Abstract
The DNA damage checkpoint, the first pathway known to be activated in response to DNA damage, is a mechanism by which the cell cycle is temporarily arrested to allow DNA repair. The checkpoint pathway transmits signals from the sites of DNA damage to the cell cycle machinery through the evolutionarily conserved ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related) kinase cascades. We conducted a genome-wide RNAi (RNA interference) screen in Drosophila cells to identify previously unknown genes and pathways required for the G2-M checkpoint induced by DNA double-strand breaks (DSBs). Our large-scale analysis provided a systems-level view of the G2-M checkpoint and revealed the coordinated actions of particular classes of proteins, which include those involved in DNA repair, DNA replication, cell cycle control, chromatin regulation, and RNA processing. Further, from the screen and in vivo analysis, we identified previously unrecognized roles of two DNA damage response genes, mus101 and mus312. Our results suggest that the DNA replication preinitiation complex, which includes MUS101, and the MUS312-containing nuclease complexes, which are important for DSB repair, also function in the G2-M checkpoint. Our results provide insight into the diverse mechanisms that link DNA damage and the checkpoint signaling pathway.
INTRODUCTION
Living organisms are constantly exposed to various DNA-damaging agents that can either kill cells or mutate their genomes. To cope with DNA damage, the cell has a number of mechanisms, collectively called the DNA damage response. One such mechanism is the G2-M DNA damage checkpoint, which temporarily arrests the cell cycle at the G2 to mitosis (M) transition to provide time for DNA repair. The G2-M checkpoint is one of at least three molecularly distinct checkpoints that include the G1-S and S-phase checkpoints. The three checkpoints are differentially activated depending on the phase of the cell cycle in which DNA damage occurs (1). The G2-M checkpoint is particularly important for protecting the genome from double-strand breaks (DSBs), because if cells enter mitosis with unrepaired DSBs, chromosomes are not properly segregated, resulting in the subsequent loss of genetic information in daughter cells. Indeed, mutations in G2-M checkpoint genes underlie several cancer pre-disposition syndromes in humans (2).
Studies in various organisms have revealed that the core components of the G2-M checkpoint pathway are conserved across eukaryotes from yeast to human. The two phosphatidylinositol 3-kinase–related protein kinase (PIKK) family kinases ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related) play a central role in transducing signals from DNA damage to the cell cycle machinery (3). ATM and ATR are activated in response to DNA damage and in turn phosphorylate more than 700 cellular proteins (4) to activate the downstream cascades. The activated ATM/ATR cascades culminate in inactivation of the cyclin-dependent kinase complex, Cyclin B–Cdc2, which controls entry into mitosis, arresting the cell cycle in G2. ATM and ATR have partially overlapping functions in the DNA damage response because they both recognize and phosphorylate serine or threonine residues in SQ/TQ motifs. In mammalian cells, the DSB-induced G2-M checkpoint is cooperatively activated by ATM and ATR, whereas in yeast and flies, it predominantly depends on ATR (5–8). Thus, combined with its genetic amenability in vivo, Drosophila offers an experimentally accessible model of the G2-M checkpoint.
Despite the wealth of knowledge accumulated over the years, the extent and the intricacies of the network comprising the G2-M checkpoint still remain obscure, and fundamental questions remain. For instance, the exact mechanism by which DNA damage activates ATR remains elusive. Comprehensive identification of relevant genes will allow a more complete picture of the system and aid in understanding the underlying molecular mechanisms. Thus, we performed a genome-wide RNA interference (RNAi) screen in Drosophila cells and confirmed several of the identified genes in vivo. Our results illustrate that checkpoint activation involves the coordinated actions of proteins involved in DNA repair, DNA replication, cell cycle control, chromatin regulation, and RNA processing. Two of the genes, mus101 and mus312, correspond to DNA damage–sensitive loci previously not implicated in the G2-M checkpoint. Using mutant animals and transgenic RNAi, we explore the in vivo functions of these genes in the G2-M checkpoint.
RESULTS
Genome-wide RNAi screen and validation of the candidate genes
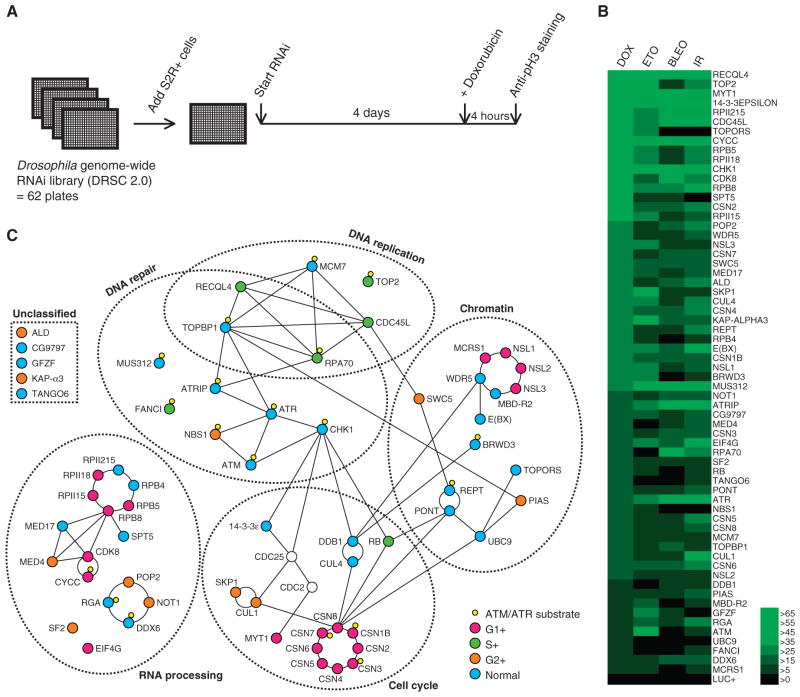
To identify genes required for the G2-M checkpoint, we performed a genome-wide RNAi screen in Drosophila S2R+ cells. S2R+ cells have an ATR-dependent G2-M checkpoint that can be activated by various DNA-damaging drugs that induce DSBs (7) (fig. S1). For the screen, we used the anticancer drug doxorubicin, which we found triggered the G2-M checkpoint in S2R+ cells. When wild-type cells were treated with doxorubicin, they arrested in G2 phase within 4 hours, as indicated by a decrease in the mitotic index (fig. S1). To identify genes required for this G2 arrest, we first treated cells with individual double-stranded RNAs (dsRNAs) targeting each of the 13,500 Drosophila genes for 4 days and then treated them with doxorubicin for 4 hours (Fig. 1A). Based on the G2 population of 30 to 40% (fig. S2) and the population doubling time of 24 hours, we estimated the length of G2 phase to be roughly 8 hours in S2R+ cells. Our assay, which measured the mitotic index at 4 hours after drug addition, therefore specifically interrogated the requirement for the G2-M checkpoint and not for the G1- or S-phase checkpoints. dsRNAs that abrogated the G2-M checkpoint were identified by the sustained presence of mitotic cells after doxorubicin treatment, which we monitored by phosphorylated histone H3 (pH3) immunostaining.
Fig. 1.
Genome-wide RNAi screen for genes required for the G2-M checkpoint. (A) Diagram of the assay. Drosophila S2R+ cells were plated in 384-well plates that contain in each well a dsRNA targeting a particular gene. Cells were incubated for 4 days and treated with 0.5 μM doxorubicin for 4 hours to induce DSBs, and then fixed and stained with an anti–phosphohistone H3 antibody to visualize mitotic cells. (B) Heat-map view of checkpoint defects induced by multiple DNA-damaging stimuli. The candidate checkpoint genes were tested with either doxorubicin (DOX; 0.5 μM), etoposide (ETO; 10 μM), Bleocin (BLEO; 50 μg/ml), or x-rays (150 Gy) for a checkpoint defect. Increase in the relative mitotic index (%) with respect to the control (no dsRNA) was colored as indicated in the right. Luciferase (LUC+) RNAi is shown at the bottom as a negative control. (C) Proteins encoded by the candidate checkpoint genes were grouped according to their cellular functions. Names of the mammalian orthologs are shown when available. A line between two proteins indicates a well-characterized interaction. Quantitative DAPI imaging was performed to determine the effect on the cell cycle profile (fig. S2), which is differentially indicated by colors in circles. CDC2 and CDC25 (clear nodes), which are cell cycle regulators that are targeted by the G2-M checkpoint, are also shown.
In the assay optimization phase, we noticed that knockdown of several positive control genes, including mei-41 (ortholog of mammalian ATR), led to only a modest checkpoint defect at the concentration of dsRNA used in our RNAi library. (Note that throughout the manuscript names of proteins or genes in parentheses are the mammalian homolog or ortholog.) We found that co-incubation with a dsRNA against a known checkpoint gene, nbs (NBS1), improved the sensitivity of the assay. Therefore, we performed the screen in this sensitized background. We used a concentration of nbs dsRNA that alone does not induce a checkpoint defect. Subsequent analysis for validation was performed without the dsRNA against nbs.
The primary screen identified 180 candidate genes, which when depleted resulted in persistent mitotic cells even after doxorubicin treatment (tables S1 to S3). Genes were considered positive if their knockdown led to more than 20 mitotic cells per well after doxorubicin treatment. The candidates contained a large number of genes that would nonspecifically affect the mitotic index owing to their role in mitotic progression. For instance, the screen identified genes required for mitotic spindle assembly, cytokinesis, and the metaphase-anaphase transition. These genes were considered potential false positives because they were likely to increase the mitotic index by delaying mitotic exit irrespective of the DNA damage checkpoint. Because it is impossible to ascertain whether they also have a function in the G2-M damage checkpoint, we removed this class of genes from our list of candidates. First, we excluded genes that have a role in mitotic progression documented in the literature (table S2). To further refine the list, we performed secondary screens to identify previously uncharacterized mitosis genes. Because suppression of such genes is expected to increase the mitotic index regardless of DNA damage, we excluded those genes whose depletion led to a more than twofold increase in mitotic index without DNA damage (table S2). As an alternative approach, we directly interrogated mitosis exit defects by monitoring mitotic cells after pharmacological block of new mitosis entry with a CDC25 inhibitor, NSC663284. We excluded those genes whose depletion led to a relative mitotic index of more than 50% after NSC663284 treatment (fig. S3 and table S2).
Another source of false positives in RNAi screens is from off-target effects, whereby a phenotype is caused by suppression of an unintended gene (9). We tested multiple dsRNAs for each of the remaining candidate genes to exclude such false positives. Of the 180 primary hits, 62 passed the secondary screens, providing a high-confidence list of genes essential for the doxorubicin-induced G2-M checkpoint (table S1).
We recovered five of the six of previously described Drosophila mutants that exhibit strong G2-M checkpoint defects in vivo: mei-41 (ATR), mus304 (ATRIP), grp (CHK1), Myt1 (MYT1), and 14-3-3ε (14-3-3ε) (8, 10–12). The functions of these genes in the G2-M checkpoint are conserved in mammals (1). Of the six known checkpoint genes, only nbs (NBS1) was not recovered in the screen. In addition, our screen did not recover tefu (ATM) and mre11 (MRE11), which lead to a minor checkpoint defect when mutated (13). In Drosophila, ATM and MRE11 play only a minor role in the G2-M checkpoint, and their phenotypes manifest only in response to low-dose x-rays or when the ATR pathway is compromised (13). Thus, our screen was efficient in recovering major components of the G2-M checkpoint but may not have been sensitive enough to identify minor contributors. This also suggests that the candidate genes we identified are major components of the pathway. To test whether robust RNAi depletion of nbs, tefu, or mre11 induces a detectable checkpoint defect, we tested them in the same assay conditions as the secondary screens, which used a higher concentration of dsRNA than was used in the primary screen. RNAi-mediated depletion of tefu or nbs led to a moderate G2-M checkpoint defect (fig. S4). We added nbs and tefu to the final list of genes, which brings the total number of our candidate genes to 64.
Doxorubicin is a topoisomerase II (TOP2) inhibitor that damages DNA by trapping TOP2 in a TOP2-DNA complex after TOP2 cuts DNA (14). Therefore, doxorubicin does not damage DNA in the absence of TOP2, raising the possibility that some of the candidate genes identified in our screen were required specifically for the response to doxorubicin. Consistent with this, TOP2 was identified as one of the strongest hits (Fig. 1B). To distinguish between genes involved in general DNA damage responses and those involved in doxorubicin-specific responses, we performed a checkpoint analysis of the candidate genes, using three additional stimuli: etoposide, bleomycin (Bleocin), and x-rays (fig. S4). Etoposide is another TOP2 inhibitor that generates DSBs through TOP2, whereas bleomycin and x-rays directly attack DNA to generate DSBs. Although the response to each stimulus was variable, 59 of the 64 genes consistently scored positive for at least three stimuli (Fig. 1B).
Notable exceptions were TOP2 and TOPORS, a SUMO (small ubiquitin-like modifier) ligase that targets topoisomerase I (TOP1) (15), both of which showed an extremely strong checkpoint defect only with doxorubicin and etoposide (Fig. 1B). Phosphorylation of histone H2Av, a marker for DNA damage, was undetectable after treatment of cells depleted for either TOP2 or TOPORS with doxorubicin (fig. S5). Therefore, we assume that these genes act upstream of doxorubicin’s damaging effect on DNA.
Systems-level view of the G2-M checkpoint
Of the 64 genes, 62 have a clear mammalian ortholog (table S1), suggesting that the molecular network containing them is conserved across higher eukaryotes. We classified the 64 genes into functional groups on the basis of their cellular functions. In the unperturbed cell cycle, the G2-M transition is driven by the kinase CDC2, which is activated by the phosphatase CDC25. The activated G2-M checkpoint targets these two proteins to induce cell cycle arrest (1). On the basis of the known physical and genetic interactions of the proteins encoded by the 64 genes, we reconstructed a protein network with CDC2 and CDC25 as a central hub (Fig. 1C). More than 90% of the genes fell into five distinct categories: DNA repair, DNA replication, cell cycle control, chromatin regulation, and RNA processing (Fig. 1C). The DNA repair and cell cycle categories included the previously known Drosophila checkpoint genes mei-41 (ATR), mus304 (ATRIP), grp (CHK1), Myt1 (MYT1), 14-3-3ε (14-3-3ε), and nbs (NBS1). Similarly, the DNA replication category included Replication Protein A 70 (RPA70), which has been implicated in the G2-M checkpoint in other organisms (16). In contrast, little is known about the role of the genes in the remaining two categories, chromatin regulation and RNA processing, in the G2-M checkpoint.
The potential involvement of chromatin regulators is particularly interesting, because various histone modifications occur around the site of DNA damage (17). We identified 14 chromatin-related genes, including ubc9 and PIAS, which encode components of a SUMO ligase that targets various chromatin proteins. In mammals, this SUMO ligase targets BRCA1 (18, 19), which is an essential component of the ATM-dependent DNA damage checkpoint in mammalian cells (20). Because Drosophila lacks an ortholog of BRCA1, our results suggest that UBC9 and PIAS target additional proteins required for the G2-M checkpoint.
We also identified 17 genes involved in RNA processing. These genes may indirectly affect the G2-M checkpoint by regulating transcription of the other checkpoint genes. Our screen identified components of the CCR4-NOT deadenylation complex, which regulates the stability of messenger RNA (mRNA) by deadenylating the poly(A) (polyadenylate) tail. Components of this complex have been identified in multiple DNA damage screens in yeast (21–23), suggesting that its function in the DNA damage checkpoint may be evolutionarily conserved.
Some of the candidate genes, such as Rb, Skp1, and DDB1, have a well-documented function in general cell cycle progression (24–26). To test whether any of the other candidate genes had a role in cell cycle, we performed cell cycle profile analysis of RNAi-treated cells by quantifying DAPI (4′,6-diamidino-2-phenylindole) intensities of nuclei in fluorescent images. Knockdown of half of the candidate genes had a detectable effect on cell cycle progression to varying degrees (Fig. 1C and fig. S2).
Genetically separable functions of TOPBP1 in DNA replication and the G2-M checkpoint
In Drosophila, zygotic mutants of known DNA checkpoint genes, including mei-41 (ATR) and grp (CHK1), are viable as long as the maternal mRNA is present (27, 28). Among our candidate genes, more than half are homozygous lethal when mutated [P-element disruption project (29)], suggesting that they also have essential functions apart from checkpoint regulation or that maternal transcripts cannot compensate for their loss. Because characterization of the G2-M checkpoint in zygotic-lethal mutants is complicated, we decided to focus on two genes, mus101 and mus312, which had not previously been implicated in the G2-M checkpoint and for which viable alleles exist.
MUS101 is an evolutionarily conserved protein; orthologs are known as TOPBP1, DPB11, and CUT5 in vertebrates, budding yeast, and fission yeast, respectively (30). TOPBP1 is an essential component of the DNA replication preinitiation complex (pre-IC). Thus, disruption of TOPBP1 in yeast and mammals results in loss of cell proliferation and organismal death (30). In addition to DNA replication, TOPBP1 has a second role in the S-phase checkpoint (30), a mechanism that retains cells in S phase until DNA replication completes. Like the G2-M checkpoint, the S-phase checkpoint is activated by the ATR pathway. TOPBP1 is believed to be the activator of ATR in the S-phase checkpoint because it directly stimulates the kinase activity of ATR invitro (31). Although its ability to activate ATR suggests that TOPBP1 may also have a role in the DSB-induced G2-M checkpoint, it has not been demonstrated in higher eukaryotes, in part because loss of TOPBP1 abrogates cell viability (32). Some of the Drosophila mus101 mutant alleles are viable and specifically sensitive to DNA damage (33), providing an opportunity to dissect its function in the context of the DNA damage response.
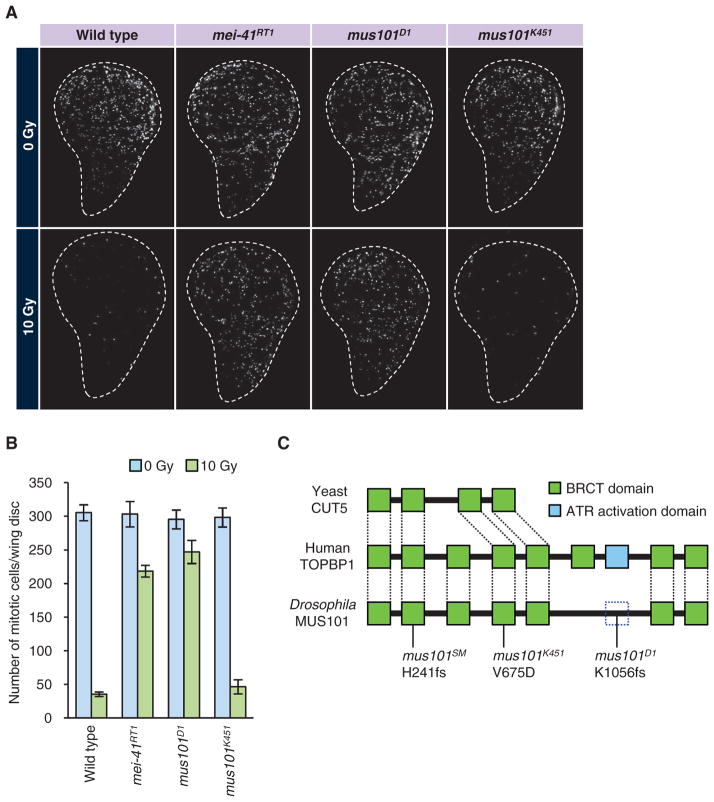
To address the in vivo function of MUS101, we used a standard immunohistochemistry assay (11) to examine the G2-M checkpoint of larval imaginal discs in mus101 mutant animals. Whereas putative null alleles, such as mus101SM, are characterized by early larval lethality resulting from loss of cell proliferation in imaginal discs (33), mus101D1 and mus101K451 are viable alleles that show distinct phenotypes. mus101D1 flies exhibit a DNA damage–sensitive phenotype and mus101K451 flies have the same phenotype, but are also female sterile due to reduced gene amplification in oocyte follicle cells (34). This gene amplification is a special form of DNA replication that locally amplifies certain regions of the genome. The impairment of this form of DNA replication caused by the mus101K451 mutation does not affect cell proliferation and animal viability in other tissues.
Cells in the imaginal disc divide rapidly and have a robust G2-M check-point that is inducible by DNA damage. Irradiation of wild-type third-instar larvae with 10 Gyof x-rays induced cell cycle arrest within 1 hour, as indicated by a more than 80% reduction in the number of mitotic cells in the wing imaginal disc (Fig. 2, A and B). In mei-41 (ATR) mutants, imaginal disc cells continued to enter mitosis after x-ray irradiation as a result of abrogation of the G2-M checkpoint (Fig. 2, A and B). Likewise, in mus101D1 mutant larvae, x-ray irradiation did not induce cell cycle arrest, demonstrating that mus101 is essential for activation oftheG2-Mcheckpointinvivo(Fig. 2, A and B). However, mus101K451 mutants underwent cell cycle arrest upon exposure to x-rays, indicating that they had an intact G2-M checkpoint (Fig. 2, A and B). These observations suggest that the functions of MUS101 in DNA replication and checkpoint activation are genetically separable.
Fig. 2.
mus101 mutants and their G2-M checkpoint phenotypes. (A) G2-M checkpoint in different mus101 alleles. Wild-type and mei-41 animals are shown as negative and positive controls, respectively. Third-instar larvae of the indicated genotype were irradiated with 10 Gy of x-rays. After 1 hour, the discs were dissected, fixed, and stained with antibody against phosphohistone H3 (white). Persistent mitosis after irradiation is an indication of a defect in the G2-M checkpoint. (B) Quantification of the data in (A). Mitotic index was calculated for nonirradiated discs and irradiated discs of each genotype. Five to eight discs were analyzed for each condition. Average and SD are shown. (C) Primary structure and mutations of MUS101. mus101SM1 is a null allele and early larval lethal. mus101K451 is DNA damage–sensitive, as well as female-sterile due to fragile egg shell resulting from compromised DNA replication in follicle cells. mus101D1 is DNA damage–sensitive and defective in the G2-M checkpoint. Dashed box represents the putative ATR activation domain in the MUS101 protein.
To gain insight into the structure-function relationship of MUS101, we determined the sequences of the mus101 mutant alleles. MUS101 and vertebrate TOPBP1 have multiple BRCA1 C-terminal (BRCT) domains, which are protein-protein interaction motifs frequently observed in DNA damage response proteins (30). MUS101 and vertebrate TOPBP1 have seven and eight BRCT domains, respectively, where the first five and the last two BRCT domains are identified as orthologous across species (Fig. 2C) (30). In vertebrate TOPBP1, an ATR activation domain has been identified between the sixth and the seventh BRCT domains. In vertebrates, a peptide containing the first six BRCT domain is sufficient to stimulate DNA replication, whereas the remainder of the peptide containing the ATR activation domain is sufficient to activate ATR (35), suggesting that the replication and checkpoint functions are encoded by separate domains. Consistent with this finding in vertebrates, we found that mus101D1, which is specifically defective in the G2-M checkpoint, carries a deletion that removes the last two BRCT domains, as well as part of the linker region in which the putative ATR activation domain is mapped (Fig. 2C). In contrast, mus101K451, which is defective in DNA replication (33), had a missense mutation in an evolutionarily conserved residue in the N-terminal fifth BRCT domain (Fig. 2C). These results corroborate the separable roles of the N- and C-terminal BRCT repeats and, together with the studies in vertebrates (35), demonstrate that the two regions are not only sufficient but also required for their respective functions.
RNAi-mediated in vivo validation of pre-IC components
Although the replication domain of MUS101 was dispensable for the G2-M checkpoint, our screen identified other components of the pre-IC, including CDC45L, RECQL4, and MCM7 (minichromosome maintenance 7), as well as the replication protein A (RPA) complex, which the pre-IC loads onto the replication fork. The pre-IC consists of the MCM2 to 7 helicase complex, TOPBP1, CDC45L, and RECQL4, as well as the GINS complex (36). The genes encoding CDC45L and RECQL4 were among the strongest hits in our screen (Fig. 1B). The fact that genes encoding components of the pre-IC were the only DNA replication genes identified in the screen suggests that the pre-IC, and not DNA replication in general, has a specific role in the G2-M checkpoint. In S2R+ cells, knockdown of the pre-IC genes had a minimal impact on the mitotic index and cell cycle profile in the absence of DNA damage (fig. S6), suggesting that the mitotic cells persisting after DNA damage resulted from failure of the G2-M checkpoint rather than failure of the S-phase checkpoint.
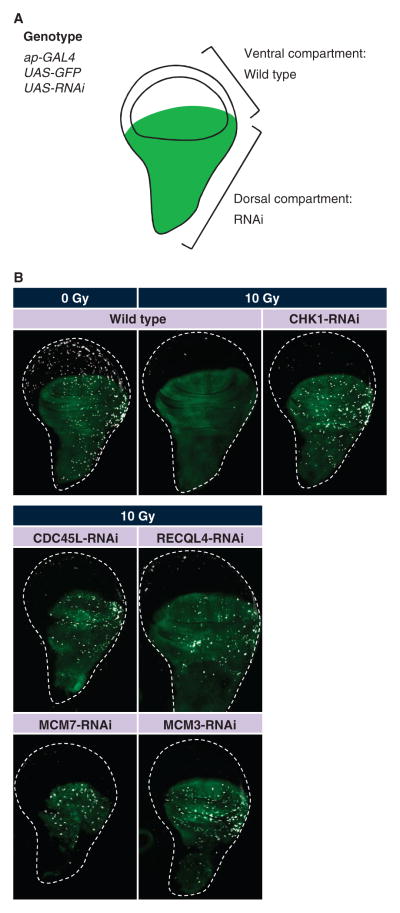
To verify that the pre-IC genes are indeed required in vivo, we examined their roles in the wing disc mitosis assay. Because mutants of the pre-IC genes are early larval lethal, we used inducible RNAi to knock down the activity of each of these genes specifically in the wing disc. To suppress gene function in larval imaginal discs, we individually knocked down the pre-IC components by overexpressing a hairpin transgene that encoded a dsRNA targeting the corresponding pre-IC component (Fig. 3A). RNAi knockdown of CDC45L, RECQL4, MCM3, or MCM7 failed to block mitosis entry upon DNA damage (Fig. 3B). Although the MCM helicase is a hexameric protein complex comprising MCM2 to 7, our screen only identified MCM7. In cultured Drosophila cells, MCM2 to 6 proteins are highly resistant to RNAi; more than 95% depletion of each of the MCM2 to 6 proteins does not induce detectable replication defects (37). However, we found that in vivo knockdown of either MCM3 or MCM7 resulted in a checkpoint defect (Fig. 3B), suggesting that the whole MCM complex is required for the G2-M checkpoint. Knockdown of MCM7 and MUS101 did not cause a strong checkpoint defect in S2R+ cells, whereas knockdown of MCM7 or a mus101 mutation did in vivo. The difference is most likely attributable to the variable efficiency of RNAi reagents in different tissue types. Together, these results confirm the in vivo function of these genes in the G2-M checkpoint and suggest that the pre-IC as a whole participates in the G2-M checkpoint.
Fig. 3.
In vivo validation by transgenic RNAi identifies the replication initiation proteins as a crucial regulator of the G2-M checkpoint. (A) Schematic representation of a wing disc depicting the dorsal compartment region (green), where RNAi transgenes were expressed with the GAL4/UAS (upstream activation sequence) system. ap-GAL4 drives expression of the UAS-RNAi hairpin transgenes, leading to compartment-specific suppression of the target gene. Transgenic animals also carry UAS-GFP (green fluorescent protein) that expresses GFP in the dorsal compartment. (B) RNAi-mediated knockdown of replication initiation proteins leads to continued mitosis after irradiation-induced DNA damage. Third-instar larvae were irradiated and the wing discs were stained with the antibody against phosphohistone H3 as described in Fig. 2.
Role of the nuclease scaffolding protein MUS312 in the Drosophila G2-M checkpoint
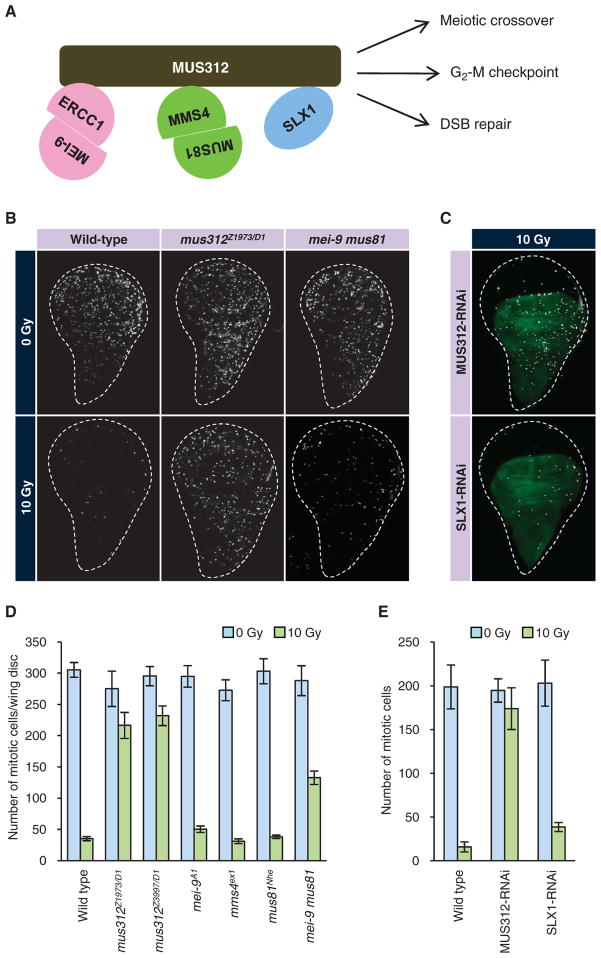
Our screen identified MUS312, a protein believed to have a direct role in DSB repair (38–42), which represents a branch of the DNA damage response that is separate from the G2-M checkpoint. mus312 mutants are sensitive to various DNA-damaging agents and exhibit reduced meiotic crossovers (43). In vitro, the mammalian ortholog of MUS312 (known as BTBD12) has resolvase activity for Holliday junction structures, an intermediate DNA structure that forms during DSB repair (38–40). The resolvase activity is mediated by the three nucleases that bind MUS312: the nuclease consisting of MEI-9 (XPF) and ERCC1, the nuclease consisting of MUS81 and MMS4, and the nuclease SLX1 (Fig. 4A). Although these observations support a direct role of MUS312 in DSB repair, they do not explain the role of MUS312 in the G2-M checkpoint because DNA repair is presumed to occur later than checkpoint activation during the DNA damage response.
Fig. 4.
Drosophila MUS312 and its binding partners are essential for the G2-M checkpoint. (A) Schematic representation of the MUS312 complex. MUS312 orthologs in vertebrates and yeast form a protein complex with three nucleases: the ERCC1–MEI-9 heterodimer, the MUS81-MMS4 heterodimer, and SLX1. (B and D) G2-M checkpoint phenotypes of mus312 or nuclease-encoding genes. Wing discs of the indicated genotype were irradiated with x-rays and stained with an antibody against phosphohistone H3 (white). Representative pictures are shown in (B). The results were quantified and shown as a bar graph in (D). (C and E) G2-M checkpoint in animals expressing a dsRNA against MUS312 or SLX1 in the dorsal compartment [green in (C)]. The animals were irradiated and stained as in (B). Mitotic cells in the dorsal compartment were quantified before and after irradiation (E). At least five discs were analyzed for each genotype. Error bars represent SDs.
To confirm the function of MUS312 in the G2-M checkpoint in vivo, we monitored the persistence of mitotic cells in response to DNA damage in three independent alleles of mus312 mutants examined in heteroallelic combinations to preclude possible artifacts caused by any second-site mutation. Each of the three alleles (D1, Z1973, and Z3997) carries a nonsense mutation in the middle of the transcript and is, therefore, considered a null allele (43). In all allelic combinations tested, we observed a strong defect in activation of the G2-M checkpoint in response to x-ray irradiation (Fig. 4, B and D). Transgenic RNAi targeting mus312 also produced a strong checkpoint defect (Fig. 4, C and E). The phenotypic strength of mus312 mutants was equivalent to that of mei-41 (ATR) (Figs. 2B and 4D), suggesting that MUS312 is a central component of the G2-M checkpoint in Drosophila.
Because MUS312 binds the three nucleases—the MEI-9–ERCC1 complex, the MUS81-MMS4 complex, and SLX1—we hypothesized that they each might play a role in the G2-M checkpoint. However, none of the genes encoding these nuclease components was recovered from our genome-wide RNAi screen. Likewise, the single mutants of mei-9, mms4, or mus81 did not exhibit a loss of checkpoint activity in response to irradiation (Fig. 4, B and D). However, we did observe an increase in mitotic cells in irradiated discs from double mutant animals lacking both mei-9 and mus81 (Fig. 4, B and D). The number of mitotic cells in these double mutants was roughly 50% that of mus312. Further, in vivo knockdown of the third nuclease, SLX1 (CG18271), led to a mild but significant increase in mitotic cells after x-ray irradiation (P < 0.001, Student’s t test; n = 6 for wild type, n = 8 for SLX1 RNAi) (Fig. 4, C and E). These results suggest that the three nucleases are redundantly required for the G2-M checkpoint and that the checkpoint function of MUS312 may depend on the three nucleases.
DISCUSSION
Here, we described a systems-level study of the G2-M DNA damage checkpoint in Drosophila. The analysis identified a high-confidence list of 64 genes, of which more than 90% were not previously known to play a role in G2 arrest in response to DNA damage. By analyzing the mitotic index of wing discs of mutant flies in response to irradiation-induced DNA damage, we confirmed the roles of two protein complexes, one containing MUS101 (TOPBP1) and the other containing MUS312 (BTBD12).
The 64 genes were subjected to rigorous secondary screens for validation. We confirmed the phenotype of each gene by multiple dsRNAs to exclude the possibility of off-target effects. We then asked whether the genes are required for general DNA damage response by testing three additional DSB-inducing agents: etoposide, Bleocin, and x-rays. We found that two genes, TOPORS and TOP2, exhibited strong checkpoint defects only for doxorubicin and etoposide. The results obtained for TOP2 are consistent with the fact that doxorubicin and etoposide are TOP2 poisons that depend on TOP2 to damage DNA. TOPORS is a chromatin-associated SUMO ligase targeting TOP1 (15). Although TOPORS has not been shown to target TOP2, both TOP1 and TOP2 are SUMOylated in response to DNA damage (44, 45), suggesting that TOPORS may also target TOP2. The mRNA expression of TOP2 in cancer cells has a negative correlation with the efficacy of doxorubicin as an antimitotic drug (46). Thus, it will be interesting to test whether there is a correlation between the presence and abundance of TOPORS and sensitivity to doxorubicin in human cancer.
Another study reported that suppression of ATM and CHK1 in mammalian cells leads to a delay in mitotic exit after DNA damage, suggesting that there is a mechanism that facilitates mitotic exit in the presence of DNA damage (47). Because the delay in mitotic exit can also result in an increased mitotic index, it is possible that some of the candidate genes identified in our screen may have a function in this process rather than in the G2-M check-point. However, the existence of this mechanism in nonmammalian species has not been reported. On the contrary, there is ample evidence in the literature that there is a mechanism that delays mitotic exit after DNA damage. For instance, Drosophila MEI-41 (ATR) delays mitotic exit after DNA damage (48). Further, it is known that the pathway involving ATR and CHK1 delays mitotic exit in yeasts (49). Thus, given these observations, we assume that most of our candidate genes have no effect on the mitotic exit delay, although we cannot formally exclude this possibility.
Extensive data mining for known physical or genetic interactions of the products of the 64 genes revealed a protein network of five major categories—DNA repair, DNA replication, cell cycle regulation, chromatin regulation, and RNA processing—with ATR at the center (Fig. 1C). A previous proteomic study in mammalian cells identified about 700 proteins that are phosphorylated by ATM or ATR in response to DNA damage, which account for 3% of the whole proteome (4). Comparison of the products of our candidate genes with these putative ATM or ATR targets revealed that 30% of our candidate proteins corresponded to ATM or ATR substrates identified in human cells (Fig. 1C). This observation lends further support to the idea that the 64 genes are involved in the ATM/ATR-dependent G2-M checkpoint signaling pathway.
MUS101 is the Drosophila ortholog of mammalian TOPBP1, a component of the DNA replication pre-IC. Previous biochemical studies have suggested that TOPBP1 plays a role in ATR activation, which is distinct from its role in DNA replication (35). Our identification of mus101D1 as an allele that is specifically defective in the G2-M checkpoint unambiguously confirms this observation in vivo. Unexpectedly, our screen identified gene encoding additional components of the pre-IC, CDC45L, RECQL4, and some components of the MCM helicase. Absence of DNA replication can force cells to skip S phase with unreplicated DNA or leave S phase with incompletely replicated DNA and enter abnormal mitosis (50). Thus, it is possible that knockdown of the pre-IC genes caused abnormal mitosis and metaphase arrest, which would lead to the persistence of mitotic cells after DNA damage regardless of the G2-M checkpoint. However, our results argue against this possibility. First, S2R+ cells depleted of each of the pre-IC genes had a mitotic index not greatly different from control (fig. S6) and exhibited normal mitotic exit in the absence of induced DNA damage (fig. S3), suggesting that mitosis was normal and the mitotic cells were not arrested in metaphase. Second, the cell cycle profile of these cells showed distinct G1 and G2 populations with a slight increase in S population (fig. S6). This suggests that the cells could complete S phase despite slowed DNA replication. Therefore, the sustained presence of mitotic cells after DNA damage was unlikely to be a secondary consequence of DNA replication defects but was caused by a bona fide G2-M checkpoint defect. Because RNAi does not completely eliminate the target protein, we presume that RNAi of the pre-IC genes in S2R+ cells resulted in a hypomorphic phenotype in which DNA replication and cell cycle progression were only moderately affected but the G2-M checkpoint was severely impaired.
Although the pre-IC has not been considered to be a component of the G2-M checkpoint, several studies have suggested its possible role in ATR activation in response to DNA damage. MCM7 is required for localization of ATR to DNA damage foci and for activation of ATR after ultraviolet irradiation, as well as controlling the S-phase checkpoint (51, 52). RECQL4 is required for the S-phase checkpoint induced by DNA damage (53, 54) and accumulates at sites of DNA damage (55). A study in zebrafish reported that TICRR, a pre-IC component without an ortholog in Drosophila, is required for the G2-M checkpoint, in addition to its roles in DNA replication and the S-phase checkpoint (56). These observations, along with our identification of multiple pre-IC components as needed for the G2-M checkpoint, raise the possibility that the entire pre-IC has a role in ATR activation and the G2-M checkpoint.
There is an interesting parallel between initiation of DNA replication forks and the cellular processing of DSBs. Once a DSB is detected by the cell, DNA around the DSB is unwound by helicase activity, which exposes single-stranded DNAs (ssDNAs). One of the DNA strands is degraded by 5′-3′ nuclease activity, whereas the other strand is loaded with the RPA complex, an ssDNA binding protein that recruits ATR and activates the G2-M checkpoint (1). At a DNA replication fork, DNA double strands are unwound by the MCM helicase, exposing ssDNAs that are loaded with RPA in a CDC45L- and RECQL4-dependent manner (36). The identity of the helicase activity during DSB processing remains obscure in higher eukaryotes. Although the BLM helicase has been suggested as a candidate (57), G2-M checkpoint defects have not been reported for BLM mutants, suggesting that there might be an additional helicase involved. We speculate that the pre-IC may play a role in the unwinding of DNA and active recruitment of RPA at DSBs.
MUS312 and its mammalian ortholog BTBD12 have emerged as important regulators of homology-directed repair (HDR) of DSBs. In vitro, mammalian BTBD12 has a Holliday junction resolvase activity that is mediated by the three associated nucleases XPF-ERCC1, MUS81-MMS4, and SLX1. Drosophila MUS312 binds at least XPF-ERCC1 and SLX1 (41), and we expect that it also binds MUS81-MMS4 because the interaction in mammals is mediated by the evolutionarily conserved SAP domain (38). We identified MUS312 as an essential component of the G2-M checkpoint. Further, we provided genetic evidence that the associated nucleases were also required for the G2-M checkpoint. This suggests that all three nucleases contributed to the checkpoint function of MUS312. The nucleases associated with MUS312 are structure-specific nucleases that are selectively active on DSB repair intermediates, including Holliday junctions and D loops (38–40), which are formed during HDR by strand invasion, a RAD51-dependent process by which single-strand protrusions of processed DSB ends invade a homologous DNA sequence of a different chromosome. Therefore, it is likely that the function of MUS312 in the G2-M checkpoint involves DSB repair intermediates.
If MUS312 requires DSB repair intermediates to activate the checkpoint, the function of MUS312 should depend on RAD51. The status of the G2-M checkpoint in Drosophila rad51 mutants has not been reported. However, because mutants of brca2 and rad54, which are required for the RAD51 function, exhibit no G2-M checkpoint defect (58, 59), rad51 mutants are likely to have an intact G2-M checkpoint. If this is indeed the case, we speculate that in rad51 mutant cells the RPA-coated ssDNA protrusion of a DSB keeps activating ATR in a MUS312-independent manner. In contrast, the ssDNA protrusion in wild-type cells is recoated with RAD51 and forms a D loop or a Holliday junction by strand invasion. We envision that once strand invasion occurs, the DSB intermediate must be somehow processed by MUS312 to continue to activate the checkpoint. Further analysis, including epistasis testing of RAD51 and MUS312, will be required to determine whether MUS312 is required for ATR activation and whether the checkpoint function of MUS312 depends on DSB repair intermediates.
In the Drosophila imaginal disc, mitotic cells completely disappear 1 hour after high-dose x-ray irradiation. In mus312 mutants, cells continue to enter mitosis at this time point, suggesting that MUS312 is required for initiation of the G2-M checkpoint. If MUS312 acts downstream of the formation of DNA repair intermediates, then MUS312 is unlikely to participate in the initial activation of ATR, which occurs before HDR. Although it may appear counterintuitive that such late ATR activation is required for initiation of the G2-M checkpoint, there is indeed a time lag between ATR activation and checkpoint activation. Full activation of the G2-M checkpoint takes 1 hour after DNA damage, whereas ATR is activated within minutes. Thus, we propose that there is a second phase of ATR activation, which involves processing of DSB repair intermediates by MUS312, and that it is critical for initiation of the G2-M checkpoint. Moreover, this mechanism may also provide an explanation for the observation that the checkpoint can be maintained for hours. Although the mechanism of ATR activation by free DSB ends has been extensively studied, less attention has been paid to how the G2-M checkpoint is maintained during subsequent DSB repair. Because the purpose of the G2-M checkpoint is to prevent entry into mitosis with unrepaired DNA, there is likely to be a mechanism by which ongoing DNA repair processes keep activating ATR until repair completes. Indeed, a single high dose of radiation can activate the checkpoint for several hours in a dose-dependent manner in Drosophila, suggesting the existence of a maintenance mechanism (60). We propose that MUS312 transduces signals from DSB repair intermediates to ATR, thereby maintaining the G2-M checkpoint until HDR completes.
The molecular mechanism of G2-M checkpoint activation by MUS312 is unclear. A study showed that Caenorhabditis elegans mutants lacking GEN1, another Holliday junction resolvase, are defective in the DNA damage checkpoint (61). This suggests that there might be an evolutionarily conserved mechanism by which nuclease-mediated processing of DSB repair intermediates activates the G2-M checkpoint. Another study suggests an alternative scenario. The yeast ortholog of MUS312, SLX4, binds TOPBP1 in response to replication stress (62). Similarly, Drosophila MUS312 may bind TOPBP1 in response to DNA damage and recruit it to DSB repair intermediates, thereby activating checkpoint signaling. Further studies, both in vivo and in vitro, will be required to uncover the mechanism by which MUS312 regulates the G2-M checkpoint.
MATERIALS AND METHODS
Primary genome-wide RNAi screen
The genome-wide dsRNA library was produced by the Drosophila RNAi Screening Center (DRSC) at Harvard Medical School (63) (http://www.flyrnai.org/). The library comprises 62 384-well plates and covers the entire genome. Each well of the plates contains roughly 0.25 μg of dsRNA. We used Drosophila S2R+ cells for the screen and all subsequent analyses. S2R+ cells were maintained in Schneider’s medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin.
The primary screen was conducted as follows. S2R+ cells were harvested from a confluent 75-cm2 flask and counted. The cells were collected in 50-ml conical tubes by centrifugation at 240g for 5 min. The cell pellet was resuspended in serum-free Schneider’s medium (with penicillin/streptomycin) at 7.5 × 105 cells/ml. A dsRNA against nbs was added at the final concentration of 12.5 ng/μl. Twenty microliters of the cell suspension was dispensed into each well of the plates containing dsRNA by the Multidrop automatic dispenser (Thermo Fisher Scientific). The cells were incubated under serum-free conditions for 1 hour at 25°C. After incubation, 20 μl of Schneider’s medium supplemented with 20% FBS and penicillin/streptomycin was added to bring the final FBS concentration to roughly 10%. A maximum of 16 plates were processed in one set of experiments. The plates were incubated at 25°C for 4 days. Doxorubicin (Sigma-Aldrich) was added at the final concentration of 0.5 μM. The cells were incubated for 4 hours and fixed with 4% paraformaldehyde for 10 min. We directly added 10 μl of 16% paraformaldehyde (Electron Microscopy Sciences) to each well of a 384-well plate by Multidrop without removing the culture medium. After fixation, the cells were washed three times in phosphate-buffered saline (PBS)/0.1% Triton X-100 (PBST) and then incubated with an antibody that recognizes phosphohistone H3 (Ser10, Millipore) at a 1:2000 dilution overnight. The next day, the cells were briefly washed once in PBST, incubated with an Alexa 488–conjugated secondary antibody (1:2000, Molecular Probes) and DAPI for 4 hours, and washed twice in PBST. Images of DAPI and FITC (fluorescein isothiocyanate) fluorescence were acquired on ImageXpress Micro (Molecular Devices) with a 4× lens. The number of phosphohistone H3–positive cells in each well was counted by a custom script with the image analysis software ImageJ (http://rsb.info.nih.gov/ij/). In control wells, fewer than 10 mitotic cells were observed after doxorubicin treatment. dsRNAs that led to more than 20 mitotic cells per well were scored as positive.
The screen was done in duplicate. The second set was processed with a slight modification. Colchicine was added to the final concentration of 5 μM at 4 hours after doxorubicin addition and incubated for a further 4 hours in an attempt to trap the cells that passed the G2-M boundary. Phenotypes were visually scored without quantification for the second set.
Secondary screens
To induce robust RNAi-mediated depletion, we performed RNAi of the candidate genes in 96-well plates with a higher concentration of dsRNA (13 ng/μl versus 5 ng/μl in the primary screen). The experiments were performed in duplicate. Briefly, S2R+ cells were harvested and resuspended at 1.0 × 106 cells/ml in serum-free Schneider’s medium. Seventy microliters of the cell suspension was dispensed into each well of a 96-well plate containing dsRNAs. After 1-hour incubation at 25°C, 70 μl of Schneider’s medium supplemented with 20% FBS was added to each well. The plates were left in the incubator at 25°C. On day 3, the cells were resuspended and split into six 96-well plates. After overnight incubation in the new plates, the cells were treated with or without 0.5 μM doxorubicin for 4 hours and fixed, and the mitotic index was determined by phosphohistone H3 immunocytochemistry.
The checkpoint phenotype was quantified as a relative mitotic index, which was calculated as (mitotic index after DNA damage)/(mitotic index before DNA damage) × 100. dsRNAs that increased the relative mitotic index by more than 50% compared to the control (luciferase RNAi) were scored as positive (fig. S4).
To identify and exclude potential false-positive genes that have a major role in mitotic progression, we monitored the mitotic index without DNA damage. Genes whose depletion led to more than a twofold increase in the mitotic index were excluded (table S2). We also directly monitored mitotic exit of RNAi-treated cells by measuring the mitotic index after blocking entry into mitosis. For this purpose, we incubated the cells with 30 μM NSC663284, a CDC25 inhibitor that blocks G2-M transition, for 8 hours and determined the mitotic index. Genes whose depletion led to a mitotic exit defect, where the relative mitotic index (before versus after drug treatment) was higher than 50%, were excluded (fig. S3 and table S2).
We used a dsRNA against firefly luciferase (luc+) as a negative control. At least two independent dsRNAs targeting nonoverlapping regions of the gene were tested to preclude the possibility of off-target effects. When multiple hits from the screen composed a known protein complex, validation by a second dsRNA was omitted, assuming that it was highly unlikely that multiple genes of the same complex scored positive because of off-target effects. We also tested only one dsRNA for known checkpoint genes. The dsRNAs obtained from the DRSC and their IDs are listed in table S1. When a second dsRNA was not available from the DRSC, a custom dsRNA was generated by invitro transcription of a DNA template PCR (polymerase chain reaction)–amplified from genomic DNA with newly designed primers (table S4).
In addition to doxorubicin, we used a variety of DSB-inducing agents to test whether the observed checkpoint defect was drug-specific. We used x-rays (150 Gy), Bleocin (50 μg/μl, single component of bleomycin family group A; Calbiochem), or etoposide (10 μM; Sigma-Aldrich). We treated the cells with these drugs for 4 hours, or irradiated the cells and let them recover for 1 hour, whereas the control cells were left untreated for 1 hour, before fixation and phosphohistone H3 immunostaining.
Image-based cell cycle profile analysis
Cells were seeded in a 96-well plate, fixed in 4% paraformaldehyde, and stained with DAPI. Fluorescent images were acquired on ImageXpress Micro with a 10× lens. The integrated fluorescence intensities of each DAPI-stained nucleus were measured with ImageJ. A histogram was generated from measurements of 1500 nuclei with Microsoft Excel.
Fly genetics
mei-41RT1, mus101D1, mus101D2, mei-9A1, and mus312D1 were obtained from the Bloomington Drosophila Stock Center. mms4ex1, mus81Nhe, mus312Z1973, and mus312Z3997 were obtained from J. Sekelsky. mus101K451 and mus101SM were obtained from D. Glover. UAS-CDC45L-RNAi (3658R-2), UAS-RecQ4-RNAi (7487R-3), and UAS-Mcm3-RNAi (9633R-1) were obtained from the National Institute of Genetics Fly Stock Center (Mishima, Japan). UAS-grp-RNAi (JF02588), UAS-mus312-RNAi (HMS00192), and UAS-slx1-RNAi (HMS00313) were obtained from the Transgenic RNAi Project at the DRSC (http://www.flyrnai.org/).
Checkpoint analysis of larval imaginal discs
Late third-instar larvae were irradiated with 10 Gy of x-rays and allowed to recover for 1 hour. Imaginal discs were dissected, fixed, and stained with an antibody against phosphohistone H3 (Ser10) (Millipore #06-570, used at 1:2000). Fluorophore-conjugated secondary antibodies were used at a 1:2000 dilution. Images were taken as described for immunostaining of cultured cells.
Mutational analysis of mus101 alleles
Genomic DNA was extracted from pools of homozygous mus101D1, mus101D2, and mus101K451 animals and heterozygous mus101SM animals. A 5-kb DNA fragment encompassing the entire mus101 open reading frame (ORF) was amplified from the genomic DNA of each allele by PCR with the primers mus101-F (GCCAGCGTTGC-CAACGGTTTGAAGGC) and mus101-R (CCAATGCATGTACAAA-GAATCTAATGATTCG) with LA-Taq (Takara) and gel-purified. The purified DNA fragment was sequenced with the following primers: mus101-F, mus101-SQF1 (AGCGCGAGGGCATCATGGCC), mus101-SQF2 (TCTGGATGGCTGCTGTGTG), mus101-SQF3 (CTCAGTGCCAGCACTCTATC), mus101-SQF4 (AGAGTCCCGAGGATTTCCCC), mus101-SQF5 (CATCAGGCAGCGCGTGATGC), and mus101-SQF6 (GCAAAGAGAAGATCCTCTGG). mus101D1 and mus101D2 had an identical deletion (c.3168-3659del, p.K1056fs), suggesting that they are derived from the same mutational event. mus101K451 had a missense mutation (c.T2024A, p.V675D). mus101SM had a 1–base pair deletion (c.722delA, p.N241fs).
Supplementary Material
Acknowledgments
We thank the Drosophila RNAi Screening Center and the Institute of Chemistry and Cell Biology-Longwood for assistance with the RNAi screen and automated microscopy, respectively; K. McKim for antibody against phosphoH2Av; and J. Sekelsky, T.-S. Hsieh, P. Gallant, D. Glover, K. Moses, H.-M. Bourbon, the Bloomington Drosophila Stock Center, the National Institute of Genetics Fly Stock Center, and the Transgenic RNAi Project for fly stocks. We thank S. Elledge for helpful discussions and M. Markstein for critical reading of the manuscript.
Funding: S.K. is supported by a Research Fellowship from the Uehara Memorial Foundation. N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions: S.K. and N.P. designed the experiments. S.K. conducted the experiments. S.K. and N.P. interpreted the results and wrote the manuscript.
Competing interests: The authors declare that they have no competing financial interests.
www.sciencesignaling.org/cgi/content/full/4/154/rs1/DC1
Fig. S1. Caffeine-sensitive G2-M checkpoint of Drosophila S2R+ cells.
Fig. S2. Cell cycle profiles of cells depleted of the candidate genes.
Fig. S3. Secondary screen for mitotic exit defects.
Fig. S4. G2-M checkpoint defect of cells depleted of the candidate genes.
Fig. S5. DNA damage–induced phosphorylation of histone H2Av in cells depleted of candidate genes.
Fig. S6. Mitotic index and cell cycle profile of cells depleted of pre-IC proteins.
Table S1. Genes required for the G2-M checkpoint induced by doxorubicin.
Table S2. Primary hits excluded from further analysis and reasons for exclusion.
Table S3. Statistics of the secondary screens.
Table S4. Sequences of primers used for synthesizing dsRNA templates.
References
REFERENCES AND NOTES
- 1.Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 2.O’Driscoll M, Jeggo PA. The role of double-strand break repair—insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 3.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 4.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 5.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakada D, Shimomura T, Matsumoto K, Sugimoto K. The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 2003;31:1715–1724. doi: 10.1093/nar/gkg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries HI, Uyetake L, Lemstra W, Brunsting JF, Su TT, Kampinga HH, Sibon OC. Grp/DChk1 is required for G2-M checkpoint activation in Drosophila S2 cells, whereas Dmnk/DChk2 is dispensable. J Cell Sci. 2005;118:1833–1842. doi: 10.1242/jcs.02309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oikemus SR, Queiroz-Machado J, Lai K, McGinnis N, Sunkel C, Brodsky MH. Epigenetic telomere protection by Drosophila DNA damage response pathways. PLoS Genet. 2006;2:e71. doi: 10.1371/journal.pgen.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 10.Hari KL, Santerre A, Sekelsky JJ, McKim KS, Boyd JB, Hawley RS. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- 11.Brodsky MH, Sekelsky JJ, Tsang G, Hawley RS, Rubin GM. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 2000;14:666–678. [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Z, Homola E, Tiong S, Campbell SD. Drosophila Myt1 is the major Cdk1 inhibitory kinase for wing imaginal disc development. Genetics. 2008;180:2123–2133. doi: 10.1534/genetics.108.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bi X, Gong M, Srikanta D, Rong YS. Drosophila ATM and Mre11 are essential for the G2/M checkpoint induced by low-dose irradiation. Genetics. 2005;171:845–847. doi: 10.1534/genetics.105.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammer E, Heilbronn R, Weger S. The E3 ligase Topors induces the accumulation of polysumoylated forms of DNA topoisomerase I in vitro and in vivo. FEBS Lett. 2007;581:5418–5424. doi: 10.1016/j.febslet.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 16.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 17.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 18.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, Ng T, Solomon E. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 19.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deshpande GP, Hayles J, Hoe KL, Kim DU, Park HO, Hartsuiker E. Screening a genome-wide S. pombe deletion library identifies novel genes and pathways involved in genome stability maintenance. DNA Repair. 2009;8:672–679. doi: 10.1016/j.dnarep.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westmoreland TJ, Wickramasekara SM, Guo AY, Selim AL, Winsor TS, Greenleaf AL, Blackwell KL, Olson JA, Jr, Marks JR, Bennett CB. Comparative genome-wide screening identifies a conserved doxorubicin repair network that is diploid specific in Saccharomyces cerevisiae. PLoS One. 2009;4:e5830. doi: 10.1371/journal.pone.0005830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett CB, Westmoreland TJ, Verrier CS, Blanchette CA, Sabin TL, Phatnani HP, Mishina YV, Huper G, Selim AL, Madison ER, Bailey DD, Falae AI, Galli A, Olson JA, Greenleaf AL, Marks JR. Yeast screens identify the RNA polymerase II CTD and SPT5 as relevant targets of BRCA1 interaction. PLoS One. 2008;3:e1448. doi: 10.1371/journal.pone.0001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du W, Dyson N. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J. 1999;18:916–925. doi: 10.1093/emboj/18.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy TD. Drosophila skpA, a component of SCF ubiquitin ligases, regulates centrosome duplication independently of cyclin E accumulation. J Cell Sci. 2003;116:2321–2332. doi: 10.1242/jcs.00463. [DOI] [PubMed] [Google Scholar]
- 26.Takata K, Yoshida H, Yamaguchi M, Sakaguchi K. Drosophila damaged DNA-binding protein 1 is an essential factor for development. Genetics. 2004;168:855–865. doi: 10.1534/genetics.103.025965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- 28.Sibon OC, Laurençon A, Hawley R, Theurkauf WE. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr Biol. 1999;9:302–312. doi: 10.1016/s0960-9822(99)80138-9. [DOI] [PubMed] [Google Scholar]
- 29.Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 32.Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto RR, Axton JM, Yamamoto Y, Saunders RD, Glover DM, Henderson DS. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics. 2000;156:711–721. doi: 10.1093/genetics/156.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orr W, Komitopoulou K, Kafatos FC. Mutants suppressing in trans chorion gene amplification in Drosophila. Proc Natl Acad Sci USA. 1984;81:3773–3777. doi: 10.1073/pnas.81.12.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto Y, Tsujimura T, Sugino A, Takisawa H. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 2006;11:993–1007. doi: 10.1111/j.1365-2443.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- 36.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: Replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 37.Crevel G, Hashimoto R, Vass S, Sherkow J, Yamaguchi M, Heck MM, Cotterill S. Differential requirements for MCM proteins in DNA replication in Drosophila S2 cells. PLoS One. 2007;2:e833. doi: 10.1371/journal.pone.0000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, III, Russell P, Fuchs RP, McGowan CH, Gaillard PH. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muñoz IM, Hain K, Déclais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, Kanaar R, Ponting CP, Lilley DM, Rouse J. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito TT, Youds JL, Boulton SJ, Colaiácovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yildiz O, Majumder S, Kramer B, Sekelsky JJ. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell. 2002;10:1503–1509. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I, A possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao Y, Desai SD, Liu LF. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J Biol Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 46.Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, Hannon GJ, Lowe SW, Hemann MT. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci USA. 2008;105:9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labib K, Kearsey SE, Diffley JF. MCM2–7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol Biol Cell. 2001;12:3658–3667. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anantha RW, Sokolova E, Borowiec JA. RPA phosphorylation facilitates mitotic exit in response to mitotic DNA damage. Proc Natl Acad Sci USA. 2008;105:12903–12908. doi: 10.1073/pnas.0803001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurençon A, Purdy A, Sekelsky J, Hawley RS, Su TT. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics. 2003;164:589–601. doi: 10.1093/genetics/164.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang F, Wang Y. DNA damage checkpoints inhibit mitotic exit by two different mechanisms. Mol Cell Biol. 2007;27:5067–5078. doi: 10.1128/MCB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsao CC, Geisen C, Abraham RT. Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J. 2004;23:4660–4669. doi: 10.1038/sj.emboj.7600463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Elledge SJ. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park SJ, Lee YJ, Beck BD, Lee SH. A positive involvement of RecQL4 in UV-induced S-phase arrest. DNA Cell Biol. 2006;25:696–703. doi: 10.1089/dna.2006.25.696. [DOI] [PubMed] [Google Scholar]
- 55.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 56.Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 2010;24:183–194. doi: 10.1101/gad.1860310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaklevic BR, Su TT. Relative contribution of DNA repair, cell cycle checkpoints, and cell death to survival after DNA damage in Drosophila larvae. Curr Biol. 2004;14:23–32. doi: 10.1016/j.cub.2003.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klovstad M, Abdu U, Schüpbach T. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet. 2008;4:e31. doi: 10.1371/journal.pgen.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol. 2006;26:7258–7268. doi: 10.1128/MCB.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailly AP, Freeman A, Hall J, Déclais AC, Alpi A, Lilley DM, Ahmed S, Gartner A. The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair. PLoS Genet. 2010;6:e1001025. doi: 10.1371/journal.pgen.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohouo PY, Bastos de Oliveira FM, Almeida BS, Smolka MB. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell. 2010;39:300–306. doi: 10.1016/j.molcel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 63.Ramadan N, Flockhart I, Booker M, Perrimon N, Mathey-Prevot B. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat Protoc. 2007;2:2245–2264. doi: 10.1038/nprot.2007.250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.