in Materials and Methods. The automated approach was tested with both spiked and naturally colonized fecal and cecal material. No cross contamination was detected with the automated approach. Furthermore, there was no detectable difference between the manual approach and the automated sample preparation approach for a set of 49 samples tested (selected from the samples presented in Table 3).
DISCUSSION
Evaluation detection systems for direct biological samples.
For evaluation of a detection system, relatively large amounts of test material should be investigated in order to obtain a sufficient sensitivity for samples containing low bacterial numbers (32). This is a challenge with direct PCR-based methods. The reaction volumes for the enzymatic reactions used are relatively small, normally in the range of 10 to 100 μl. For zoonotic pathogens such as C. jejuni, however, the bacteria exist in high concentrations in the intestines of colonized animals. For instance, the expected concentration of C. jejuni in the cecal content is 6 to 8 log10 CFU/g (26). High numbers are also expected in feces. These concentrations are clearly within the range of what is obtainable for direct PCR-based methods.
Samples from the intestines and feces contain high amounts of compounds that are inhibitory for PCR. This limits the amount of material that is tolerated in a PCR. Common inhibitors are DNases, polysaccharides, and proteases (31). The level of PCR inhibitors was found to be higher in the cecal content than in the fecal samples. The inhibition in both sample types was partially overcome through the addition of BSA, which may act as an alternative substrate for the proteases and as an alternative binding site for other inhibitory compounds. An effect of PEG on PCR inhibitors was only obtained for the fecal samples. Interestingly, the addition of facilitators also seemed to have an effect on the assay sensitivity. The effect of PEG is to bind water. This could facilitate the first rounds of the PCR if, for example, the DNA is associated with polysaccharides. A possible sensitivity effect of BSA could be to scavenge DNA binding compounds that reduce the efficiency of the PCR.
The quality of samples is important for the analytical results. We ensured the uniformity of the fecal samples analyzed in the main experimental series by collecting them from transport cages. These feces were no more than 2 h old. The effect of fecal sample quality was evaluated in an independent experiment. The empirical data indicated that sample quality is very important. The consideration of sample quality is particularly important in screening programs for which farmers collect fecal materials themselves. It is often recommended that cecal droppings be used as test materials in screening programs. Our experience is that the identification of cecal droppings is more or less impossible at the farm level and that farmers collect normal fecal samples with high contents of urate crystals.
Comparison of alternative sample preparation methods.
A major limitation for the wide application of DNA-based methods in routine diagnostics is the complicated execution of most of the methods applied. A prerequisite for such methods is simplicity and automation (A. Holmberg, A. Deggerdal, and F. Larsen, AMS ’95: 3rd Int. Conf. Autom. Mapp. DNA Seq., abstr. A10, 1995). To our knowledge, the work presented here describes the first fully automated method for both detecting and quantifying C. jejuni directly from chicken cecal contents and feces without enrichment.
A comparison with standard DNA purification methods was conducted in order to evaluate the potential for further improvements. A centrifugation-based pretreatment for the other methods was included since no other methods integrate cell isolation and DNA purification (see Materials and Methods). The detection limit, amplification efficiency, and accuracy were approximately similar for the Bugs'n Beads, Prepman, and DNeasy methods. The DNA DIRECT approach, however, gave a higher detection limit and lower amplification efficiency than the other approaches. Recovery by the DNA DIRECT system is seemingly dependent on the amount of bacteria present in the test sample. This is probably due to the fact that DNA DIRECT is based on a coaggregation of DNA and beads and is designed for approaches with relatively high amounts of DNA in the samples (20, 21). The smaller amount of short PCR products (primer dimers) for the Bugs'n Beads method could be due to a “hot-start DNA” effect. The DNA is attached to the beads when added to the PCR and released in the initial denaturation phase. This may reduce the amount of false priming of PCR amplification.
The evaluation of different methods is very difficult due to the large amounts of data to be compared. We used PCA to investigate patterns in the performance of the methods evaluated. PCA give a visual overview of the performances of the different methods. The covariance between different parameters is also revealed. For instance, in our comparison, there was a clear negative correlation between the detection limit and amplification efficiency (Fig. 4B).
Screening of naturally colonized poultry.
There was a high qualitative correspondence (of positive and negative determinations of flock colonization) among the different testing approaches applied in this work. Enrichment and the qualitative PCR gave identical results. The benefits of using direct DNA testing instead of traditional testing are that the time required for the analysis can be reduced from >2 days to <4 h and that the process can be automated. This may help poultry producers to adapt a production system that prevents foods containing C. jejuni from reaching the consumer. With traditional testing schemes, up to 50% of poultry flocks can become C. jejuni positive in the interval between testing and slaughter, resulting in an unacceptably high number of false-negative flocks (16).
We were unable to make quantitative comparisons between the enrichment-based testing and the direct 5′-nuclease PCR test employed. However, previous estimates have indicated that the C. jejuni content in the cecum is in the range of 6 to 8 log10 CFU/g (1). This corresponds well to the range estimated with our direct DNA quantification method.
In our study, we found a relatively uniform distribution of Campylobacter within the infected flocks, while the differences between the flocks were quite large (Fig. 5). This may indicate that within flocks the colonization of poultry is extremely rapid and that the differences between flocks reflect that the flocks are in different stages of the colonization process. Other possibilities are that different strains of C. jejuni have different abilities to colonize poultry and that the flocks have different competing floras in their intestines, influencing the colonization ability of C. jejuni (19). These issues have not yet been properly addressed (due to limitations with traditional diagnostics). Understanding the colonization of poultry, however, is crucial for the control of C. jejuni (12).
Implications of the ability to colonize poultry on the potential for causing human disease.
The virulence of microorganisms is an important area of research (25), while the ability of zoonotic microorganisms to colonize animal hosts has received relatively little attention (26). From a disease-causing perspective, however, the combined effect of both the amount of bacteria present in the animal host and the virulence of these bacteria is important.
The contents of C. jejuni in the ceca ranged from 4.5 to >8 log10 CFU/g for the flocks tested in this work. This is more than a 1,000-fold difference. If the strain colonizing flock 29 has an infective dose that is 0.1% that of the strain colonizing flock 3, then from a human infection perspective these two strains would be equally likely to cause disease through food contamination. Thus, the whole chain from the number of organisms colonizing production animals to infectious doses needs to be considered in determining the potential of a pathogen to cause human disease.
Acknowledgments
This work was supported by grants 139782/30, 153088/110, and 157596/130 from the Norwegian Research Council.
We give special thanks to Signe M. Drømtorp for excellent technical assistance. Furthermore, we thank Ellen S. Tronrud and Helga Næs for carefully reading and commenting on the manuscript.
REFERENCES
- 1.Berndtson, E., M. L. Danielsson-Tham, and A. Engvall. 1996. Campylobacter incidence on a chicken farm and the spread of Campylobacter during the slaughter process. Int. J. Food Microbiol. 32:35-47. [DOI] [PubMed] [Google Scholar]
- 2.Berndtson, E., U. Emanuelson, A. Engvall, and M. Danielsson-Tham. 1996. A 1-year epidemiological study of Campylobacter in 18 Swedish chicken farms. Vet. Med. 26:167-185. [Google Scholar]
- 3.Bovill, R. A., and B. M. Mackey. 1997. Resuscitation of “non-culturable” cells from aged cultures of Campylobacter jejuni. Microbiology 143:1575-1581. [DOI] [PubMed] [Google Scholar]
- 4.Campyforum. 2000. Campylobacter control in poultry production. A challenge to industry and scientists. Danish Veterinary Laboratory, Copenhagen, Denmark.
- 5.Chuma, T., K. Yano, H. Omori, K. Okamoto, and H. Yugi. 1997. Direct detection of Campylobacter jejuni in chicken cecal contents by PCR. J. Vet. Med. Sci. 59:85-87. [DOI] [PubMed] [Google Scholar]
- 6.Hazeleger, W., C. Arkesteijn, A. Toorop-Bouma, and R. Beumer. 1994. Detection of the coccoid form of Campylobacter jejuni in chicken products with the use of the polymerase chain reaction. Int. J. Food Microbiol. 24:273-281. [DOI] [PubMed] [Google Scholar]
- 7.Lazaro, B., J. Carcamo, A. Audicana, I. Perales, and A. Fernandez-Astorga. 1999. Viability and DNA maintenance in nonculturable spiral Campylobacter jejuni cells after long-term exposure to low temperatures. Appl. Environ. Microbiol. 65:4677-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Logan, J. M., K. J. Edwards, N. A. Saunders, and J. Stanley. 2001. Rapid identification of Campylobacter spp. by melting peak analysis of biprobes in real-time PCR. J. Clin. Microbiol. 39:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martens, H., and M. Martens. 2001. Multivariate analysis of quality. An introduction. John Wiley & Sons Ltd., West Sussex, United Kingdom.
- 10.Nachamkin, I. 1997. Campylobacter jejuni, p. 159-170. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers. American Society for Microbiology, Washington, D.C.
- 11.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barre syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4334-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogva, H. K., A. Bergh, A. Holck, and K. Rudi. 2000. Application of the 5′-nuclease PCR assay in the evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nogva, H. K., K. Rudi, K. Natersad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure culture, water skim milk and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordic Committee on Food Analysis (NMKL). 1990. Campylobacter jejuni detection in foods. Report no. 119. National Veterinary Institute, Oslo, Norway.
- 16.Norwegian Food Control Authority. 2002. Analyses and results 2000-2002. Norwegian Food Control Authority, Oslo, Norway. (In Norwegian.)
- 17.On, S. 1997. Identification methods for Campylobacter, Helicobacter, and related organisms. Clin. Microbiol. Rev. 9:405-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oyofo, B. A., S. M. Abd el Salam, A. M. Churilla, and M. O. Wasfy. 1997. Rapid and sensitive detection of Campylobacter spp. from chicken using the polymerase chain reaction. Zentbl. Bakteriol. 285:480-485. [DOI] [PubMed] [Google Scholar]
- 19.Reid, G., and R. Friendship. 2002. Alternatives to antibiotic use: probiotics for the gut. Animal Bio/Technol. 13:97-112. [DOI] [PubMed] [Google Scholar]
- 20.Rudi, K., F. Larsen, and K. S. Jakobsen. 1998. Detection of toxin-producing cyanobacteria by use of paramagnetic beads for cell concentration and DNA purification. Appl. Environ. Microbiol. 64:34-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudi, K., M. Kroken, O. J. Dahlberg, A. Deggerdal, K. S. Jakobsen, and F. Larsen. 1997. Rapid, universal method to isolate PCR-ready DNA using magnetic beads. BioTechniques 22:506-511. [DOI] [PubMed] [Google Scholar]
- 22.Rudi, K., O. M. Skulberg, R. Skulberg, and K. S. Jakobsen. 2000. Application of sequence-specific labeled 16S rRNA gene oligonucleotide probes for genetic profiling of cyanobacterial abundance and diversity by array hybridization. Appl. Environ. Microbiol. 66:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sails, A. D., A. J. Fox, F. J. Bolton, D. R. A. Wareing, and D. L. A. Greenway. 2003. A real-time PCR assay for the detection of Campylobacter jejuni in foods after enrichment culture. Appl. Environ. Microbiol. 69:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shane, S. 1992. The significance of Campylobacter jejuni infection in poultry: a review. Avian Pathol. 21:189-213. [DOI] [PubMed] [Google Scholar]
- 25.Solomon, E., and D. Hoover. 1999. Campylobacter jejuni: a bacterial paradox. J. Food Safety 19:121-136. [Google Scholar]
- 26.Stern, N. 1992. Reservoir for Campylobacter jejuni and approaches for intervention in poultry, p. 49-60. In L. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, D.C.
- 27.Tveit, I., H. Ulvesæter, and A. Walde. 1997. Prevalence of Campylobacter in poultry meat in Norway, 1995-1997. Norwegian Food Control Authority, Oslo, Norway. (In Norwegian.)
- 28.van de Giessen, A., S. I. Mazurier, W. Jacobs-Reitsma, W. Jansen, P. Berkers, W. Ritmeester, and K. Wernars. 1992. Study on the epidemiology and control of Campylobacter jejuni in poultry broiler flocks. Appl. Environ. Microbiol. 58:1913-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waller, D. F., and S. A. Ogata. 2000. Quantitative immunocapture PCR assay for detection of Campylobacter jejuni in foods. Appl. Environ. Microbiol. 66:4115-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weissensteiner, T., and J. S. Lanchbury. 1996. Strategy for controlling preferential amplification and avoiding false negatives in PCR typing. BioTechniques 21:1102-1108. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winters, D. K., and M. F. Slavik. 1995. Evaluation of a PCR based assay for specific detection of Campylobacter jejuni in chicken washes. Mol. Cell. Probes 9:307-310. [DOI] [PubMed] [Google Scholar]