Abstract
Cr(VI) (chromate) is a toxic, soluble environmental contaminant. Bacteria can reduce chromate to the insoluble and less toxic Cr(III), and thus chromate bioremediation is of interest. Genetic and protein engineering of suitable enzymes can improve bacterial bioremediation. Many bacterial enzymes catalyze one-electron reduction of chromate, generating Cr(V), which redox cycles, generating excessive reactive oxygen species (ROS). Such enzymes are not appropriate for bioremediation, as they harm the bacteria and their primary end product is not Cr(III). In this work, the chromate reductase activities of two electrophoretically pure soluble bacterial flavoproteins—ChrR (from Pseudomonas putida) and YieF (from Escherichia coli)—were examined. Both are dimers and reduce chromate efficiently to Cr(III) (kcat/Km = ∼2 × 104 M−1 · s−1). The ChrR dimer generated a flavin semiquinone during chromate reduction and transferred >25% of the NADH electrons to ROS. However, the semiquinone was formed transiently and ROS diminished with time. Thus, ChrR probably generates Cr(V), but only transiently. Studies with mutants showed that ChrR protects against chromate toxicity; this is possibly because it preempts chromate reduction by the cellular one-electron reducers, thereby minimizing ROS generation. ChrR is thus a suitable enzyme for further studies. During chromate reduction by YieF, no flavin semiquinone was generated and only 25% of the NADH electrons were transferred to ROS. The YieF dimer may therefore be an obligatory four-electron chromate reducer which in one step transfers three electrons to chromate and one to molecular oxygen. As a mutant lacking this enzyme could not be obtained, the role of YieF in chromate protection could not be directly explored. The results nevertheless suggest that YieF may be an even more suitable candidate for further studies than ChrR.
Cr(VI) (chromate) is a serious environmental pollutant due to the wide use of chromium compounds in industries such as tanning, corrosion control, plating, pigment manufacture, and nuclear weapons production (2). At the Department of Energy (DOE) waste sites, for example, it is the second most common heavy-metal contaminant, ranging in concentration between 0.008 and 173 μM in groundwater and 98 nM and 76 mM in soil and sediments (30); since soil water is stored in small capillary spaces, the last-mentioned concentrations are very high. Chromate is toxic, mutagenic, and carcinogenic (14, 35). Several factors contribute to its toxicity. Because of its structural similarity to SO42−, it is readily taken up by both bacterial and eukaryotic cells through the sulfate transport system (5, 35). Inside the cell, it is reduced nonenzymatically, as well as by various enzymes. Its partial reduction, particularly by the cellular one-electron reducers, generates Cr(V) and reactive oxygen species (ROS); the latter may be a major factor in causing cellular damage (5, 35, 39). Chromate is soluble and thus readily spreads beyond the site of initial contamination. Bacteria can convert chromate to Cr(III), which is much less toxic and less soluble, and thus bacterial bioremediation of chromate is of considerable interest, especially given the fact that alternative chemical means are prohibitively expensive for large-scale cleanup (5).
Field studies have shown that biostimulation is a promising approach for bioremediation (19, 24). This method entails addition of nutrients to the environment, such as aquifers, to stimulate the growth of indigenous bacteria. Although enhanced growth does promote bioremediation, the resulting large amount of biomass can result in clogging of subsurface pores and confining of remediation to a narrow zone (19). Moreover, polluted environments, especially the DOE sites (30), contain multiple pollutants. Biostimulation of such sites is likely to have limited effectiveness, since the remediating bacteria, as well as the enzymes involved, are inhibited by the mixed waste present in such environments (M. Keyhan and A. Matin, unpublished data). Chromate remediation in such environments is further complicated by the fact that its reduction involves the generation of toxic intermediates, which are detrimental to the remediating bacteria (5).
One way to address these problems is to use genetic- and protein engineering approaches. For instance, the use of appropriate promoters can ensure maximal expression of desired genes in slowly growing bacteria, thereby minimizing biomass formation and clogging (21, 22). Also, protein engineering of bacterial chromate reductases can generate improved enzymes that reduce chromate more efficiently, that minimize chromate toxicity to the remediating bacteria, and that can function in the presence of other pollutants. Some bacteria can evidently use chromate as the terminal electron acceptor, employing membrane-bound enzymes (23, 25, 41). However, several others reduce it using soluble enzymes (4, 12, 28, 39). The functional aspects of the latter group have been little studied, and they are more amenable to protein engineering. We have therefore focused on characterizing the soluble chromate reductases to identify suitable candidates for further work.
Using classical biochemical techniques, a novel soluble enzyme (ChrR) with chromate reductase activity was previously purified to homogeneity from Pseudomonas putida (28). N-terminal and internal amino acid sequence determination of the enzyme allowed the design of appropriate primers to clone the chrR gene (27). BLAST searching of protein databases with the derived ChrR amino acid sequence revealed a conserved family of proteins whose members are present in a wide range of organisms. Over 40 of these homologs, including the predicted product of a previously uncharacterized open reading frame (yieF) from Escherichia coli, show >30% amino acid identity with ChrR. All contain the characteristic signature of the NADH_dh2 family of proteins, which consists of bacterial and eukaryotic NAD(P)H oxidoreductases (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi). Using appropriate primers, the E. coli yieF gene was cloned as well, and it was shown that this enzyme also can reduce chromate (27). The availability of the cloned genes has enabled us to obtain large quantities of the two enzymes in electrophoretically pure form and to compare several of their characteristics: chromate reduction kinetics, generation of ROS during chromate reduction, and induction patterns. While both enzymes are suitable for further work to improve bacterial chromate bioremediation, YieF appears to be a better candidate.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) liquid medium was used for bacterial growth unless otherwise specified. Recombinant and lysogenic strains were selected on solid media with appropriate antibiotics: ampicillin (100 μg/ml), kanamycin (25 μg/ml for E. coli; 50 μg/ml for P. putida), chloramphenicol (20 μg/ml), or streptomycin (400 μg/ml).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| CC118λpir | λpir phage lysogen; RifT | 24 |
| BL21(DE3) | Novagen | |
| DH5α | 27 | |
| AMS6 | 27 | |
| AMS6λY | AMS6 (λpCLA-3) | This study |
| P. putida | ||
| KT2440 | 12 | |
| CRK3 | KT2440 transconjugant bearing a cointegrate of the plasmid pCHP8 | This study |
| CRK4 | KT2440 chrR::Cm mutant; SmS/Sucr | This study |
| Plasmids | ||
| pUC 19 | Cloning vector; Ampr | 27 |
| pET28a(+) | Translation vector with T7 lac promoter and His tag sequence; Kmr | Novagen |
| pRK600 | Helper plasmid with ori ColE1 RK2-Mob+ Rk2-Tra+; Cmr | 31 |
| pMRS101 | Suicide vector; Ampr | 31 |
| pCG-1 | pET28a(+) with EcoRI-NdeI yieF gene; Kanr | This study |
| pCHP4 | pET28a(+) containing the P. putida chrR gene (NdeI/EcoRI); Kmr | This study |
| pCHP5 | pUC 19 containing P. putida 1.5-kb PCR fragment (BamHI/XbaI); Apr | This study |
| pCHP6 | Identical to pCHP5 but Cm cassette in the middle of the chrR gene at ClaI site; Apr Cmr | This study |
| pCHP7 | pMRS101 with 2.3-kb fragment containing 1.5-kb PCR fragment and 0.8-kb Cm cassette at BamHI/XbaI site; Apr Cmr Smr | This study |
| pCHP8 | Identical to pCHP7 but oriE1 and Apr were removed by NotI digestion; Cmr Smr | This study |
| pRS415 | lacZ fusion vector; Apr | 34 |
| pCLA-3 | pRS415 with yieF fusion | This study |
| RS45 | Helper phage | 34 |
Growth conditions and assays.
Cell growth was followed by A660 measurements. Chromate reduction rates were quantified in growing cultures, as well as cell suspensions. Residual chromate was measured by the diphenyl carbazide method (27, 28) after removing the cells by microcentrifugation. Cell suspensions were prepared from overnight cultures which had been washed and resuspended (A660, 4 to 6; 1.3 to 1.9 mg · ml of total protein−1) in fresh LB medium containing the specified concentration of potassium chromate. The suspensions were incubated with shaking at 30°C and sampled at appropriate intervals to measure residual chromate. At these cell concentrations, little change in culture density occurred during the experiment.
Cell extract preparation and chromate reductase assays have been described previously (10, 28). Kinetic measurements of enzyme activity were performed at the optimal pH and temperature, which were determined in separate experiments. For determinations of optimal pH, 50 mM citrate and phosphate buffers, whose ranges overlap in part, were used. Protein concentrations were determined with the Bio-Rad Dc protein assay kit, using bovine serum albumin as a standard. H2O2 formation was quantified using the Amplex Red kit (Molecular Probes).
Protein overproduction and purification.
The plasmid pCHP4 (Table 1), containing the chrR gene (27) in a pET-28a+ vector, was used to overproduce the ChrR protein in the E. coli BL21(DE3) strain. To prevent inclusion body formation, the cells were grown at 18°C in LB medium containing 1 M d-sorbitol and 2.5 mM glycine betaine, induced with 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an A660 of 0.4, and harvested 8 h later (1). Recombinant YieF was produced in a similar fashion. The coding region of the gene was amplified from E. coli AMS6 genomic DNA using appropriate primers (27) and cloned in the corresponding restriction sites of pET-28a+ (plasmid pCG-1; Table 1). Overproduction of YieF in E. coli BL21(DE3) did not lead to significant inclusion body formation, and soluble recombinant protein was purified without the addition of d-sorbitol and glycine betaine to the LB culture, as was required for ChrR purification (see above).
The enzymes were purified using a Ni-nitrilotriacetic acid binding column (Novagen) and incubated with 10 mM EDTA at room temperature for 10 min; the EDTA treatment markedly enhanced enzyme activity. The enzyme preparations were desalted with an Amersham Biosciences HiTrap desalting column and stored in 10 mM Tris- HCl, pH 7.5. A biotinylated thrombin capture kit (Novagen) was used in thrombin cleavage. Polyacrylamide gel electrophoresis (PAGE) under denaturing conditions was performed using 12% Criterion precast acrylamide gels and a MiniProtean II electrophoresis cell (Bio-Rad). The gels were stained with Coomassie brilliant blue R-250 (28).
TLC.
The enzyme-bound cofactor was released by boiling 2 mg of purified protein ml−1 for 10 min, followed by centrifugation at 12,000 × g for 10 min. Fifty microliters of the supernatant was applied to 0.2 mm Kieselgel 60 thin-layer chromatography (TLC) plates (Merck), using butanol-acetic acid-water (4:1:1) as a solvent. Commercial flavin mononucleotide (FMN) and flavin adenine dinucleotide (Sigma) were used as standards, and the fluorescent spots were visualized under UV light.
Construction of P. putida KT2440 mutant with chrR gene disrupted.
A 1.5-kb DNA fragment containing the chrR gene plus upstream (627-bp) and downstream (342-bp) sequences was flanked with BamHI and XbaI restriction sites and cloned in pUC19 (pCHP5) (Table 1). The ClaI site in the middle of the chrR gene was used to insert a ClaI-flanked chloramphenicol cassette, generating the plasmid pCHP6. The BamHI-XbaI fragment was excised from pCHP6 and cloned in pMRS101 (31) (pCHP7), followed by oriE1 deletion from the plasmid (NotI digestion [pCHP8]). pCHP8 was transferred from E. coli CC118 λpir to P. putida KT2440 by triparental mating using the helper plasmid pRK600 (6). Since E. coli cannot grow on benzoate and KT2440 can, selection for plasmid cointegrates of KT2440 was accomplished using benzoate (10 mM)-M9 minimal medium supplemented with streptomycin. The Smr colonies were unable to grow on LB medium containing 5% sucrose, confirming that the plasmid pMRS101 (with its sacB gene) had integrated into these strains. The transconjugant CRK3 was grown overnight in streptomycin-free LB medium, diluted 1,000-fold, and following incubation for 12 h, serially diluted and plated on LB medium with or without sucrose. PCR analysis of one of the Sucr Sms clones, CRK4, confirmed that gene replacement had occurred.
Rapid-scan kinetic measurements.
Rapid-scan kinetic measurements were performed using a stopped-flow spectrophotometer (8) that consisted of a mixing chamber equipped with an OLIS-RSM data acquisition and analysis system (On Line Instrument Systems, Inc., Bogart, Ga.). The dual-beam optical system used is capable of achieving 1,000 spectral scans per s. Interchangeable grating monochromators (400 lines blazed at 550 nm) permitted the acquisition of repeated kinetic scans of the contents of the observation cell over a 230-nm span in the visible region. The samples of ChrR and YieF (3.0 μM) and substrates were prepared in 0.05 M citrate buffer, pH 5.0, and added to separate syringes of the stopped-flow spectrophotometer. The temperature of the driving syringes was maintained at 30 ± 1°C; room temperature solutions were allowed to equilibrate for 10 min in the driving syringes. Reactions were initiated by rapidly mixing 0.10 ml of the solution from each driving syringe.
For reductive half reactions, oxidized enzyme and different concentrations of NADH were added to separate syringes of the instrument. For oxidative half reactions, different concentrations of chromate were added to one syringe, while the other syringe received 3.0 μM ChrR or YieF that was amended with NADH as specified. The enzyme solution was gently bubbled with nitrogen gas to lower the concentration of dissolved molecular oxygen. Reduced enzyme prepared under these conditions exhibited air oxidation with a half-life of ∼15 min, a reaction that was sufficiently slow to permit stopped-flow observations of the reduced enzyme without imposing strict anaerobic conditions.
Western analysis.
Western analysis was performed as described previously (42), using a polyclonal antibody against the ChrR protein raised in rabbits by Josman Laboratories. The antibody was purified using an AminoLink Plus Immobilization kit (Pierce) followed by nitrocellulose membrane immunoaffinity purification (11) and desalting (Amersham Biosciences HiTrap desalting column). The purified antibody in 10 mM Tris-HCl, pH 8.0, remained stable for ∼2 weeks at 4°C, but not upon freezing. The secondary antibody used was horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Sigma), and the bands were detected using an ECL Plus Western Blotting detection system (Amersham Pharmacia). Cells were taken from an overnight culture grown without Cr(VI), diluted to an A660 of 0.1 in LB with or without Cr(VI), and incubated with aeration at 30°C. Samples were collected by centrifugation at the appropriate time points and resuspended in sodium dodecyl sulfate (SDS) lysis buffer (20), and 26 μg of total protein was loaded per lane. A negative control from CRK4 (not shown) was used to confirm the identity of the ChrR bands. Relative band intensities were estimated using the program ImageQuant (Molecular Dynamics).
Construction of the yieF-lacZ fusion.
The yieF upstream region was PCR amplified using the following primers: forward (−690), 5′-ATCGAATTCTCCCCTCCCAGTGTCATC-3′; reverse (+183), 5′-ATCGCGGATCCCGTTGCTGGAAAACCTTC-3′. (The numbers in parentheses indicate positions in relation to the translational start site.) Appropriate flanking restriction sites (EcoRI and BamHI; underlined sequences) were added to obtain directional cloning. The fragments were cloned into the fusion vector pRS415 (pCLA-3) (Table 1) and transformed into E. coli AMS6, and the colonies were screened on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates. The resulting plasmid was recombined in the phage RS45 and used to lysogenize AMS-6 to obtain a single-copy fusion strain (AMS6λY) (18, 34). To determine the induction pattern of the yieF-lacZ fusion, cells were grown in LB broth with or without chromate, and aliquots (250 μl) were lysed using BugBuster solution (Novagen). Protein was measured with a Bio-Rad protein assay kit, and β-galactosidase activity was assayed as described previously (18); the activity is expressed in Miller units.
RESULTS
ChrR and YieF are dimeric flavoproteins.
As stated in Materials and Methods, the ChrR and YieF proteins were expressed and purified as N-terminal His-tagged fusions. The purification products each gave a single band on SDS-PAGE, running at ∼22 kDa. Size exclusion chromatography confirmed that the purity of these preparations was >95% and also indicated that the native molecular masses of the enzymes were ca. 50 kDa.
Both ChrR and YieF had typical flavoprotein absorption spectra and a yellow color that was resistant to dialysis. The cofactor was released by boiling and identified by TLC as FMN (data not shown). Several regions of conservation in the derived amino acid sequences of the two proteins were evident, in particular, the signature sequence of the NADH_dh2 protein family (LFVTPEYNXXXXXXLKNAIDXXS) and the 18 residues shown in Fig. 1. A clustal W alignment showed that these residues are highly conserved (>95% identity) in 44 homologs of ChrR. These amino acids may be involved in FMN binding, since residues of the same identity and arrangement bind this cofactor in the Vibrio fischeri enzyme flavin reductase P, whose crystal structure has been solved (40).
FIG. 1.
Clustal W alignment of YieF and ChrR. The asterisks indicate identical residues, the colons indicate residues with a high level of similarity, and the periods indicate residues with a lower level of similarity. The characteristic signature of the NADH_dh2 family of proteins is boxed, and the 18 residues that are particularly highly conserved within ChrR homologs are shaded.
The extra mass in the native proteins probably arises from the bound cofactor and potential possession of an intersubunit disulfide cross-linkage; such linkages can cause nonproportionate movement in SDS-PAGE and gel filtration (36).
Chromate reduction kinetics and range of electron acceptors.
In a previous work (28), which relied on classical purification methods, it was not possible to generate sufficient electrophoretically pure ChrR protein to conduct kinetic studies; they were carried out with a partially pure preparation. The kinetics reported here are for electrophoretically pure proteins.
ChrR and YieF displayed very similar kinetics for chromate reduction (Table 2) (kcat/Km, 2.2 × 104 M−1 · s−1 and 1.9 × 104 M−1 · s−1, respectively), indicating that they are effective agents for chromate reduction. Both had the same optimal pH for this reaction, but ChrR functioned most efficiently at 70°C while YieF had an optimal temperature of 35°C. Both enzymes, however, displayed >40% of maximal activity across a broad range of pH (4.0 to 9.5) and temperature (25 to 80°C). At limiting cofactor concentrations, ChrR displayed an eightfold preference for NADH over NADPH, while YieF appeared to be able to utilize both cofactors equally well.
TABLE 2.
Kinetics for chromate reduction of ChrR and YieF proteinsa
| Protein | Vmax (μmol · min−1 · mg−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 · s−1) | Optimal temp (°C) | Optimal pH |
|---|---|---|---|---|---|---|
| ChrR | 8.8 | 260 | 5.8 | 2.2 × 104 | 70 | 5.0 |
| YieF | 5.0 | 200 | 3.7 | 1.9 × 104 | 35 | 5.0 |
All kinetic parameters were determined at optimal temperature and pH.
Inclusion of an EDTA incubation step in the enzyme purification protocol resulted in ca. fivefold increase in activity compared to that previously reported (27). This suggested that ChrR and YieF might be inhibited by divalent metal ions. Subsequent incubation of the EDTA-treated enzymes with up to 20 mM Cu2+, Ni2+, Mg2+, Ca2+, Fe2+, Zn2+, or Mn2+, however, had no significant effect on activity. This phenomenon is under further investigation. As stated in Materials and Methods, proteins purified by a process including the EDTA step were used in the studies reported in this paper, as opposed to a previous report from this laboratory (27). Removal of the His tag by thrombin cleavage had no effect on the activity of either enzyme.
The end product of chromate reduction was determined using the X-ray absorption near-edge structure (XANES) spectrum, as described previously (28). In this method, Cr(VI) and Cr(III) can be distinguished by the pronounced preedge feature of the former (28). The fraction of Cr(VI) was calculated by dividing the height of the Cr(VI) preedge peak by the total absorption; that of Cr(III) was calculated from the difference between the amount of chromium represented by the preedge peak and the total absorption jump. Chromate was quantitatively converted to Cr(III) by both ChrR and YieF (data not shown). Neither enzyme had any activity with Cr(III) as a substrate.
Both of the enzymes also reduced quinones, potassium ferricyanide, and 2,6-dichloroindophenol, but while ChrR did not appear to have activity with any of the other high-valence metals tested [U(VI), V(V), Mn(IV), and Mo(VI)], YieF reduced V(V) and Mo(VI). YieF also reduced methylene blue and cytochrome c and thus appears to have a broader substrate range than ChrR.
Detection of flavin semiquinone forms of enzymes during chromate reduction.
Many bacterial flavoenzymes, e.g., glutathione reductase (32, 33, 39), reduce chromate by a one-electron transfer process that leads to the generation of flavin semiquinone forms of the enzymes and the Cr(V) radical. Both are subject to redox cycling, resulting in the generation of large amounts of ROS, and this is thought to be a central mechanism of chromate toxicity (14, 35, 39). Since, as noted above, the end product of both ChrR- and YieF-catalyzed reactions was Cr(III), it is clear that these enzymes did not catalyze merely a one-electron reduction of Cr(VI) to Cr(V). Nevertheless, since the XANES analysis at the sensitivity level used cannot detect Cr(V) (28), the results do not rule out transient formation of Cr(V) and the flavin semiquinone forms of the enzymes during the reaction. We therefore performed rapid-scan kinetic measurements using a stopped-flow spectrophotometer (8) to determine if the flavin semiquinone was generated during ChrR- or YieF-catalyzed chromate reduction, since the appearance of this enzyme form would imply Cr(V) formation. The time resolution of this technique permits dissection of the kinetics of the reductive and oxidative half reactions resulting in the transfer of electrons from NADH to chromate.
The absorbance at 450 nm exhibited by flavoproteins disappears upon their reduction. When either of the oxidized enzymes was rapidly mixed with different molar excess NADH concentrations, each kinetic change in absorbance at this wavelength conformed to a single exponential function of time (data not shown). The values of the individual pseudo-first-order rate constants for enzyme reduction were directly proportional to the concentration of excess NADH, yielding values for the corresponding second-order rate constants of 1.0 × 107 M−1 · s−1 for ChrR and 5.2 × 106 M−1 · s−1 for YieF. (The rate constant is referred to as “pseudo” because it is assumed that the concentration of the excess NADH does not change during the reaction.) The direct proportionality indicates that the formation of a kinetically significant complex between oxidized ChrR or YieF and NADH did not occur, thus validating this technique for following the kinetics of the oxidation of the reduced enzyme by chromate.
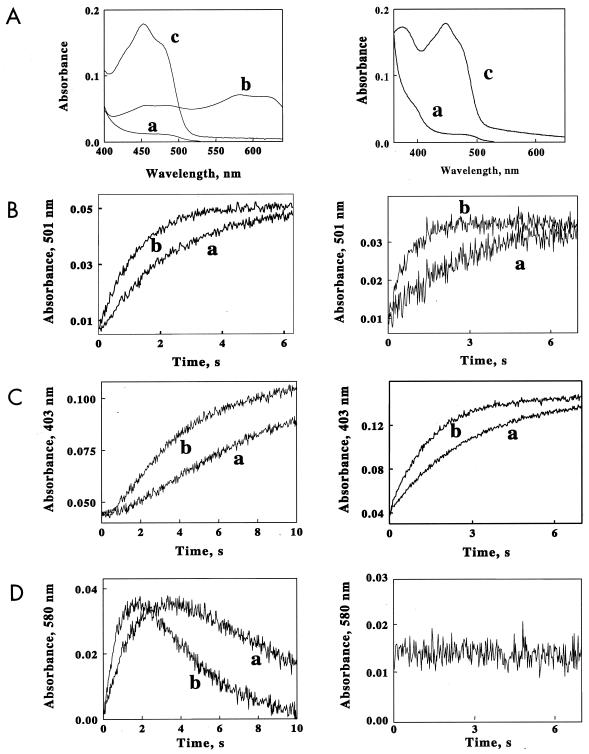
The oxidation of ChrR or YieF was studied, using limiting NADH and excess chromate concentrations. The resulting changes in the absorbances of the enzymes were a complex function of the wavelength. Time-dependent absorbance changes in ChrR or YieF were collected at each of 230 wavelengths between 400 and 630 nm. A global fit of the resulting three-dimensional data set indicated that the observed spectral changes at all wavelengths were described by a sum of two exponential time functions. The spectra shown in Fig. 2A were obtained by the global fit of all of the absorbance-versus-time data for the two enzymes to a biphasic kinetic mechanism. Spectrum a was obtained in the dead time of the instrument and represents the fully reduced enzyme. Spectrum c is the final absorbance recorded in the mixture after the reaction had ceased and represents the oxidized enzyme. Spectrum b is the transient intermediate that formed and disappeared during the overall oxidation reaction and represents the semiquinone form of the flavin. It is evident that this intermediate is formed only during the reaction catalyzed by ChrR.
FIG. 2.
Rapid-mixing studies with ChrR and YieF. The left column presents data for the ChrR protein; the right column presents data for the YieF protein. (A) Spectra extracted from the data sets collected when ChrR or YieF (3 μM) was rapidly mixed with limiting NADH (10 μM) and an excess of chromate (40 μM). The spectra were obtained from a global fit of all the absorbance-versus-wavelength-versus-time data to a biphasic kinetic mechanism. Spectrum a, reduced enzyme; spectrum b, semiquinone form of FMN; spectrum c, oxidized enzyme. (B to D) Time courses of absorbance changes at A501 (B, showing the disappearance of the reduced enzyme), A403 (C, showing the appearance of the oxidized enzyme), and A580 (D, showing flavin semiquinone formation) when oxidized ChrR or YieF was mixed with NADH and different molar excesses of chromate. The final concentrations after mixing were as follows: ChrR, 3.0 μM; NADH, 10 μM; chromate, 20 (trace a) or 40 (trace b) μM.
Further clarification of the difference between the chromate reduction characteristics of the two enzymes is provided by the kinetics of change in their electronic states at limiting NADH (10 μM) and excess chromate (20 [trace a] or 40 [trace b] μM) concentrations. While for both enzymes the disappearance of the reduced enzyme (followed at A501 [Fig. 2B]) upon exhaustion of NADH exhibited monophasic kinetics, the two enzymes differed with regard to the appearance of the oxidized enzyme (measured at A403 [Fig. 2C]). For ChrR, but not for YieF, the appearance of oxidized enzyme exhibited a lag. Furthermore, in the case of the former, but not the latter, the disappearance of the reduced enzyme was coupled with transient production of a flavin semiquinone enzyme intermediate (Fig. 2D).
The spectrum for the flavin semiquinone shown in Fig. 2A, trace b, is that calculated if all the ChrR flavin were converted to that form; this concentration of semiquinone was never actually realized in the rapid-mixing experiment. The values of the apparent rate constants for both phases of the ChrR biphasic reaction were directly proportional to the chromate concentration (data not shown). These proportionalities yielded second-order rate constants for the fast and slow phases of ChrR oxidation of 2.1 × 104 and 1.1 × 104 M−1 · s−1, respectively.
ROS generation during chromate reduction by the two enzymes.
No flavin semiquinone form of YieF was detected during the reduction of chromate catalyzed by the enzyme in the rapid-mixing experiments described above. This implies that YieF may be an “obligatory” two-electron reducing enzyme, as is the case with the mammalian enzyme DT-diaphorase, which reduces electrophiles by such obligatory divalent transfer of electrons (3, 7, 17). Full chromate reduction [to Cr(III)] can utilize only three electrons from a four-electron-reduced dimeric enzyme, such as YieF. Thus, if YieF is an obligatory two-electron reducer for chromate reduction, only one electron per molecule of chromate reduced would be available to generate ROS. Direct measurements showed a stoichiometry of 37:17:8 nmol at 1 min and 90:44:24 nmol at 2 min of the reaction among NADH consumed, chromate reduced, and H2O2 generated, respectively. At each time point, the ratio corresponds to three electrons being passed to chromate and one electron being passed to molecular oxygen (subsequently generating H2O2) for every four electrons donated by NADH. The results are consistent with the obligatory four-electron reduction of chromate by the YieF dimer, in which the enzyme simultaneously transferred three electrons to chromate and one to molecular oxygen.
The appearance of the flavin semiquinone form of ChrR, on the other hand, implies a nonsimultaneous transfer of electrons, which would generate Cr(V). Both the flavin semiquinone and Cr(V) have a high probability of reacting with O2 and generating ROS through redox cycling. Measurements revealed that at 1 and 2 min of chromate reduction by ChrR, of the 76 and 150 nmol of NADH electrons consumed, 40 (53%) and 59 (40%), respectively, were utilized in H2O2 formation, i.e., significantly more than the 25% that would have been consumed in the absence of redox cycling. The amounts of chromate that disappeared at corresponding times were 32 and 51 nmol, respectively, which for full reduction to Cr(III) would require some 96 and 153 nmol of electrons, i.e., more than the total NADH electrons consumed. The results are consistent with partial chromate reduction and redox cycling during the above-mentioned reaction. The numbers given for YieF and ChrR represent averages of two independent measurements with each enzyme with <2% variation.
Roles in chromate transformation and detoxification.
The amounts of the toxic ROS generated by ChrR and YieF during chromate reduction differ. We were therefore interested in comparing their roles in affording protection against chromate toxicity. Several attempts employing different suicide vectors (pMRS101 and pGP704), homology regions, antibiotic markers, and culture conditions, however, failed to generate a yieF mutant. yieF is probably essential to E. coli viability, and we are attempting to isolate conditional mutants in this gene.
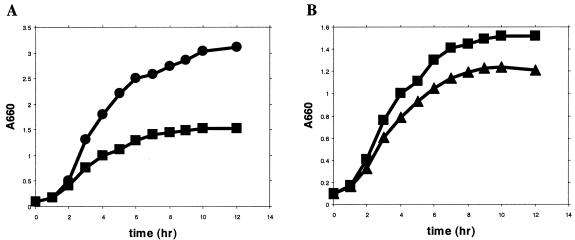
A P. putida strain with a disrupted chrR gene (CRK4) was obtained, enabling us to compare the growth kinetics, chromate transformation, and chromate sensitivity of the wild type and the CRK4 mutant. Chromate (400 μM) inhibited the growth of the wild-type P. putida (Fig. 3A). CRK4 was not affected in growth in the absence of chromate, but it was inhibited to a greater extent than the wild type in its presence (Fig. 3B). The greater toxicity of chromate to CRK4 became more marked at higher chromate concentrations. After 24 h of incubation, the final A660s for the wild type and the mutant at 5, 15, and 20 mM chromate were 0.70 and 0.40, 0.40 and 0.16, and 0.14 and no growth, respectively. Chromate disappearance from the medium occurred during the growth of the culture but was slower for the mutant. After 24 h of incubation in medium containing 400 μM chromate, the wild-type and the mutant strains transformed some 38 and 25% of the chromate, respectively.
FIG. 3.
(A) Growth of wild-type P. putida KT2440 in LB medium with (▪) or without (•) 400 μM Cr(VI). (B) Growth of wild-type P. putida KT2440 (▪) and the CRK4 mutant (▴) in LB medium with 400 μM Cr(VI). The data are the mean points of three independent measurements; the standard error of the mean was <7%.
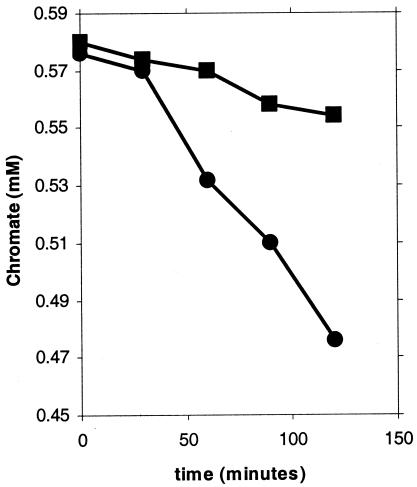
The initial rates of chromate conversion were quantified using dense cell suspensions: the mutant exhibited a 70% decrease in this rate (Fig. 4). Two controls were run to test the possibility that cell surface adsorption influenced chromate disappearance. The first involved the use of autoclaved cells but an otherwise identical protocol. No chromate disappearance was seen, suggesting that surface adsorption did not contribute to the observed results. The findings also eliminated the possible contribution of Fe3+, which is present in significant amounts in the LB broth, in abiotic conversion of chromate. Since autoclaving could have altered the surface properties of the bacteria, a second control involved cells suspended in buffer. Such suspensions removed chromate at some 2% of the rate seen in LB medium, indicating little surface adsorption of chromate; these results also show that the availability of an energy source is essential for chromate reduction. ChrR thus contributes significantly to the chromate-transforming ability of P. putida and protects against its toxicity. How the latter may be affected is considered in Discussion below.
FIG. 4.
Initial rates of chromate removal from LB medium by wild-type (KT2440; •) and mutant (CRK4; ▪) cell suspensions. The results are averages of five independent measurements; the standard error of the mean was <5%.
Induction pattern.
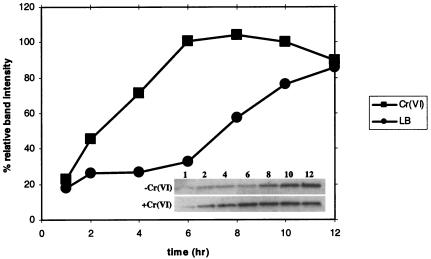
The above results indicate that ChrR may have a functional role in detoxifying chromate in P. putida. Induction patterns can be indicative of function. We therefore determined if chromate induced ChrR. Western analysis showed low levels of the ChrR protein in the exponential phase, with a fourfold induction at the onset of the stationary phase in P. putida cells grown in LB broth without chromate supplementation (Fig. 3A and 5). Chromate (400 μM)-supplemented cultures exhibited increased levels, compared to the unsupplemented cultures, at all time points except 12 h. In these cultures also, the greatest increase was seen between 4 and 6 h, i.e., at the onset of the stationary phase (Fig. 3A). Thus, ChrR is induced by both chromate and the stationary phase; the implication of the latter is considered in Discussion below.
FIG. 5.
Relative band intensities of Western immunoblot detection of ChrR in P. putida KT2440 grown in LB medium with (▪) or without (•) 400 μM Cr(VI). The 8-h (darkest) band intensity, seen in cultures containing chromate, was taken as 100%. The inset shows Western bands for the challenged and unchallenged samples. The numbers indicate the collection time points in hours; 26 μg of total protein was loaded in each lane. Qualitatively similar results were seen in two independent measurements.
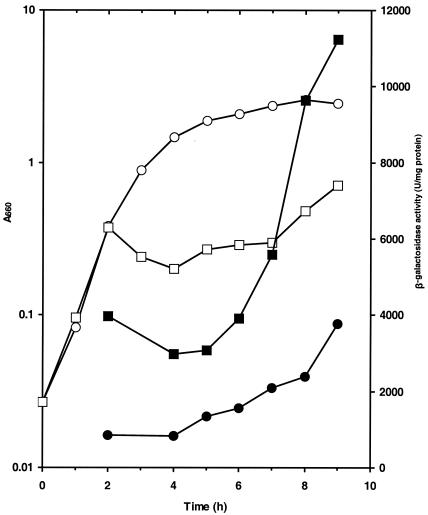
Our inability to obtain a yieF E. coli mutant precluded direct investigation of whether YieF protected the bacterium against chromate toxicity. We nevertheless examined whether chromate induced the enzyme in E. coli for the reason stated above. The ChrR antibody did not recognize the YieF protein, and we used E. coli strain AMS6λY, bearing a single-copy yieF-lacZ transcriptional fusion (Table 1), to address this question. The fusion exhibited induction in the stationary phase in cultures without chromate, while chromate induced the fusion in all growth phases (Fig. 6). In the presence of chromate, a biphasic growth pattern was seen, with growth arrest after two to three generations, followed by partial recovery. The recovery coincided with maximal yieF induction, which may be because the induction of the fusion reflected induction of the protein. Thus, the induction pattern of YieF appears to be consistent with the possibility that it, too, protects against chromate toxicity.
FIG. 6.
β-Galactosidase activity determined during growth of AMS-6λY in LB medium without (•) or with (▪) 200 μM chromate. The open symbols show culture growth monitored at A660 in the absence (○) or presence (□) of chromate. The results represent averages of three experiments; the standard error of the mean was <5%.
DISCUSSION
Many soluble flavoproteins with unrelated metabolic functions have been examined for chromate reductase activity. Examples are glutathione reductase, lipoyl dehydrogenase, and ferredoxin-NADP oxidoreductase. These catalyze one-electron reduction of chromate, generating flavin semiquinones and Cr(V), which redox cycle and generate large quantities of ROS. These studies were conducted to elucidate the biological basis of chromate toxicity, and such enzymes are clearly not appropriate for chromate bioremediation. Their activity damages the cell, and Cr(III) is not their end product (14, 32, 33).
Besides previous (27, 28; C. F. Gonzalez, D. F. Ackerley, M. Keyhan, R. Blake, and A. Matin, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. O-123, 2003.) and the present work, two soluble bacterial enzymes have been studied with respect to Cr(VI) reduction beyond Cr(V). One is the chromate reductase purified from Pseudomonas ambigua, which was shown to generate Cr(V) only transiently. Indirect evidence suggested that the end product of the reaction was Cr(III) (39). Whether the enzyme activity enabled the organism to remediate and/or detoxify chromate was not addressed. That the E. coli Fre protein reduced Cr(VI) to Cr(III) was shown directly (29), and since this enzyme has superior chromate reduction kinetics, it too may be a suitable candidate for further work. Fre does not contain bound flavin but reduces free flavins (FMN, flavin adenine dinucleotide, and riboflavin) that are then able to pass electrons to chromate. Free flavins are highly reactive, particularly in generating ROS; this may limit the usefulness of this enzyme in chromate bioremediation. Whether Fre contributes to the E. coli capacity to detoxify and remediate chromate has also not been investigated.
Both ChrR and YieF, in electrophoretically pure form, converted Cr(VI) to Cr(III). This is shown by the XANES spectrum measurements, as well as by stopped-flow experiments, which demonstrated that each of the reduced enzymes was fully oxidized by chromate, albeit with some delay in the case of ChrR. Further, we show that ChrR not only contributes to the chromate-remediating ability of P. putida but also reduces chromate toxicity to the bacterium, since the mutant CRK4 lacking this enzyme exhibited significant impairment in both respects.
ChrR belongs to the NADH_dh2 family of proteins, which consists of obligatory two-electron reducers of electrophiles. Thus, a possible reason for the protective role of ChrR against chromate toxicity could have been that the dimeric enzyme reduced chromate by a simultaneous four-electron transfer. In such a scenario, the four-electron-reduced enzyme dimer would catalyze a one-step, three-electron reduction of Cr(VI) to Cr(III), and the remaining one flavin semiquinone would simultaneously react with another electron acceptor, such as molecular oxygen, so that no more than 25% of the electrons consumed would form ROS. However, the fact that the semiquinone form of the enzyme was detected in stopped-flow experiments, together with the finding that >25% of the NADH electrons were transferred to ROS, indicates that ChrR is not a pure two-electron reducer of chromate and that Cr(V) is likely to be an intermediate in the reaction it catalyzes. Whether Cr(V) is formed during ChrR-catalyzed reduction of Cr(VI) is under investigation.
Regardless, and notwithstanding considerable ROS generation, ChrR does protect P. putida against chromate toxicity. What accounts for this effect? As stated above, several cellular enzymes are pure one-electron reducers of chromate. Indeed, cellular metabolites, such as glutathione and ascorbate, can also do the same (9, 16). Since the resulting redox cycling generates extensive ROS, an enzyme that generates Cr(V) only transiently can in principle have a protective role. During ChrR-catalyzed chromate reduction, ROS formation diminished considerably with time. This is consistent with the possibility that Cr(V) generation occurs only transiently during this reaction. If so, the protective role of ChrR may lie in its ability to preempt reduction of chromate by cellular one-electron reducers, thereby minimizing ROS formation. Competition studies between ChrR and one-electron reducers for chromate reduction are now in progress to test this notion.
In contrast to ChrR, YieF did not show semiquinone flavoprotein generation during chromate reduction and consumed only some 25% of NADH electrons to molecular oxygen in ROS generation. Thus, this enzyme produces only the minimal amount of ROS, involving a reaction in which a four-electron-reduced enzyme catalyzes a three-electron-requiring reduction. To our knowledge this is the first enzyme for which stoichiometric production of ROS associated with Cr(VI) reduction has been demonstrated. YieF should thus prove to be particularly effective in protecting against chromate toxicity; our present attempts to obtain a conditional E. coli yieF mutant are aimed at testing this possibility.
Have ChrR and YieF evolved specifically to protect against chromate toxicity? Given that chromium in nature exists primarily as chromite ore (FeCr2O4), in which it is present in the 3+ state (2), and that the introduction of chromate in the environment is a relatively recent event, this is unlikely. Instead, our ongoing work strongly suggests that ChrR and YieF may be bacterial counterparts of the mammalian DT-diaphorase (7, 15, 17) with the main biological role of detoxifying quinonoid compounds, which are a constant threat to bacteria (37). Their protection against chromate toxicity may be a reflection of a more generalized role of divalent reduction of electrophiles that are prone to one-electron reduction and ROS generation. As YieF can reduce cytochrome c, other metals, and K3FeCN6, it may have additional roles, e.g., in metal assimilation. These and other possibilities are presently under investigation.
As stated in the introduction, our long-term goal is to improve bacterial bioremediation through genetic and protein engineering, and one of the objectives of the present work was to identify suitable enzymes for this end. Several findings make ChrR and YieF appropriate for further investigation. Both convert Cr(VI) to Cr(III); ChrR protects against chromate toxicity, and it is likely that YieF will prove to be even more effective in this respect; and both enzymes appear to be naturally under the control of starvation promoters, as they are induced in the stationary phase. Such promoters are activated at low growth rates (13), and this should prove advantageous. By permitting effective gene expression without the need for rapid growth, these promoters minimize biomass formation and can enhance the efficacy of biostimulation (21, 22). The fact that yieF and chrR belong to a large class of homologous genes will facilitate the application of techniques like DNA shuffling (38) for evolving enzymes with improved capacities for chromate bioremediation.
Acknowledgments
This research was funded by grants DE-FG03-97 ER-624940, DE-FG02-03 ER63627, and NAG2-1 to A.M. from the Natural and Accelerated Bioremediation (NABIR) program, Biological and Environmental Research (EBER), DOE, and NASA, respectively, and grant DE-FG02-96 ER20228 from the DOE to R.B. C.F.G. and D.F.A. were supported by grants from Conicet, Argentina, and STAX0101 from FRST New Zealand, respectively.
REFERENCES
- 1.Ackerley, D. F., T. T. Caradoc-Davies, and I. L. Lamont. 2003. Substrate specificity of the nonribosomal peptide synthetase PvdD from Pseudomonas aeruginosa. J. Bacteriol. 185:2848-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barceloux, D. G. 1999. Chromium. J. Toxicol. Clin. Toxicol. 37:173-194. [DOI] [PubMed] [Google Scholar]
- 3.Cadenas, E. 1995. Antioxidant and prooxidant functions of DT-diaphorase in quinone metabolism. Biochem. Pharmacol. 49:127-140. [DOI] [PubMed] [Google Scholar]
- 4.Campos-Garcia, J., G. Martinez-Cadena, R. Alvarez-Gonzalez, and C. Cervantes. 1997. Purification and partial characterization of a chromate reductase from Bacillus. Rev. Lat. Microbiol. 39:73-81. [PubMed] [Google Scholar]
- 5.Cervantes, C., J. Campos-Garcia, S. Devars, F. Gutierrez-Corona, H. Loza-Tavera, J. C. Torres-Guzman, and R. Moreno-Sanchez. 2001. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25:335-347. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernster, L. 1967. DT-diaphorase. Methods Enzymol. 10:309-317. [Google Scholar]
- 8.Gibson, Q. H., B. E. P. Swoboda, and V. Massey. 1964. Kinetics and mechanism of action of glucose oxidase. J. Biol. Chem. 239:3927-3934. [PubMed] [Google Scholar]
- 9.Goodgame, D. M., and A. M. Joy. 1986. Relatively long-lived chromium(V) species are produced by the action of glutathione on carcinogenic chromium(VI). J. Inorg. Biochem. 26:219-224. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg, A. E., J. J. Connors, D. Jenkins, and M. A. Franson (ed.). 1981. Standard methods for the examination of water and wastewater, 15th ed., p. 187-190. American Public Health Association, Washington, D.C.
- 11.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Ishibashi, Y., C. Cervantes, and S. Silver. 1990. Chromium reduction in Pseudomonas putida. Appl. Environ. Microbiol. 56:2268-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins, D. E., E. A. Auger, and A. Matin. 1991. Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival. J. Bacteriol. 173:1992-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein, C., E. Snow, and K. Frenkel. 1998. Molecular mechanisms in metal carcinogenesis: role of oxidative stress, p. 79-137. In O. Aruoma and B. Halliwell (ed.), Molecular biology of free radicals in human diseases. Academic Press, New York, N.Y.
- 15.Landi, L., D. Fiorentini, M. C. Galli, J. Segura-Aguilar, and R. E. Beyer. 1997. DT-diaphorase maintains the reduced state of ubiquinones in lipid vesicles thereby promoting their antioxidant function. Free Radic. Biol. Med. 22:329-335. [DOI] [PubMed] [Google Scholar]
- 16.Lefebvre, Y., and H. Pezerat. 1992. Production of activated species of oxygen during the chromate(VI)-ascorbate reaction: implication in carcinogenesis. Chem. Res. Toxicol. 5:461-463. [DOI] [PubMed] [Google Scholar]
- 17.Lind, C., P. Hochstein, and L. Ernster. 1982. DT-diaphorase as a quinone reductase: a cellular control device against semiquinone and superoxide radical formation. Arch. Biochem. Biophys. 216:178-185. [DOI] [PubMed] [Google Scholar]
- 18.Lomovskaya, O., K. Lewis, and A. Matin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty, P. L. 1994. Ground-water treatment for chlorinated solvents, p. 87-116. In J. E. Matthews (ed.), Handbook of bioremediation. Lewis Publishers, Ann Arbor, Mich.
- 20.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Matin, A. 1994. Starvation promoters of Escherichia coli. Their function, regulation, and use in bioprocessing and bioremediation. Ann. N. Y. Acad. Sci. 721:277-291. [DOI] [PubMed] [Google Scholar]
- 22.Matin, A., C. D. Little, C. D. Fraley, and M. Keyhan. 1995. Use of starvation promoters to limit growth and selectively enrich expression of trichloroethylene- and phenol-transforming activity in recombinant Escherichia coli. Appl. Environ. Microbiol. 61:3323-3328. (Erratum, 61:4140.) [DOI] [PMC free article] [PubMed]
- 23.Michel, C., M. Brugna, C. Aubert, A. Bernadac, and M. Bruschi. 2001. Enzymatic reduction of chromate: comparative studies using sulfate-reducing bacteria. Key role of polyheme cytochromes c and hydrogenases. Appl. Microbiol. Biotechnol. 55:95-100. [DOI] [PubMed] [Google Scholar]
- 24.Norris, R. D. 1994. In-situ bioremediation of soils and groundwater contaminated with petroleum hydrocarbons, p. 17-37. In J. E. Matthews (ed.), Handbook of bioremediation. Lewis Publishers, Ann Arbor, Mich.
- 25.Ohtake H., K. Komori, C. Cervantes, and K. Toda. Chromate-resistance in a chromate-reducing strain of Enterobacter cloacae. FEMS Microbiol. Lett. 55:85-88. [DOI] [PubMed]
- 26.Pandza, S., M. Baetens, C. H. Park, T. Au, M. Keyhan, and A. Matin. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 36:414-423. [DOI] [PubMed] [Google Scholar]
- 27.Park, C. H., C. F. Gonzalez, D. F. Ackerley, M. Keyhan, and A. Matin. 2002. Molecular engineering of soluble bacterial proteins with chromate reductase activity, p. 103-111. In R. E. Hinche et al. (ed.), Remediation and beneficial reuse of contaminated sediments. Batelle Press, Columbus, Ohio.
- 28.Park, C. H., M. Keyhan, B. Wielinga, S. Fendorf, and A. Matin. 2000. Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl. Environ. Microbiol. 66:1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puzon, G. J., J. N. Petersen, A. G. Roberts, D. M. Kramer, and L. Xun. 2002. A bacterial flavin reductase system reduces chromate to a soluble chromium(III)-NAD(+) complex. Biochem. Biophys. Res. Commun. 294:76-81. [DOI] [PubMed] [Google Scholar]
- 30.Riley, R. G., J. M. Zachara, and F. J. Wobber. 1992. Chemical contaminants on DOE lands and selection of contaminants mixtures for subsurface science research. Report DOE/ER-0547T. U.S. Department of Energy, Washington, D.C.
- 31.Sarker, M. R., and G. R. Cornelis. 1997. An improved version of suicide vector pKNG101 for gene replacement in gram-negative bacteria. Mol. Microbiol. 23:410-411. [DOI] [PubMed] [Google Scholar]
- 32.Shi, X. L., and N. S. Dalal. 1990. One-electron reduction of chromate by NADPH-dependent glutathione reductase. J. Inorg. Biochem. 40:1-12. [DOI] [PubMed] [Google Scholar]
- 33.Shi, X. L., and N. S. Dalal. 1990. NADPH-dependent flavoenzymes catalyze one electron reduction of metal ions and molecular oxygen and generate hydroxyl radicals. FEBS Lett. 276:189-191. [DOI] [PubMed] [Google Scholar]
- 34.Simmons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 35.Singh, J., D. L. Carlisle, D. E. Pritchard, and S. R. Patierno. 1998. Chromium-induced genotoxicity and apoptosis: relationship to chromium carcinogenesis. Oncol. Rep. 5:1307-1318. [DOI] [PubMed] [Google Scholar]
- 36.Smith, B. J. 1994. SDS polyacrylamide gel electrophoresis of proteins. Methods Mol. Biol. 32:23-34. [DOI] [PubMed] [Google Scholar]
- 37.Soballe, B., and R. K. Poole. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145:1817-1830. [DOI] [PubMed] [Google Scholar]
- 38.Stemmer, W. P. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, T., N. Miyata, H. Horitsu, K. Kawai, K. Takamizawa, Y. Tai, and M. Okazaki. 1992. NAD(P)H-dependent chromium(VI) reductase of Pseudomonas ambigua G-1: a Cr(V) intermediate is formed during the reduction of Cr(VI) to Cr(III). J. Bacteriol. 174:5340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanner, J. J., B. Lei, S. C. Tu, and K. L. Krause. 1996. Flavin reductase P: structure of a dimeric enzyme that reduces flavin. Biochemistry 35:13531-13539. [DOI] [PubMed] [Google Scholar]
- 41.Wang P. C., T. Mori, K. Toda, and H. Ohtake. 1990. Membrane-associated chromate reductase activity from Enterobacter cloacae. J. Bacteriol. 172:1670-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zgurskaya, H., M. Keyhan, and A. Matin. 1997. The σs level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24:643-651. [DOI] [PubMed] [Google Scholar]