Abstract
Chitin synthase (CHS) represents an attractive target site for combating insect pests as insect growth and development are strictly dependent on precisely tuned chitin biosynthesis and this pathway is absent in humans and other vertebrates. Current knowledge on CHS in insects, especially their structures, functions, and regulations is still very limited. We report the identification and characterization of two chitin synthase genes, AgCHS1 and AgCHS2, in African malaria mosquito, Anopheles gambiae. AgCHS1 and AgCHS2 were predicted to encode proteins of 1,578 and 1,586 amino acid residues, respectively. Their deduced amino acid sequences show high similarities to other insect chitin synthases. Transcriptional analysis indicated that AgCHS1 was expressed in egg, larval, pupal and adult stages whereas AgCHS2 appeared to be expressed at relatively low levels, particularly during the larval stages as examined by reverse transcription (RT)-PCR and real-time quantitative PCR. Relatively high expression was detected in the carcass followed by the foregut and hindgut for AgCHS1, and the foregut (cardia included) followed by the midgut for AgCHS2. Fluorescence in situ hybridization (FISH) and immunohistochemical analysis revealed new information including the localization of the two enzymes in the ommatidia of the compound eyes, and AgCHS2 in the thoracic and abdominal inter-segmental regions of pupal integument.
Keywords: Anophele gambiae, chitin synthases, gene expression pattern, fluorescent in situ hybridization, immunohistochemistry
1. Introduction
Chitin, a linear polysaccharide of N-acetyl-β-D-glucosamine residues joined by β-1,4 glycosidic linkages, is the second most abundant biological polymer after cellulose (Merzendorfer, 2006; Kramer and Muthukrishnan, 2005). It is widely distributed in arthropods, fungi and nematodes. In arthropods, chitin is a vital component of the cuticular exoskeleton and thus is crucial for growth and development (Merzendorfer and Zimoch, 2003). Chitin is also found in internal structures of many insects and other arthropods, including the cuticular linings of trachea and in the peritrophic matrix (PM) lining the gut epithelium (Richards, 1951; Hunt, 1970; Cohen, 2001).
Chitin synthase is a crucial enzyme catalyzing the transfer of sugar moieties from activated sugar donors to specific acceptors. The first cDNA encoding insect chitin synthase was isolated and sequenced from the sheep blowfly (Lucilia cuprina) in 2000 (Tellam et al., 2000). Since then, several cDNAs encoding chitin synthases have been isolated and sequenced from at least ten other insect species including the Anopheles gambiae, Aedes aegypti (Ibrahim et al., 2000), Drosophila melanogasler (Gagou et al., 2002), Manduca sexta (Zhu et al., 2002; Hogenkamp et al., 2005), Tribolium castaneum (Arakane et al., 2004), Spodoptera frugiperda (Bolognesi et al., 2005), An. quadrimaculatus (Zhang and Zhu, 2006), Plutella xylostella (Ashfaq et al., 2007), S. exigua (Chen et al., 2007; Kumar et al., 2008), Locusta migratoria manilensis (Zhang et al., 2010), and Ostrina funracalis (Qu and Yang, 2011). Furthermore, the completion of several insect genome sequencing projects has provided comprehensive information for identifying and characterizing these genes in different insect species.
Insects are known to possess two chitin synthases encoded by two different genes, CHS1 and CHS2 (also known as CHS A and CHS B, respectively). These genes are closely related but can be clearly grouped into two different phylogenetic classes (Merzendorfer, 2006). CHS1 is exclusively expressed in the epidermis underlying the cuticular exoskeleton and related ectodermal cells such as tracheal cells, whereas CHS2 is known to be expressed in gut epithelial cells and its coding enzyme is responsible for the synthesis of the PM-associated chitin (Merzendorfer and Zimoch, 2003; Arakane et al., 2004, 2005; Hogenkamp et al., 2005; Zimoch et al., 2005; Ashfaq et al., 2007). The insect CHS1 contains alternative exons which lead to the production of two splicing mRNA variants. These variants are differentially expressed in the epidermis and tracheae during insect development (Arakane et al., 2004, Hogenkamp et al., 2005, Zimoch et al., 2005, Qu and Yang, 2011). In contrast, alternative splicing variants have not been reported for CHS2 in insects.
Current knowledge on the function and regulation of chitin synthases in insects is rather limited. Functional analysis of two chitin synthases using RNA interference (RNAi) in different insect species such as T. castaneum, S. exigua and L. migratoria mannilensis showed that chitin synthases are required for survival, fecundity and egg hatching, and molting processes (Arakane et al., 2005, 2008; Merzendorfer, 2006; Tian et al., 2009; Zhang et al., 2010). Chitin synthase presents an attractive target for combating insect pests and fungi-born diseases as insect and fungus growth and development is dependent on precisely tuned expression of chitin synthase genes and this process is absent in vertebrates (Merzendorfer, 2006). For example, peptidyl nucleosides including polyoxins and nikkomycins are anti-fungi agents which competitively inhibit chitin synthases in fungi. Benzylphenolureas (BPUs) such as diflubenzuron are potent insecticides that inhibit chitin biosynthesis. However, it remains controversal whether chitin synthases are the direct targets for this group of insecticides. Interestingly, a recent study from our lab showed that up-regulation of CHS1 at the transcriptional level is associated with the exposure to diflubenzuron in An. quadrimaculatus (Zhang and Zhu, 2006).
An. gambiae is an important arthropod-borne disease vector in Africa (Hay et al., 2004). To date, a very limited number of insecticides are available for control of mosquitoes and other human health-related arthropods. The BPU insecticides including diflubenzruon and lufenuron have shown a great potential for control of the mosquito populations (Moreira et al., 2007; Zhu et al., 2007). In this paper, we reported the identification and characterization of two chitin synthase genes (AgCHS1 and AgCHS2) of An. gambiae. The study is expected to better understand the molecular properties of insect chitin synthase genes and potentially help the development of new insecticides targeting on chitin metabolic pathways in insects.
2. Materials and Methods
2.1. Mosquito rearing
A colony of An. gambiae obtained from the Malaria Research and Reference Reagent Resource Center (MR4) (Manassas, VA) was maintained in the Department of Entomology at Kansas State University (Manhattan, KS) since 2007 by using the same methods as described by Zhang and Zhu (2006). Briefly, the larvae were fed with slurry of brewer’s yeast and TetraMin Baby-E fish food, whereas adults were fed with a 10% sucrose solution soaked into cotton balls. Two-day-old females were fed with pre-warmed, defibrinated horse blood (Colorado Serum Company, Denver, Colorado) in a membrane feeder made of a lubricated Naturalamb brand condom (Church and Dwight Co., Inc., Princeton, NJ) and allowed to lay eggs.
2.2. Sequence analysis of AgCHS1 and AgCHS2
By searching the NCBI databases, three cDNA sequences putatively derived from two different genes including AgCHS1 (VectorBase gene no. AGAP001748-PA) and AgCHS2 (VectorBase gene no. AGAP001205-PA, NCBI RefSeq no. XP_321951.2) were identified in An. gambiae. AgCHS1 appeared to transcribe two splicing variants including AgCHS1A (NCBI RefSeq no. XM_321336.5) and AgCHS1B (NCBI RefSeq no. XM_321336.4). It should be mentioned that AgCHS2 was previously referred to as AgCHS1 (Arakane et al., 2004) and has been amended to its current nomenclature by Merzendorfer (2006). Sequence analysis was performed using the computer software suite Lasergene (DNAStar, WI). Genomic organizations of AgCHS1 and AgCHS2 were obtained by blast search of genome sequence of An. gambiae using UCSC Genome Bioinformatics program (http://genome.ucsc.edu/). Other software available from online servers was described in the results.
2.3. RT-PCR analysis
Total RNA was isolated from insect sample representing each of seven developmental stages, including egg; first-, second-, third- and fourth-instar larva; pupa; and adult, by using the TRIzol Total RNA Isolation kit (Invitrogen, Carlsbad, CA) for studying stage-specific expressions of AgCHS genes. The same method was used to isolate total RNA from insect sample representing each of five egg developmental times, including 12, 24, 36, 48, and 60 hour after egg deposition, and five pupal developmental times, including 0, 10, 20, 30, and 34 hour after pupation, respectively. Similarly, total RNA was also isolated from tissue samples representing each of four tissue types, including foregut, midgut, hindgut and carcass (whole larva after the gut was removed), for studying tissue-specific expressions. In brief, fourth-instar larvae were chilled on ice and dissected in cold 1×PBS to get tissues. Mosquito larva was longitudinally opened by carefully cutting the integument from one side of the larva without damaging the gut. Then the whole intestine was gently removed and detached from adhering tissues including Malpighian tubules, trachea, and fat bodies. The midgut, foregut, and hindgut were carefully separated and immediately placed in the TRIzol reagent. The foregut and midgut were separated in the junction of the gastric caecum. All remaining body tissues excluding the whole intestine were collected as the carcass.
After total RNA was isolated, the concentration was measured using the NanoDrop ND-1000 instrument (NanoDrop Technologies, Inc., Wilmington, DE). Aliquots of 2.5 μg of total RNA were firstly treated with DNase using the DNase I kit (Fermentas, Glen Burnie, MD) and then used for the first-strand cDNA synthesis with the First Strand cDNA Synthesis kit (Fermentas, Glen Burnie, MD) using an oligo (dT)12–18 primer in a 20-μl reaction volume following the manufacturer’s protocol. Beacon Designer software from Primer Biosoft (http://www.premierbiosoft.com) was used to design the gene-specific primers for AgCHS1A, AgCHS1B, and AgCHS2 that are shown in Table 1. PCR was performed using the PCR Master Mix (Fermentas) with a thermal cycle program consisting of an initial denaturation at 94°C for 2 min followed by 29 cycles of 94°C for 30s, 55°C for 30s and 72°C for 45s, and a final extension at 72°C for 10 min. The PCR products were resolved on a 1.8% agarose gel and visualized by staining with ethidium bromide. If the gene expression level was low and the PCR products were not detected, five more cycles were run and the products were checked again. The mosquito ribosomal protein S3 (AgRPS3) gene was used as a reference for RT-PCR analysis. At least three biological replicates with independent preparations of total RNA from each developmental stage or each tissue type, were performed for each of the two AgCHS genes and each of the two AgCHS1 splicing variants, AgCHS1A and AgCHS1B.
Table 1.
Sequences and relevant parameters of the primers used for RT- PCR and qPCR.
| Gene | Primer name | primer sequence (5′-3′) | Length | Tm (°C) |
|---|---|---|---|---|
| AgCHS1 | AgCHS1F | ACCAACTGTCTGTGTGTTATAC | 22 | 54.9 |
| AgCHS1R | CCGCAAGATGTTAGAAGAGC | 20 | 54.6 | |
| AgCHS1A | AgCHS1AF | ACGAGCGCGACTTCCTCAC | 19 | 55.4 |
| AgCHS1AR | GAGTCGCGCAACTCCTTGAG | 20 | 55.9 | |
| AgCHS1B | AgCHS1BF | ACGAGCGCGACTTCCTCAC | 19 | 55.4 |
| AgCHS1BR | GTCGACGGCGATTCTCGC | 18 | 54.9 | |
| AgCHS2 | AgCHS2F | CACCAGCAACGCCATCATC | 19 | 53.2 |
| AgCHS2R | GAACACCAGCAGCAGAGTAAC | 21 | 54.4 | |
| AgRPS3 | AgRPS3F | GCTGGGCATCAAGGTCAAG | 19 | 55.4 |
| AgRPS3R | ATCTCATCCTTCGGCTCAAC | 20 | 54.4 |
2.4. Real-time quantitative PCR (qPCR) analysis
To profile the developmental stage- and tissue-specific expression patterns of each AgCHS gene, cDNA prepared from above mentioned samples representing each of four developmental stages, including egg, forth-instar larva, pupa, and adult, and from each of four tissues, including foregut, midgut, hindgut, and carcass, was used for qPCR analysis. The qPCR was performed in a 25-μl reaction containing 10.5 μl of 10-fold diluted cDNAs, 0.4 μM of each primer, and 1× Maxima SYBR Green qPCR Master Mix (Fermentas) using the iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA). The optimized qPCR program used for quantification of transcripts for both AgRPS3 and AqCHS1 consisted of an initial denaturation step at 95°C for 5 min followed by 40 cycles of 95°C for 15s, 55°C for 30s and 70°C for 30s. At the end of the PCR, amplification specificity was verified by obtaining the dissociation curve, in which the samples were cooled to 55°C after denaturing, and then the melting curves were obtained by increasing 0.5°C/10 s for each cycle with a total of 80 cycles until reaching 95°C to denature the double-stranded DNA. The specificity of each reaction was evaluated based on the melting temperatures of the PCR products. The amplification efficiency of primer pairs was determined from the slope of the curve generated by amplification from serially diluted cDNA. The primer efficiency had to be at least 0.9 for a primer pair to be accepted. Relative expression values (REVs) for the tissue-specific gene expressions were then determined by dividing the quantity of the target sequence of interest with the quantity obtained for AgRPS3 as an internal reference gene. We found that expression of AgRPS3 fluctuated across the developmental stages that were tested. Other genes including RpS7, ribosomal protein L32, elongation factor 2, and the ubiquitin-ribosomal protein L40 fusion protein were also tested. However, none of these was suitable as a “housekeeping gene” to normalize our data across the developmental stages in An. gambiae as also noted in other insect species (Togawa et al., 2008). Therefore, we did not normalize the stage-specific gene expression using AgRPS3, but adopted very careful quantification of RNA using NanoDrop measurements to standardize our samples. The qPCR for each gene was conducted with three biological replicates, each with two technical repeated measurements.
2.5. Heterologous expression and purification of antigens and antibody preparations
cDNA fragments encoding partial protein sequences of AgCHS1 and AgCHS2 were amplified by RT-PCR with primers 5′-CCATGGGCAAAACGACGGACGAGAAGGCGCA and 5′-
 GTGCGCAATACGTGCCTGTTCCTC, and 5′-CCATGGCCAGCGCCGAAAAGGAGCAAATCGCA and 5′-
GTGCGCAATACGTGCCTGTTCCTC, and 5′-CCATGGCCAGCGCCGAAAAGGAGCAAATCGCA and 5′-
 CTTCATCTCTTCCTTCTGCTTTTCCG, respectively, using total RNA (RNeasy Mini Kit, QIAGEN) isolated from the fourth-instar larvae of An gambiae. The underlining, double underlining, and dashed lines below the primer sequences indicate the Nco I, EcoR I site, and stop codon, respectively. Each amplified fragment (StrataScript® one-tube RT-PCR system with Easy-A® High-fidelity PCR cloning enzyme, Stratagene) (about 400bp) was inserted into PCR 2.1 vector of Topo TA cloning kit (Invitrogen) and sequenced to confirm their sequences and orientaions. The plasmid DNA was digested with Nco1 and EcoR1 and the resultant DNA fragment was ligated into pET-32a (+) vector (Novagen, Madison, WI) that had been digested with Nco1 and EcoR1 to obtain the plasmid for expression of recombinant AgCHS1 and AgCHS2 proteins (rAgCHS1 and rAgCHS2) according to the manufacturer’s instructions. In brief, the plasmid encoded the fusion protein with a His-tag and a GST-tag was used to transform the BL21 (DE3) cells. The transformant cells were cultured at 37°C for 16 h in LB medium containing 100 μg/ml ampicillin. Two milliliters of the culture was added to 400 ml of LB medium and incubated at 37°C until OD600 reached 0.5 followed by addition of IPTG at a final concentration of 0.4 mM and incubation of 4 h at 37°C. The cells were pelleted and resuspended in 10 ml lysis buffer (8 M urea, 0.1 M sodium phosphate buffer, 0.01 M Tris-CI, pH 8.0). The cells were lysed by gently shaking for 45 min at room temperature. After cell debris was removed by centrifugation at 5,000×g for 15 min, the recombinant proteins in the supernatant were purified by NTA-Ni2+-resin (Novagen) following the manufacturer’s protocol. The eluted proteins were concentrated by using centriprep centrifugal YM-3 (Millipore, Billerica, MA), then digested with enterokinases (S·Tag™ rEK Purification Kit, Novagen) to cut the GST and His tag. The digested proteins were separated by 4–20% gradient SDS-PAGE. The bands of the recombinant proteins without the tag were cut and the proteins were eluted by using Tris-NaCI buffer. The resultant recombinant proteins, rAgCHS1 and rAgCHS2, were used to immunize mice (Interdisciplinary Center for Biotechnology Research, University of Florida). The serum of the immunized mice was collected as the anti-AgCHS1 and AgCHS2 serums.
CTTCATCTCTTCCTTCTGCTTTTCCG, respectively, using total RNA (RNeasy Mini Kit, QIAGEN) isolated from the fourth-instar larvae of An gambiae. The underlining, double underlining, and dashed lines below the primer sequences indicate the Nco I, EcoR I site, and stop codon, respectively. Each amplified fragment (StrataScript® one-tube RT-PCR system with Easy-A® High-fidelity PCR cloning enzyme, Stratagene) (about 400bp) was inserted into PCR 2.1 vector of Topo TA cloning kit (Invitrogen) and sequenced to confirm their sequences and orientaions. The plasmid DNA was digested with Nco1 and EcoR1 and the resultant DNA fragment was ligated into pET-32a (+) vector (Novagen, Madison, WI) that had been digested with Nco1 and EcoR1 to obtain the plasmid for expression of recombinant AgCHS1 and AgCHS2 proteins (rAgCHS1 and rAgCHS2) according to the manufacturer’s instructions. In brief, the plasmid encoded the fusion protein with a His-tag and a GST-tag was used to transform the BL21 (DE3) cells. The transformant cells were cultured at 37°C for 16 h in LB medium containing 100 μg/ml ampicillin. Two milliliters of the culture was added to 400 ml of LB medium and incubated at 37°C until OD600 reached 0.5 followed by addition of IPTG at a final concentration of 0.4 mM and incubation of 4 h at 37°C. The cells were pelleted and resuspended in 10 ml lysis buffer (8 M urea, 0.1 M sodium phosphate buffer, 0.01 M Tris-CI, pH 8.0). The cells were lysed by gently shaking for 45 min at room temperature. After cell debris was removed by centrifugation at 5,000×g for 15 min, the recombinant proteins in the supernatant were purified by NTA-Ni2+-resin (Novagen) following the manufacturer’s protocol. The eluted proteins were concentrated by using centriprep centrifugal YM-3 (Millipore, Billerica, MA), then digested with enterokinases (S·Tag™ rEK Purification Kit, Novagen) to cut the GST and His tag. The digested proteins were separated by 4–20% gradient SDS-PAGE. The bands of the recombinant proteins without the tag were cut and the proteins were eluted by using Tris-NaCI buffer. The resultant recombinant proteins, rAgCHS1 and rAgCHS2, were used to immunize mice (Interdisciplinary Center for Biotechnology Research, University of Florida). The serum of the immunized mice was collected as the anti-AgCHS1 and AgCHS2 serums.
Specificity of two antibodies was examined by western blotting analysis. Briefly, each of the two recombinant proteins and mosquito homognenate were treated with 6 × SDS sample buffer containing β-mercaptoethanol at 95°C for 5 min and then separated by 15% SDS-PAGE followed by transferring the proteins to a PVDF membrane. After blocking in 5% milk in 1× Tris Buffer Saline Tween20 (TBST) buffer (TBST) (Tris-Buffered Saline/TWEEN®-20) for 1 h, the membrane was immunoblotted with 1:1000-diluted anti-serum in 1× TTBS containing 0.5% milk for 1h at room temperature (23–25 °C). After 3 times of washing with 1×TBST (10 min each), goat anti-mouse IgG HRP-conjugated secondary antibodies (Promega) were used at a 1:20,000 dilution 1×TBST containing 0.5% milk for 1h at room temperature. After 3 times washing with 1×TBST (10 min each), primary antibody binding was visualized by western Lightning Chemiluminescence Reagent Plus Kit (Perkin Elmer).
2.6. Immunohistochemistry
Paraffin-embedded thin sections were used for immunohistochemical analysis of two chitin synthases. In brief, 12 to 24 h pupae were fixed in 4% paraformaldehyde at 4°C overnight and then washed three times (5 min each) with PBST (PBS and 0.1% Triton X-100). The samples were then dehydrated through a series of ethanol solutions (2× 30 min in 70 and 96%, 2× 20 min in 100%), followed by 2× 1 h in chloroform. The dehydrated samples were finally embedded in paraplast (56°C, Tyco Healthcare) after overnight penetration. Histological sections (8 μm) were prepared by using a microtome (Richard-Allan Scientific Microm) with a low profile microtome blade (Richard-Allan), straightened on Fisherbrand ColorFrost Plus microscope slides with 0.5% gelatin, and allowed to dry for 2 d at 40°C on the top of a slide warmer. The sections were deparaffinized with two washes of 10 min xylene, rehydrated through successive baths of ethanol (100%, 96%, and 70% in water, 1× 5 min each), two water baths for 5 min each, and finally PBST for 10 min.
To determine the localization of AgCHS1 and AgCHS2, the sections were blocked in 1% BSA (bovine serum albumin) in PBST for 15 min followed by incubation with a 1:100 dilution of anti-AgCHS1 and anti-AgCHS2 sera in PBST at 4°C overnight, respectively. Paraffin-embedded thin sections immunostained with a preimmune serum were used as negative controls. The sections were then washed with PBST three times for 2 min each. The sections were incubated with Alexa 488-conjugated goat anti-mouse IgG (1:500 dilution in PBST) at 4°C overnight. After four washes in PBS for 10 min each, the sections were mounted in glycerol on a glass side and the fluorescence was observed using Nikon Eclipse E800 fluorescence compound microscope equipped with appropriate filters. Photograph was taken with a Cool SNAP digital camera.
2.7. Fluorescence in situ hybridization (FISH)
To label the RNA probes, partial cDNA fragments of AgCHS1 and AgCHS2 were amplified by PCR using the sequence-specific primers with built-in restriction sites Xho I and Xba I as underlined: 5′-GTACACTCGAGATGTTGGTGGGTGCGTTC -3′ (forward) and 5′-CTGCATCTAGAGATGATGGAGTAGAGGATGAGC -3′ (reverse) for AgCHS1, and 5′-GTACACTCGAGCATGAAGAAGTACACTACCAAGTC -3′ (forward) and 5′-CTGCATCTAGAGGAAGGAGGTCCAAATGTCG -3′ (reverse) for AgCHS2. The PCR product was inserted into pBlueScript SK (+) vector containing the same restriction sites. The plasmid DNA was then linearized with either Xho I or Xba I and used for transcription with T7 and SP6 RNA polymerases (Invitrogen) to generate anti-sense and sense probes, respectively. The anti-sense and sense (as negative control) probes were labeled with Fluorescence in situ hybridization (FISH) Tag™ RNA Multicolor Kit (Catalog No. F32956, Invitrogen) following the manufacturer’s protocol. The probes were labeled with Alexa Fluor®488.
For the whole mount in situ hybridization, the 4th-instar larvae were dissected in 1× PBS to get the whole gut and whole cuticle. Only half integument was used by longitudal cutting of the whole integument into two equal sections. The tissues were fixed in 4% paraformaldehyde at 4 °C overnight, washed in PBST (0.1 M PBS + 0.2% Triton X-100) for three times (5 min each), treated with proteinase K (50 microgram/ml in PBST) for 10 min at room temperature. The reaction was stopped with PBST-glycin (2 mg Glycin/ml PBST) for 5 min followed by two washes in PBST (5 min each). The tissues were re-fixed in 4% formaldehyde for 1 h followed by three washes in PBST, PBST:hybridization buffer (1:1), and hybridization buffer (5 min each) at room temperature. The tissues were pre-hybridized in hybridization buffer at 48 °C for 20 min, and then hybridized in the same hybridization buffer containing 10 μg/ml anti-sense probes for 20–30 h at 48 °C. Control samples were hybridized with the same amount of sense probes under the same conditions. After hybridization, the samples were washed with hybridization buffer for 2–4 hr at 48 °C, then three times with PBST at room temperature. The samples were mounted in a glycerol and visualized under a Leica M205 FA fluorescence stereomicroscope equipped with GFP2 filter set. The images were obtained by using a Leica DFC 400 digital camera.
2.8. Data analysis for qPCR
For qPCR analysis, the percentage data of the relative AgCHS expression were obtained by dividing the REV of each developmental time point or tissue for each gene by the sum of REV throughout development or all tissues for that gene. The percentage data were then transformed using arcsine square root transformation before one-way ANOVA. Fisher’s least significant difference (LSD) multiple comparisons were then used to separate the means among the samples.
3. Results
3.1. cDNA and deduced amino acid sequences
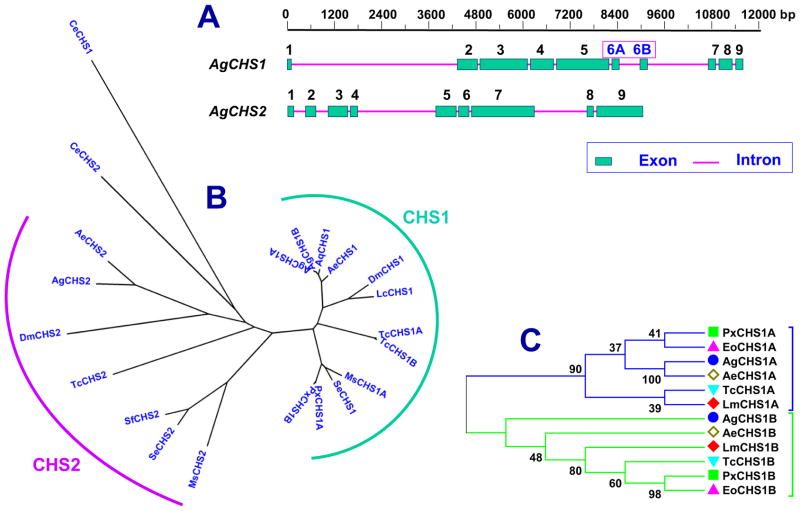
Two chitin synthase genes, AgCHS1 and AgCHS2, were identified from An. gambiae. AgCHS1 has two alternative splicing variants, AgCHS1A and AgCHS1B. AgCHS1A contains exon 6A, whereas AgCHS1B contains 6B (Fig. 1A). The two alternative spliced exons are the same in size and each encodes a peptide of 59 amino acid residues. These alternative exons are highly conserved both in size and amino acid identity in all CHS1 genes from several insect species. AgCHS1 and AgCHS2 can be clearly grouped into two different phylogenetic groups (Fig. 1B), whereas the exon 6A or 6B further separates AgCHS1 into two distinct phylogenetic groups (Fig. 1C). The open reading frame (ORF) of AgCHS1A cDNA is 4734 bp in length, encoding chitin synthase 1A of 1578 amino acid residues. Its predicted molecular mass and isoelectric point was 179.55 kDa and 6.43, respectively. The ORF of AgCHS1B cDNA is 4734 bp in length, encoding the chitin synthase 1B with 1578 amino acid residues. Its predicted molecular mass and isoelectric point are 179.64 kDa and 6.50, respectively. The ORF of AgCHS2 cDNA is 4758 bp in length, encoding the chitin synthase 2 with 1586 amino acid residues. Its predicted molecular mass and isoelectric point are 181.76 kDa and 6.37, respectively. The ORF of each chitin synthase gene contains 9 exons and 8 introns.
Fig. 1.
Sequence analysis of AgCHS genes. A) Gene structures of AgCHS1 and AgCHS2. AgCHS1 has two splice variants with alternative splicing exons (6A and 6B). B) Phylogenetic tree of insect chitin synthases. C) Phylogenetic tree of the alternative exons of insect CHS1. Ag, Anopheles gambiae; Ae, Aedes aegypti; Aq, A. quadrimaculatus; Dm, Drosophila melanogaster; Lc, Lucilia cuprina; Lm, Locusta migratoria; Ms, Manduca sexta; Px, Plutella xylostella; Tc, Tribolium castaneum; Se, Spodoptera exiqua; Sf, Spodoptera frugi; Eo, Ectropis obliqua; Ce, Caenorhabditis elegans. GenBank Accession numbers are listed in Table 2. Phylogenetic tree was generated using the software MEGA 5 after alignment by using ClustalW method.
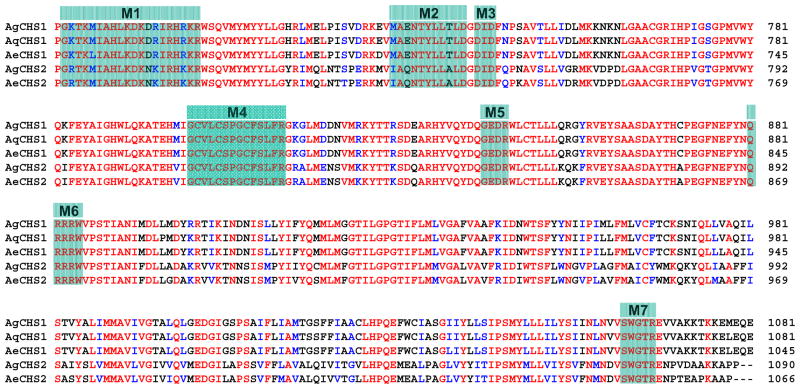
The deduced amino acid sequences of these two chitin synthases exhibited seven highly conserved motifs (Fig. 2) which are the characteristic features of family 2 glycosyltransferases (GTF2) from fungi, insects and other organisms (Merzerdorfer, 2006). Each Anopheles chitin synthase contains 10 transmembrane helices at the N-terminal and seven transmembrane helices at the C-terminal, five of which are located immediately next to the predicted central catalytic domain (Supplementary data 1). These characters match the typical tripartite domain structure of insect chitin synthases with a central catalytic region that is flanked by two transmembrane domains (Merzendorfer, 2006). The deduced amino acid sequences of two chitin synthases from An. gambiae showed high identities to those from two other mosquito species An. quadrimaculatu and Ae. aegypti (Fig. 2; Table 2). The identities between AgCHS1A/AgCHS1B and other insect CHS1s are much higher than the identities between AgCHS2 and other insect CHS2s (Table 2).
Fig. 2.
Alignment of the conserved catalytic domain of the chitin synthases from three mosquito species: Ag, Anopheles gambiae; Aq, An. quadrimaculatus; Ae, Aedes aegypti. Seven characteristic motifs (M1–7) for insect chitin synthases are highlighted. Dashes are used to denote gaps introduced for a maximum alignment.
Table 2.
Pairwise comparison of An. gambiae chitin synthases with chitin synthases from other insect species and C. elegans.
| Name | Identity (%)
|
Accession No. | ||
|---|---|---|---|---|
| AgCHS1A | AgCHS1B | AgCHS2 | ||
| AgCHS1A | - | 99.2 | 47.5 | AGAP001748-PA (XP_321336.5) |
| AgCHS1B | 99.2 | - | 47.6 | AGAP001748-PA (XP_321336.4) |
| AqCHS1 | 97.0 | 97.8 | 47.7 | DQ415985 |
| AeCHS1 | 91.8 | 91.9 | 46.4 | AAEL002718 |
| LcCHS1 | 79.2 | 78.9 | 45.6 | AF221067 |
| DmCHS1 | 78.5 | 78.1 | 45.3 | NM_079509 |
| TcCHS1A | 73.4 | 73.6 | 47.9 | AY291475 |
| TcCHS1B | 73.3 | 73.5 | 47.4 | AY291476 |
| PxCHS1A | 73.3 | 73.3 | 47.5 | AB271784 |
| PxCHS1B | 72.9 | 73.2 | 47.2 | AB281490 |
| SeCHS1 | 72.7 | 72.7 | 48.5 | DQ062153 |
| MsCHS1 | 72.1 | 71.8 | 47.6 | AY062175 |
| CeCHS1 | 30.6 | 30.8 | 29.2 | NP_492113 |
| AgCHS2 | 47.5 | 47.6 | - | AGAP001205 |
| AeCHS2 | 46.3 | 46.4 | 81.9 | AAEL005618 |
| DmCHS2 | 42.3 | 42.5 | 48.1 | NM_079485 |
| SeCHS2 | 52.5 | 52.6 | 43.3 | EU622827 |
| SfCHS2 | 51.3 | 51.4 | 43.3 | AY525599 |
| TcCHS2 | 48.0 | 48.0 | 43.0 | AY291477 |
| MsCHS2 | 52.6 | 52.6 | 42.8 | AY82156 |
| CeCHS2 | 40.9 | 40.9 | 35.2 | NP_493682 |
3.2. Stage-specific expression of AgCHS1 and AgCHS2
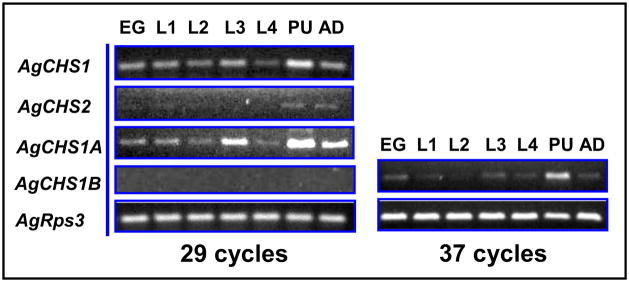
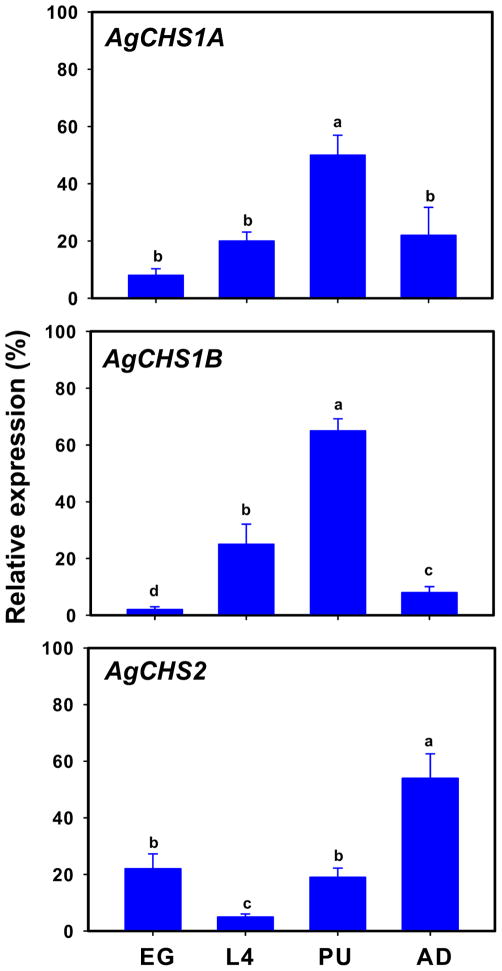
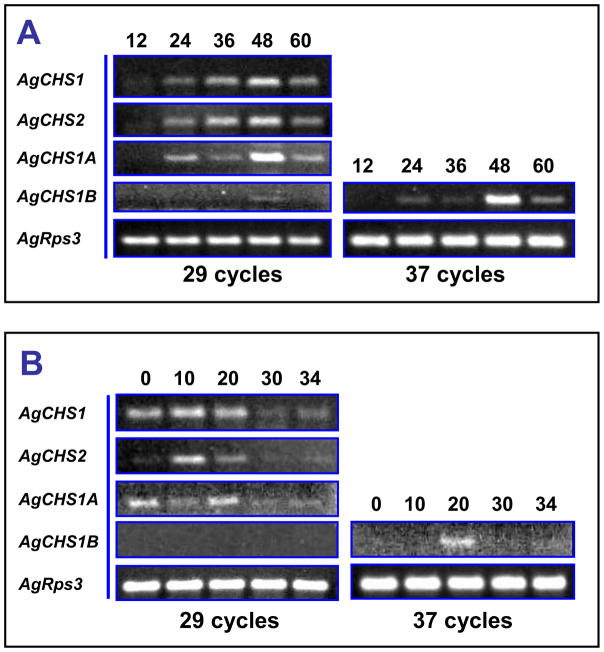
Stage-specific expression patterns of two An. gambiae CHS genes were determined in eggs, four different larval instars (1st, 2nd, 3rd and 4th), pupae and adults by using RT-PCR (Fig. 3). AgCHS1 was expressed in all seven life stages. There were some apparent variations in its expression level but the peak expression appeared to be in pupal stage. We further examined the expression of the two alternative splicing variants of AgCHS1 and revealed that the expression pattern of AgCHS1A was consistent with AgCHS1, whereas the expression of AgCHS1B was non-detectable by normal PCR cycles (29) (Fig. 3). These RT-PCR results were also confirmed by qPCR (Fig. 4). As the primer efficiencies were similar between AgCHS1A and AgCHS1B (more than 95% for each variant), these results suggested that AgCHS1A might be the predominant form of AgCHS1 at all the seven stages examined. In contrast, both RT-PCR and qPCR revealed that AgCHS2 was expressed at low level during the larval stages and high level in adult stage (Figs. 3 and 4).
Fig. 3.
Analysis of stage-specific expression patterns of AgCHS genes in eggs (EG), larvae from first to fourth instars (L1–4), pupae (PU) and adults (AD) of An. gambiae by RT-PCR.
Fig. 4.
Analysis of stage-specific expressions of AgCHS genes in eggs (EG), fourth-instar larvae, pupae (PU) and adults (AD) of An. gambiae by qPCR. Same letters on the error bars indicate no significant differences in the expression of the same gene or alternative splicing variant among the four developmental stages based on Fisher’s LSD (P≥0.05).
We further examined the stage-specific expression patterns of the AgCHS genes in eggs and pupae at different times by RT-PCR. Both AgCHS1 and AgCHS2 expressed in 24-h eggs and gradually increased their expressions after that and reached the maximum at 48 h (Fig. 5A). Expression of AgCHS1B was rather low and was only detected at 48-h eggs under normal RT-PCR cycles. In pupae, relatively high expressions of two AgCHS genes were found at early stages, for example, from 0 to 20 h, and then gradually decreased afterwards (Fig. 5B). Similarly, AgCHS1A might be the predominant form of AgCHS1 as AgCHS1B was almost non-detectable under normal PCR cycles. When the cycle number increased to 37, expression of AgCHS1B was detected only in 20-h pupae (Fig. 5B).
Fig. 5.
RT-PCR analysis of stage-specific expression patterns of AgCHS genes in eggs (A) at 12, 24, 36, 48, and 60 h, respectively, and in pupae (B) at 0, 10, 20, 30, and 34 h, respectively.
3.3. Tissue-specific expression profiles of AgCHS1A, AgCHS1B and AgCHS2
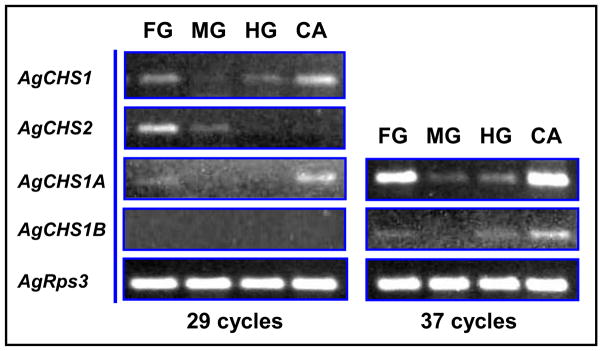
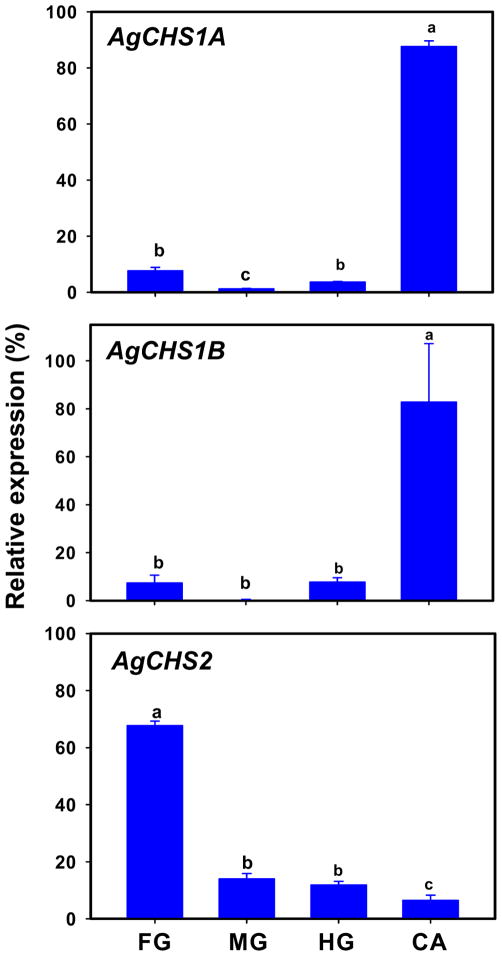
We examined the tissue-specific expression patterns of two exon-specific variants of AgCHS1 by using RT-PCR and qPCR in each of four different tissues, including the foregut, midgut, hindgut, and carcass (the insect body after its digestive canal was removed), dissected from the fouth-instar larvae. AgCHS1A and AgCHS1B showed the same expression patterns and were predominantly expressed in the carcass. Low expressions of AgCHS1A and AgCHS1B were detected in the foregut and hindgut, and almost no expression of these variants was found in the midgut as evaluated by both RT-PCR (Fig. 6) and qPCR (Fig. 7). However, more than 60% of transcripts of AgCHS2 were found in the foregut (Fig. 7). The qPCR analysis of AgCHS2 showed the consistent results with our RT-PCR analysis (Figs. 6 and 7).
Fig. 6.
Analysis of tissue-specific expression patterns of AgCHS genes in foregut (FG), midgut (MG), hindgut (HG), and carcass (CA) dissected from the fouth-instar larvae of An. gambiae by RT-PCR.
Fig. 7.
Analysis of tissue-specific expressions of AgCHS genes in foregut (FG), midgut (MG), hindgut (HG), and carcass (CA) dissected from the fouth-instar larvae of An. gambiae by qPCR. Same letters on the error bars indicate no significant differences in the expression of the same gene or alternative splicing variant among the four tissues based on Fisher’s LSD (P≥0.05). (P≥0.05).
3.4. Localization of AgCHS1 and AgCHS2 transcripts and proteins by FISH and immunohistochemistry
We performed FISH in adult and larval gut by using AgCHS1 and AgCHS2 probes, respectively. In the guts of the female adults 24 h after a blood meal, the AgCHS2 transcript was evenly distributed in all midgut epithelium cells (Fig. 8A), whereas the AgCHS1 transcript was not detected in the midgut epithelium cells (Fig. 8B). In a fourth-instar larval gut, positive immunoreactive signals of AgCHS2 was mainly detected in cardia and posterior midgut (Fig. 8C), whereas no positive signals of AgCHS1 was detected in the gut (Fig. 8D).
Fig. 8.
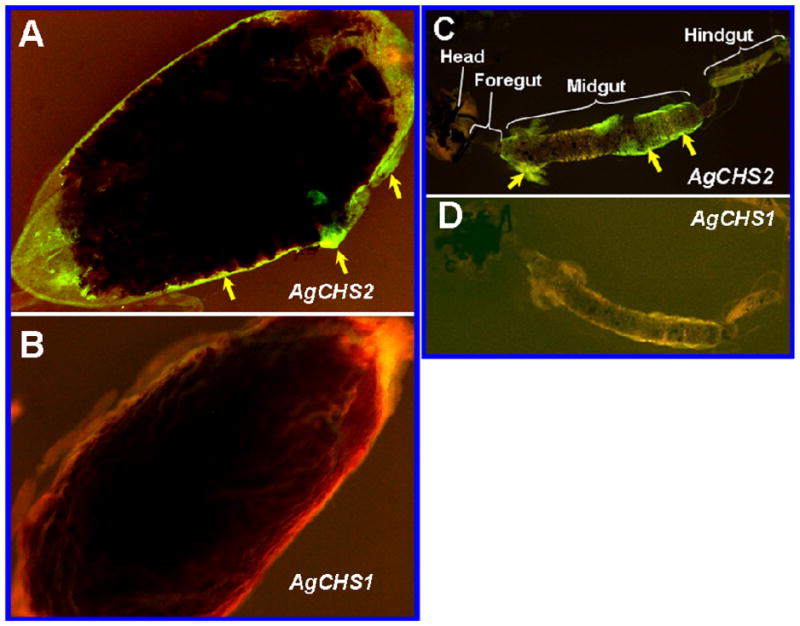
Fluorescence in situ hybridization (FISH) analysis of AgCHS genes in isolated mosquito gut. Localization of AgCHS gene transcripts in the midgut of the adult mosquito 24 h after a bloodmeal by using AgCHS2 (A) and AgCHS1 (B) probes, respectively. Localization of the transcripts in the isolated whole larval gut stained by AgCHS2 (C) and AgCHS1 (D) probes, respectively. The green color indicated by arrows shows positive staining.
Western blot analysis showed that each antibody of AgCHS1 and AgCHS2 can specifically recognize its own recombinant protein and there was no cross recognization (Supplementary data 2), indicating high specificity of each antibody. By using these antibodies, immunohistochemical analysis was performed in the paraffin-embedded thin sections of the 12- to 24-h pupae and revealed that AgCHS1 protein was localized in the ommatidia of the compound eyes and epidermal cells of the adult integument that newly developed underneath of the pupal integument (Fig. 9A, B). In contrast, AgCHS2 protein was localized in the ommatidia of the compound eyes, thoracic and abdominal inter-segmental regions of pupal integument (Fig. 9C, D).
Fig. 9.
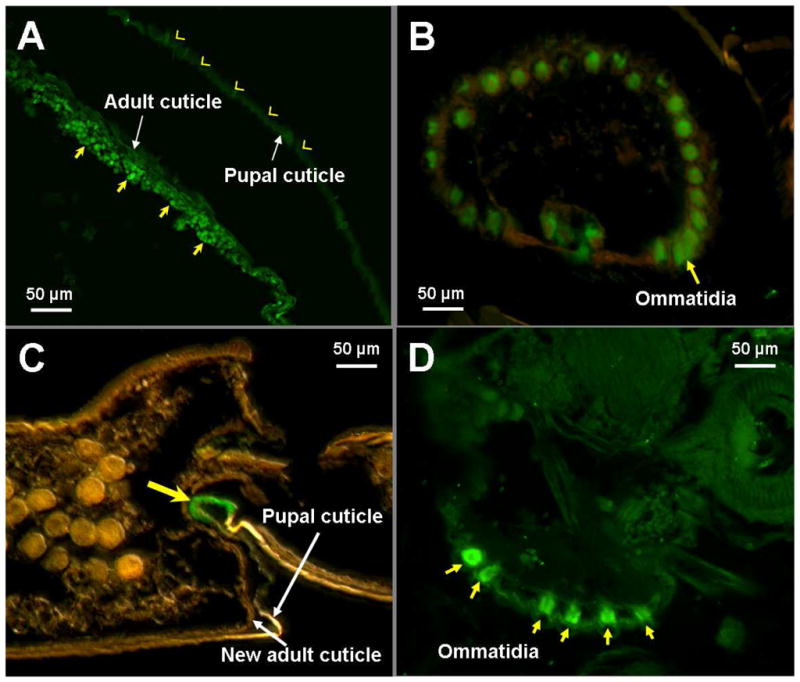
Immunohistochemistry of anti-AgCHS1 (A, B) and anti-AgCHS2 serum (C, D) in the pupal stage of An. gambiae. Paraffin-embedded thin sections of the 12- to 24-hour-old pupae were immunostained with primary antibodies and visualized by the reaction with Alexa 488-conjugated goat anti-mouse IgG. The epidermal cells in the adult cuticle (A) and the eyes (B) were immunoreactive (arrows indicate positive staining, arrow heads indicate negative staining). The abdominal inter-segmental region of the pupal cuticle (C) and the eyes (D) were immunoreactive (arrows indicate positive staining).
4. Discussion
The presence of chitin in the exoskeleton, peritrophic matrix, and trachea has been well documented in insects. However, recent advances in research of chitin and chitin synthases in insects have demonstrated that the distribution of either chitin or chitin synthases is beyond these tissues. For example, a recent study documents that the putative chitin-like material was identified in the eggs, ovaries, and egg shells from Ae. Aegypti (Moreira et al., 2007). The chitin-like material in the egg shells possibly comes from the female parents, whereas the chitin-like material from the eggs might be synthesized during embryogenesis. A more recent study reported that chitin was synthesized in the serosal cuticle (SC) 11–13 h after egg deposition and played an important role the desiccation tolenrence for Ae. aegypti eggs. Very possibly, the chitin in the SC was synthesized by AaCHS1A as it was the sole variant specifically expressed during the SC formation (Rezende et al., 2008). In An. gambiae, we found both AgCHS1 and AgCHS2 started to express 24 h after egg deposition (Fig. 5A), which may imply their roles in embryonic development for both of these enzymes in An. gambiae eggs. However, the exact function for both enzymes remains elusive. Firstly, An. gambiae eggs are not as tolerant to desiccation as Aedes eggs (Clements, 1999). To some extent, this implies the difference of the profiles in chitin structure and functions between Anopheles eggs and Aedes eggs. Secondly, why Anopheles eggs need chitin synthase 2 as chitin synthase 2 is well documented to make PM-associated chitin? Is it possible that the typical cycles of chitin biosynthesis and degradation occurring in larvae also occur in the eggs? A parallel study did show that several chitinase genes were expressed in 24-h eggs and the followup time frames before egg hatching (Zhang et al., 2011). Such expression profiles are nearly identical to those for two chitin synthase genes. Embryonic molts have been documented in hemimetabolous insects such as grasshopper and crickets (Erezyilmaz et al., 2004). However, similar process has never been reported in dipterans to date.
Furthermore, we detected both AgCHS1 and AgCHS2 proteins in the newly formed compound eyes of the pupae by immnohistochemical analysis (Fig. 9B, D). This result suggests the roles of chitin synthases in the structure and function in Anopheles visual system. This contention was supported by the fact that chitin was detected in both the ommatidial lenses and ocellar lenses in Drosaphila melanogaster (Yoon et al., 1997; Faschinger, 2010). More interestingly, we detected the expression of AgCHS2 protein in the pupal stage. Indeed, relatively high level of AgCHS2 transcript was found in the pupal stage as compared with that in the larval stages (Figs. 3 and 4). Apparently, mosquitoes do not need chitin synthases to make PM-associated chitin in the pupal stage as the PM will not be produced until a blood meal in the adults (Shao et al., 2001; Hegedus et al., 2009). In addition to the localization in the compound eyes, immuohitochemical analysis also revealed the distribution of AgCHS2 in the abdominal inter-segmental regions of the pupal cuticle (Fig. 9C). All these new findings suggest that further study is needed to understand potentially new functions of AgCHS2 during the pupal stage.
As discussed above, current knowledge supports the notion that CHS2 enzymes are responsible for biosynthesis of the PM chitin (Arakane et al., 2005). Specially, two types of PM have been found in mosquitoes and other blood-feeding insects. The type 1 PM is thick (2–20 μm) and is produced from all midgut epithelial cells and is produced in direct response to a blood meal, whereas type 2 PM is thin (less than 1 μm) and produced continuously by a small group of highly specialized cells in the cardia in mosquito larvae (Shao et al., 2001; Kato et al., 2006). Consistent with this convention, FISH analysis in this study showed that AgCHS2 transcripts are evenly distributed in all adult midgut epithelial cells (Fig. 8A). In the larval gut, we did find high level of AgCHS2 transcripts in the cardia. However, the detection of relatively high level of AgCHS2 transcript in the posterior midgut (Fig. 8C) has challenged the convention as PM is thought to be exclusively produced in the special cells localized in the cardia. Further work is needed to address whether the AgCHS2 protein could be translated from these transcripts. If so, what is the potential function of these enzymes localized at the posterior part of the midgut? Current studies support the notion that different parts of the midgut serve different physiological functions in mosquito larvae. Direct supporting evidence comes from the pH gradients in the gut contents. The pH of the luminal contents in mosquito larvae is near neutrality in the foregut, reaches to 10 in anterior midgut, and drops to 7.5 in the posterior midgut (Dadd, 1975; Okech et al., 2008). Further work is needed to address how AgCHS2 involves in the physiological function of the posterior midgut. One issue to clarify here is that high level of AgCHS2 transcript detected by RT-PCR and qPCR in the foregut (Figs. 6 and 7) possibly is an artifact caused by the difficulty to separate the cardia from the foregut in dissection as the foregut is tiny and the cardia always tightly attached to the foregut. FISH analysis has revealed the high level expression of AgCHS2 in the cardia but not in the foregut (Fig. 8C).
Current understanding of the structure and molecular constituents of the type 2 PM is rather limited. The chitin content was really low (7.2% of the weight of the matrix) in the type 2 PM from Lucilia cuprina, indicating chitin is a minor structural component of the type 2 PM in this insect (Tellam and Eisemann, 2000). The PM associated chitin was not detectable in the dissected mosquito larval midgut by using high specificity chitin binding reagent FITC-CBD even after the midgut were pretreated with either proteinases or alkalines to release the PM associated proteins (data not shown). Consistently, we found that the transcript level of AgCHS2 during the larval stage was also pretty low. However, the low level of AgCHS2 transcript and the possible low chitin content do not necessarily mean that chitin is not important for PM structure and its physiological functions. Our recent study on the function of AgCHS2 via RNA interference has revealed that AgCHS2 is critical to maintain the integrity of type 2 PM in An. gambiae larvae (Zhang et al., 2010). Type 1 PM has been considered as a potential barrier for malarial protozonan parasite before reaching the midgut epithelial cells (Shen and Jacobs-Lorena, 1998). The integrity of the adult PM is an important factor in regulating the parasite’s passthrough of the PM barrier and the parasite development inside the gut (Dessens et al., 2001; Li et al., 2005). Disruption of the PM formation by RNA interference of chitin synthase gene results in the decreased infectivity of Plasmodium gallinaceum in Aedes aegypti (Kato et al., 2008). Thus, the study on chitin synthase and the PM structure and function in mosquitoes may provide new insight on blocking the malaria parasites in mosquitoes.
Supplementary Material
Research Highlight.
We identified and characterized two chitin synthase genes (AgCHS1 and AgCHS2) in Anopheles gambiae.
Both AgCHS1 and AgCHS2 were expressed in egg, larval, pupal and adult stages but AgCHS2 was expressed at low levels.
AgCHS1 was mainly expressed in the carcass whereas AgCHS2 was expressed in the foregut including cardia and the midgut.
Immunohistochemical analysis localized the two enzymes in the ommatidia of the compound eyes and other tissues.
Acknowledgments
The authors thank Dr. Chitvan Khajuria for his helpful comments on an earlier draft of this manuscript, Dr. Ludek Zurek for providing access to the real-time PCR facility, and Ms. Sharon Starkey for rearing mosquito colonies and other technical assistance. This research was supported in part by Kansas Agricultural Experiment Station and NIH (P20 RR016475) from the INBRE Program of the National Center for Research Resources. This manuscript is contribution No. 12-206-J from the Kansas Agricultural Experiment Station, Kansas State University, Manhattan, Kansas, USA. The Anopheles gambiae voucher specimens (voucher No. 211) are located in the Kansas State University Museum of Entomological and Prairie Arthropod Research, Manhattan, Kansas.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arakane Y, Hogenkamp DG, Zhu YC, Kramer KJ, Specht CA, Beeman RW, Kanost MR, Muthukrishnan S. Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochem Mol Biol. 2004;34:291–304. doi: 10.1016/j.ibmb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Muthukrishnan S, Kramer KJ, Specht CA, Tomoyasu Y, Lorenzen MD, Kanost M, Beeman RW. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix, respectively. Insect Mol Biol. 2005;14:453–463. doi: 10.1111/j.1365-2583.2005.00576.x. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Specht CA, Kramer KJ, Muthukrishnan S, Beeman RW. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:959–962. doi: 10.1016/j.ibmb.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Ashfaq M, Sonoda S, Tsumuki H. Developmental and tissue-specific expression of CHS1 from Plutella xylostella and its response to chlorfluazuron. Pestic Biochem Physiol. 2007;89:20–30. [Google Scholar]
- Bolognesi R, Arakane Y, Muthukrishnan S, Kramer KJ, Terra WR, Ferreira C. Sequences of cDNAs and expression of genes encoding chitin synthase and chitinase in the midgut of Spodoptera frugiperda. Insect Biochem Molec Biol. 2005;35:1249–1259. doi: 10.1016/j.ibmb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Chen X, Yang X, Senthil Kumar N, Tang B, Sun X, Qiu X, Hu J, Zhang W. The class A chitin synthase gene of Spodoptera exigua: molecular cloning and expression patterns. Insect Biochem Mol Biol. 2007;37:409–17. doi: 10.1016/j.ibmb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Clements AN. The Biology of Mosquitoes. CABI Publishing; UK: 1999. [Google Scholar]
- Cohen E. Chitin synthesis and inhibition: a revisit. Pest Manag Sci. 2001;57:946–950. doi: 10.1002/ps.363. [DOI] [PubMed] [Google Scholar]
- Dadd RH. Alkalinity within the midgut of mosquito larvae with alkaline-active digestive enzymes. J Insect Physiol. 1975;21:1847–1853. doi: 10.1016/0022-1910(75)90252-8. [DOI] [PubMed] [Google Scholar]
- Dessens JT, Mendoza J, Claudianos C, Vinetz JM, Khater E, Hassard S, Ranawaka GR, Sinden RE. Knockout of the rodent malaria parasite chitinase pbCHT1 reduces infectivity to mosquitoes. Infect Immun. 2001;69:4041–4047. doi: 10.1128/IAI.69.6.4041-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erezyilmaz DF, Riddiford LM, Truman JW. Juvenile hormone acts at embryonic molts and induces the nymphal cuticle in the direct-developing cricket. Dev Genes Evol. 2004;214:313–323. doi: 10.1007/s00427-004-0408-2. [DOI] [PubMed] [Google Scholar]
- Faschinger C. Astonishing evolution of the lenses in different eyes. Spektrum Der Augenheilkunde. 2010;24:174–180. [Google Scholar]
- Gagou ME, Kapsetaki M, Turberg A, Kafetzopoulos D. Stage-specific expression of the chitin synthase DmeChSA and DmeChSB genes during the onset of Drosophila metamorphosis. Insect Biochem Mol Biol. 2002;32:141–146. doi: 10.1016/s0965-1748(01)00101-1. [DOI] [PubMed] [Google Scholar]
- Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 2004;4:327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D, Erlandson M, Gillott C, Toprak U. New insights into peritrophic matrix synthesis, architecture, and function. Annu Rev Entomol. 2009;54:285–302. doi: 10.1146/annurev.ento.54.110807.090559. [DOI] [PubMed] [Google Scholar]
- Hogenkamp DG, Arakane Y, Zimoch L, Merzendorfer H, Kramer KJ, Beeman RW, Kanost MR, Specht CA, Muthukrishnan S. Chitin synthase genes in Manduca sexta: Characterization of a gut-specific transcript and differential tissue expression of alternately spliced mRNAs during development. Insect Biochem Mol Biol. 2005;35:529–540. doi: 10.1016/j.ibmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Hunt S. Polysaccharide-protein complexes in invertebrates. Academic Press; New York, NY: 1970. [Google Scholar]
- Ibrahim GH, Smartt CT, Kiley LM, Christensen BM. Cloning and characterization of a chitin synthase cDNA from the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2000;30:1213–1222. doi: 10.1016/s0965-1748(00)00100-4. [DOI] [PubMed] [Google Scholar]
- Kato N, Mueller CR, Fuchs JF, McElroy K, Wessely V, Higgs S, Christensen BM. Evaluation of the function of a type I peritrophic matrix as a physical barrier for midgut epithelium invasion by mosquito-borne pathogens in Aedes aegypti. Vector Borne Zoonotic Dis. 2008;8:701–712. doi: 10.1089/vbz.2007.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Mueller CR, Fuchs JF, Wessely V, Lan Q, Christensen BM. Regulatory mechanisms of chitin biosynthesis and roles of chitin in peritrophic matrix formation in the midgut of adult Aedes aegypti. Insect Biochem Mol Biol. 2006;36:1–9. doi: 10.1016/j.ibmb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Kramer KJ, Muthukrishnan S. Chitin metabolism in insects. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 4. Elsevier; New York: 2005. pp. 111–144. [Google Scholar]
- Kumar NS, Tang B, Chen X, Tian H, Zhang W. Molecular cloning, expression pattern and comparative analysis of chitin synthase gene B in Spodoptera exigua. Comp Biochem Physiol B Biochem Mol Biol. 2008;149:447–453. doi: 10.1016/j.cbpb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Li F, Patra KP, Vinetz JM. An anti-chitinase malaria transmission-blocking single-chain antibody as an effector molecule for creating a Plasmodium falciparum-refractory mosquito. J Infect Dis. 2005;192:878–887. doi: 10.1086/432552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzendorfer H. Insect chitin synthases: a review. J Comp Physiol B. 2006;176:1–15. doi: 10.1007/s00360-005-0005-3. [DOI] [PubMed] [Google Scholar]
- Merzendorfer H, Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 2003;206:4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- Moreira MF, Dos Santos AS, Marotta HR, Mansur JF, Ramos IB, Machado EA, Souza GH, Eberlin MN, Kaiser CR, Kramer KJ, Muthukrishnan S, Vasconcellos AM. A chitin-like component in Aedes aegypti eggshells, eggs and ovaries. Insect Biochem Mol Biol. 2007;37:1249–1262. doi: 10.1016/j.ibmb.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Okech BA, Boudko DY, Linser PJ, Harvey WR. Cationic pathway of pH regulation in larvae of Anopheles gambiae. J Exp Biol. 2008;211:957–68. doi: 10.1242/jeb.012021. [DOI] [PubMed] [Google Scholar]
- Qu M, Yang Q. A novel alternative splicing site of class A chitin synthase from the insect Ostrinia furnacalis – Gene organization, expression pattern and physiological significance, Insect Biochem. Mol Biol. 2011;41:923–931. doi: 10.1016/j.ibmb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Rezende GL, Martins AJ, Gentile C, Farnesi LC, Pelajo-Machado M, Peixoto AA, Valle D. Embryonic desiccation resistance in Aedes aegypti: presumptive role of the chitinized serosal cuticle. BMC Dev Biol. 2008;8:82. doi: 10.1186/1471-213X-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AG. The integument of arthropods. University of Minnesota Press; Minneapolis, MN: 1951. [Google Scholar]
- Shao L, Devenport M, Jacobs-Lorena M. The peritrophic matrix of hematophagous insects. Arch Insect Biochem Physiol. 2001;47:119–125. doi: 10.1002/arch.1042. [DOI] [PubMed] [Google Scholar]
- Shen Z, Jacobs-Lorena M. A type I peritrophic matrix protein from the malaria vector Anopheles gambiae binds to chitin. J Biol Chem. 1998;273:17665–17670. doi: 10.1074/jbc.273.28.17665. [DOI] [PubMed] [Google Scholar]
- Tellam RL, Eisemann C. Chitin is only a minor component of the peritrophic matrix from larvae of Lucilia cuprina. Insect Biochem Mol Biol. 2000;30:1189–1201. doi: 10.1016/s0965-1748(00)00097-7. [DOI] [PubMed] [Google Scholar]
- Tellam RL, Vuocolo T, Johnson SE, Jarmey J, Pearson RD. Insect chitin synthase: cDNA sequence, gene organization and expression. Eur J Biochem. 2000;267:6025–6042. doi: 10.1046/j.1432-1327.2000.01679.x. [DOI] [PubMed] [Google Scholar]
- Tian H, Peng H, Yao Q, Chen H, Xie Q, Tang B, Zhang W. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One. 2009;4:e6225. doi: 10.1371/journal.pone.0006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Dunn WA, Emmons AC, Nagao J, Willis JH. Developmental expression patterns of cuticular protein genes with the R&R Consensus from Anopheles gambiae. Insect Biochem Mol Biol. 2008;38:508–519. doi: 10.1016/j.ibmb.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon CS, Hirosawa K, Suzuki E. Corneal lens secretion in newly emerged Drosophila melanogaster examined by electron microscope autoradiography. J Electron Microsc. 1997;46:243–246. doi: 10.1093/oxfordjournals.jmicro.a023515. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Zhu KY. Characterization of a chitin synthase cDNA and its increased mRNA level associated with decreased chitin synthesis in Anopheles quadrimaculatus exposed to diflubenzuron. Insect Biochem Mol Biol. 2006;36:712–725. doi: 10.1016/j.ibmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu X, Zhang J, Li D, Sun Y, Guo Y, Ma E, Zhu KY. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen) Insect Biochem Mol Biol. 2010;40:824–833. doi: 10.1016/j.ibmb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang X, Arakane Y, Muthukrishnan S, Kramer KJ, Ma E, Zhu KY. Comparative genomic analysis of chitinase and chitinase-like genes in the African malaria mosquito (Anopheles gambiae) PLoS ONE. 2011;6:e19899. doi: 10.1371/journal.pone.0019899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae) Insect Mol Biol. 2010;19:683–693. doi: 10.1111/j.1365-2583.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- Zhu KY, Heise S, Zhang JZ, Anderson TD, Starkey SR. Comparative studies on effects of three chitin synthesis inhibitors on common malaria mosquito (Diptera: Culicidae) J Med Entomol. 2007;44:1047–1053. doi: 10.1603/0022-2585(2007)44[1047:csoeot]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Zhu YC, Specht CA, Dittmer NT, Muthukrishnan S, Kanost MR, Kramer KJ. Sequence of a cDNA and expression of the gene encoding a putative epidermal chitin synthase of Manduca sexta. Insect Biochem Molec Biol. 2002;32:1497–1506. doi: 10.1016/s0965-1748(02)00070-x. [DOI] [PubMed] [Google Scholar]
- Zimoch L, Hogenkamp DG, Kramer KJ, Muthukrishnan S, Merzendorfer H. Regulation of chitin synthesis in the larval midgut of Manduca sexta. Insect Biochem Mol Biol. 2005;35:515–527. doi: 10.1016/j.ibmb.2005.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.