Abstract
Existing transgenic RNAi resources in Drosophila melanogaster based on long double-stranded hairpin RNAs are powerful tools for functional studies, but they are ineffective in gene knockdown during oogenesis, an important model system for the study of many biological questions. We show that shRNAs, modeled on an endogenous microRNA, are extremely effective at silencing gene expression during oogenesis. We also describe our progress toward building a genome-wide shRNA resource.
Current Drosophila transgenic RNAi resources use long hairpins as silencing triggers1. However, for reasons unknown, long hairpins are ineffective for gene silencing in the female germline, a conclusion that we reached after extensive testing of various construct designs (Supplementary Fig. 1 and Supplementary Note 1). In Drosophila, RNAi can be triggered via distinct routes, each of which generates small silencing RNAs via discrete processing and loading machineries2. In particular, artificial microRNAs, referred to as shRNAs, have been shown to trigger effective silencing in somatic cells3 and in the female germline in one case4. As shRNAs have not been used extensively and compared to long hairpins for their efficacies, we systematically evaluated their use as a transgenic trigger of RNAi in Drosophila.
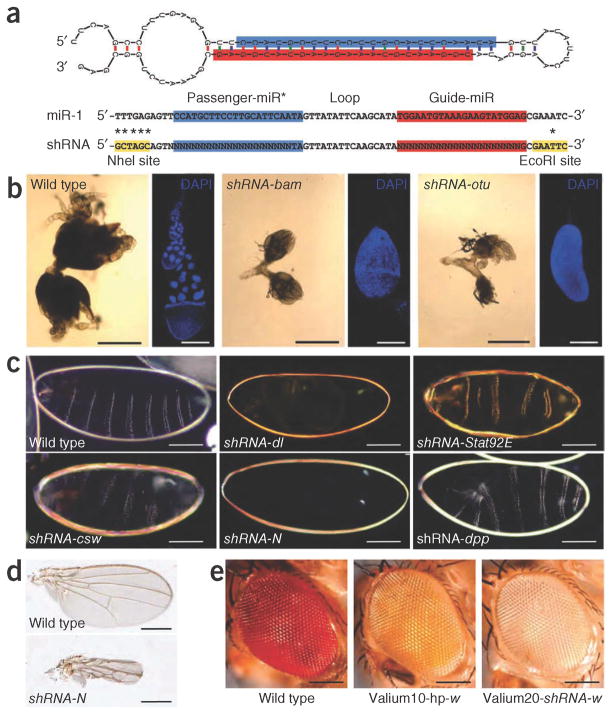
We constructed Valium20, a vector that combines the optimized expression features of the previously reported Valium10 for somatic RNAi5 with a modified scaffold of the microRNA miR-1 (Fig. 1a and Supplementary Fig. 2). Unique cloning sites allow the generation of shRNAs that accommodate the desired sequences, leading to a hairpin with perfect duplex structure, which favors shRNA loading into AGO2, the principal effector of RNAi in flies (Fig. 1a). To test the effectiveness of Valium20, we generated several fly lines containing shRNA constructs that target genes associated with either distinctive germline or maternal effect phenotypes. We induced shRNA expression specifically in the germline with MTD-Gal4 (ref. 6), a line that carries three Gal4 drivers expressed at various stages during oogenesis (Online Methods). For all examined lines, we recovered the expected oogenesis and maternal effect mutant phenotypes (Supplementary Note 1), indicating that shRNAs triggered potent gene knockdown during oogenesis (Fig. 1b,c). To determine whether maternal expression of shRNAs can also block expression of zygotically expressed genes, we generated shRNAs to a few zygotic genes that result in embryonic lethality when mutated. In all cases, we observed the expected phenotypes as shown for decapentaplegic (dpp; Fig. 1c). Finally, we tested the effectiveness of shRNAs expressed from Valium20 in somatic tissues. In general, the obtained phenotypes were stronger than those obtained with the long-hairpin–based vector Valium10 and resembled genetic null mutations for the respective genes (Fig. 1d,e and Supplementary Fig. 3).
Figure 1.
Design of shRNA constructs and phenotypes of shRNA-mediated gene silencing. (a) Structure of the Drosophila miR-1 and shRNA hairpins (miR-1 nucleotides replaced by the sequence of interest are indicated by N). (b) Phase contrast images showing ovary phenotypes associated with knockdown of bag-of-marbles (shRNA to bam, labeled shRNA-bam) and ovarian tumor (shRNA-otu) in MTD-Gal4/UAS-shRNA females (using Valium20). DAPI images show single tumorous egg chamber and wild-type egg chamber. Scale bars, 500 μm (phase contrast) and 200 μm (DAPI). (c) Dark field images of the cuticle of wild-type embryo and embryos derived from MTD-Gal4/UAS-shRNA females. Scale bars, 100 μm. (d) Knockdown of Notch in the wing using C96-Gal4/UAS-shRNA-N. Scale bars, 400 μm. (e) Knockdown of white using GMR-Gal4. In the labels, hp stands for hairpin. Scale bars, 100 μm.
Whereas shRNAs expressed from Valium20 generated effective knockdown phenotypes in germline and soma, the phenotypic penetrance in the germline was influenced by temperature and maternal age, indicating room for improvement (Supplementary Note 1). We therefore generated Valium22 (Supplementary Fig. 2) based on the UASp vector7, which is optimized for transgene expression in the female germline. Indeed, Valium22- mediated knockdowns in the germline were overall stronger than those generated using Valium20. We note, however, that Valium22 did not allow robust transgene expression in the soma, leading to incomplete somatic knockdowns (data not shown).
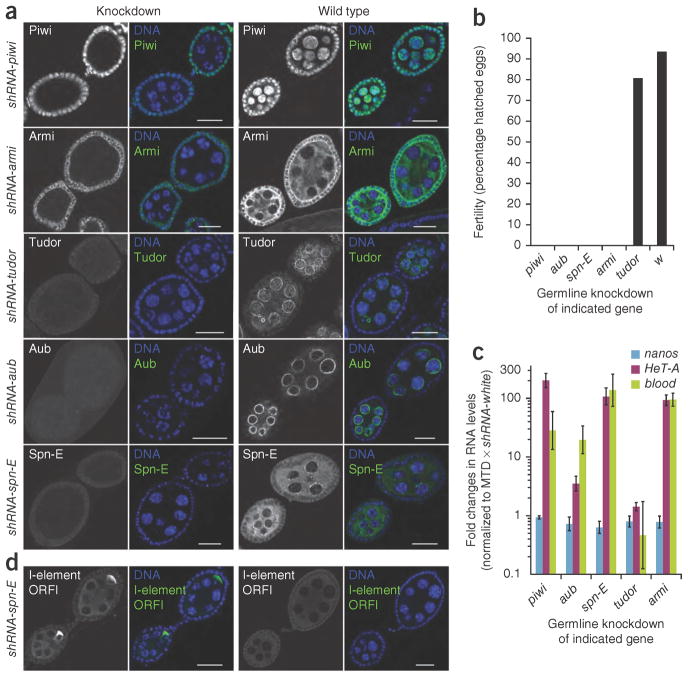
We chose the ovarian Piwi-interacting RNA (piRNA) pathway as a system to compare the efficacies and specificities of Valium20 and Valium22. Both somatic and germline cells of the Drosophila ovary produce piRNAs to silence transposable elements, but the pathway architecture differs in both cell types8. We generated multiple shRNA lines targeting proteins with a role in the piRNA pathway. Consistent with the strong knockdown observed for each target (Fig. 2a), RNAi phenotypes generated using the maternal MTD-Gal4 line and Valium22 were highly reminiscent of each published null mutant. Depletion of the proteins Piwi, Aub, Spn-E or Armi resulted in complete sterility (Fig. 2b), and we observed strong derepression of three transposable elements known to be targets of the germline piRNA pathway by quantitative reverse transcription–PCR (RT-PCR) or by antibody staining (Fig. 2c,d). Loss-of-function mutations in tudor are known to have no impact on transposon silencing in the germline or on fertility, but these mutants are defective in Aub localization to the posterior end of the growing oocyte. This leads to a failure in the specification of primordial germ cells and results in a ‘grandchild-less’ phenotype. We observed all of these characteristics in the shRNA-tud (shRNA to the tudor gene) transgenic flies (data not shown). Typically, we observed similar phenotypes when we tested the same shRNAs with Valium20. However, Valium20-mediated knockdown of Piwi or Armi led to more severe phenotypes. This is in full agreement with data for their respective genetic null mutants, as it has been shown that the function of Piwi and Armi in the somatic support cells of the ovary is critical for germline development and therefore ovarian morphology. This indicates leaky shRNA expression from Valium20 transgenes in somatic ovarian cells (Supplementary Table 1, Supplementary Fig. 4 and Supplementary Note 1). We suspect that a temperature-sensitive element present in the hsp70 minimal promoter of Valium20 causes low-level expression that, in combination with basal Gal4 levels, leads to shRNA expression sufficient for gene knockdowns in the soma. Altogether, our results indicate that Valium22 is not only more effective for knockdowns in the germline but is also superior in terms of tissue specificity as the effects appear to be strictly restricted to the female germline.
Figure 2.
Analysis of the piRNA pathway during oogenesis. (a) Immunofluorescence staining of early egg chambers showing depletion of the indicated piRNA pathway components using specific antibodies (green) upon shRNA expression via MTD-Gal4 (using Valium22). DNA was visualized with DAPI (blue). Black and white images are of the antibody staining only. Scale bars, 20 μm. (b) Fertility rates of females in which the indicated genes were knocked down with shRNAs in the germline via MTD-Gal4 (using Valium22). For each knockdown 300–500 eggs were counted. (c) Fold changes in steady-state RNA levels of the transposable elements HeT-A and blood in comparison to the germline-specific nanos transcript upon knockdown of the indicated genes via shRNAs. The data were compared to a control sample in which the white gene was knocked down (rp49 transcript levels were used for normalization). Data are averages of three independent biological replicates; error bars, s.d. (d) Immunofluorescence staining of early egg chambers with an antibody to the I-element ORF1p. Left two images are of flies expressing shRNA-spn-E with MTD-Gal4 (using Valium22); right two images are of wild-type flies. DNA was visualized with DAPI (blue).
It was important to understand the biogenesis requirements, processing accuracy and loading characteristics of shRNAs, which are designed to take advantage of different aspects of the mostly distinct fly miRNA and siRNA pathways to favor loading into AGO2, the major RNAi machinery in Drosophila. Our analysis indicated that shRNAs are processed by the sequential action of Drosha-Pasha and Dicer-1-LoqsPB complexes followed by loading into AGO1 and AGO2 proteins (Supplementary Figs. 5a,b and 6 and Supplementary Note 1). Loading into AGO2 required Dcr-2 and presumably also R2D2 and perfect base-pairing at positions 9 and 10 of the siRNA, as has been shown for endogenous siRNAs. Immunopurification experiments indicated that shRNAs are efficiently loaded into AGO2 as intended but that a fraction is also associated with AGO1. Small RNA sequencing efforts from cultured Drosophila cells transfected with three different Valium20 constructs showed that shRNAs are accurately processed at the intended 5′ ends. Furthermore, the amount of mature shRNA strands accumulated at levels comparable to those of the most abundant cellular microRNA (miR) strands and exceed miR complementary strand (miR*)-passenger strand strand levels several fold as predicted from the differences in thermodynamic stability of guide and passenger strand 5′ ends.
RNAi approaches can suffer from unspecific targeting of genes exhibiting sufficient sequence complementarity to the experimental siRNA (referred to as off-target effects)9. In comparison to long-hairpin constructs, shRNA constructs are expected to be advantageous as they give rise to only two siRNA species, the guide and passenger strands (Supplementary Fig. 6 and Supplementary Note 1). We minimized off-target effects caused by extended complementarity by filtering out shRNAs whose guide or passenger strands have complementary matches of 16 nucleotides or more to the fly transcriptome. As shRNAs are partially loaded into AGO1, they could also produce off-target effects through fortuitous recognition of mRNAs via the ‘seed’ region leading to miRNA-like repression10. To address the problem of off-target effects experimentally, we generated several shRNA lines targeting genes that are not required for viability. Only one line out of eight tested was associated with unexpected lethality (Supplementary Table 2). This is similar to what has been observed with long hairpins5,11. Thus, off-target effects with shRNAs are still a matter of concern, and we suggest verification of obtained phenotypes using independent shRNA constructs to the same gene.
Encouraged by the remarkable silencing potency of shRNAs, we started to generate a large-scale community resource for transgenic RNAi in Drosophila (http://www.flyrnai.org/TRiP-HOME.html). As of March 2011, we have constructed over 2,900 fly stocks (an updated list is available at the Transgenic RNAi Project (TRiP) homepage), and they are available from the Bloomington Drosophila Stock Center. We aim to construct shRNAs to all 14,208 annotated Drosophila protein-coding genes (genome release 5). Toward this end, we predicted shRNA sequences for all genes using designer of small interfering RNA (DSIR)12, an algorithm trained on effective siRNAs (http://biodev.extra.cea.fr/dsir/dsir.html). DSIR has proven quite reliable for the prediction of shRNAs for effective knockdown in transgenic flies, and the vast majority of the shRNAs described in this paper were designed using the DSIR algorithm. We synthesized 83,256 unique shRNA oligonucleotides in situ on four custom glass-slide microarrays13. We amplified these as pools, and inserted them into Valium20 and Valium22. We analyzed ~160,000 individual clones per vector, and identified accurate clones through either conventional sequencing or a two-step process involving DNA Sudoku14 compression followed by Illumina sequencing. We anticipate that at least 8,000 constructs per year will become available from the TRiP for distribution to the community.
ONLINE METHODS
Drosophila strains
The maternal triple driver (MTD)-Gal4 stock6 was a gift from L. Cooley (Yale University). The stock contained homozygous insertions of three Gal4 constructs, which together provide robust germline and maternal Gal4 expression. The genotype was P{COG-Gal4:VP16}; P{Gal4-nos.NGT}40; P{nos-Gal4-VP16} (Bloomington stock 31777). P{COG-Gal4:VP16}7 contained a promoter from the otu gene and the 3′ untranslated region (UTR) from the K10 gene. Gal4:VP16 expression from this transgene was weak or absent in the germarium and robust beginning in stage-1 egg chambers. P{nos-Gal4-VP16} contained both the promoter and 3′ UTR from the nanos gene15 and was expressed throughout the germarium and in all stages of egg chambers, with lower expression in young egg chambers (~stages 2–6)7. P{Gal4-nos.NGT}40 contained the nanos promoter and αTub84E 3′ UTR16, and was made for maternal loading of Gal4 to drive expression during embryogenesis.
GMR-Gal4 and C96-Gal4 were used to drive expression in the eye and wing, respectively, as described previously5. Their descriptions are available from FlyBase (http://flybase.org/). Details on the full genotype of all the lines used in this study are available on the TRiP website (http://www.flyrnai.org/TRiP-HOME.html).
Phenotypic analyses
For DAPI staining, ovaries were dissected in PBS and fixed in 4% electron microscopy (EM)-grade para-formaldehyde (Electron Microscopy Sciences) diluted in PBS for 30 min. Ovaries were counterstained with DAPI (Invitrogen) for 10 min. Embryonic cuticles and wings were prepared as described previously17,18. For immunofluorescence, ovaries were dissected from 3–5-day-old flies into ice-cold PBS and subsequently fixed in 4% formaldehyde (Thermo Scientific) containing 0.15% Triton X 100 (Sigma-Aldrich), diluted in PBS, for 25 min. After three rinses with PBT (PBS with 0.3% Triton X 100) ovaries were blocked in BBX (PBS containing 0.3% Triton X 100 and 0.1% BSA) for 30 min at room temperature (20–22 °C). Ovaries were incubated with primary antibodies over night at 4 °C diluted in BBX (antibodies to Piwi, Aub and Ago3, 1:500; antibodies to Armi and I element, 1:1,000; antibodies to Tudor, 1:10; antibodies to Spn-E, 1:50). After four PBT washes secondary antibodies were incubated 5 h at room temperature diluted in BBX (1:500; Molecular Probes). Ovaries were stained with DAPI for 10 min in the second of four PBT washes. Antibodies used were: antibody to Piwi, antibody to Aub and antibody to AGO3 (ref. 19); antibody to Tudor, antibody to Spn-E20; antibody to Armi21 and antibody to I element (gift from D. Finnegan; University of Edinburgh). For the sterility test, ten 3–5-day-old female flies were pre-mated with wild-type males overnight in small cages on apple juice plates with yeast paste. Apple juice plate was changed without anesthetizing flies. After 18 h at 25 °C, the flies were removed and the number of laid eggs was counted (typically ~200 eggs). Forty-eight hours later hatched and non-hatched eggs were determined.
Additional information on the phenotypic analyses of RNAi reagents is available in Supplementary Figures 7 and 8 as well as in Supplementary Table 3.
Vector construction
For descriptions of vector construction, see Supplementary Note 2.
β-elimination
The chemical structure of the 3′ termini of small RNAs was analyzed as described previously22. In brief, RNA from immunoprecipitates or 25 μg of total RNA from S2 cells treated with the indicated dsRNAs (17.5 μl total volume for each sample) was incubated at room temperature for 30 min with 5 μl 5× borate buffer (148 mM borax and 148 mM boric acid; pH 8.6) supplemented with 3.125 μl freshly prepared 200 mM NaIO4. We added 5 μl of 50% glycerol to quench nonreacted sodium periodate by incubating for an additional 15 min at room temperature. Samples were then vacuum-dried and dissolved in 60 μl 1× borax buffer (30 mM borax, 30 mM boric acid and 50 mM NaOH; pH 9.5). β-elimination was carried out by incubation for 2 h at 45 °C. RNAs were ethanol-precipitated and resolved in 1× gel loading buffer.
Northern blotting
Northern blotting was carried out as described previously23,24. In brief, total RNAs from knockdown cells were isolated using TRIzol (Invitrogen). We separated 30 μg total RNAs from cultured cells (with or without β-elimination) or RNAs from immunoprecipitations on 15% denaturing poly-acrylamide gels and transferred to Hybond-N+ membranes (Amersham Biosciences) in 1× TBE buffer. Small RNAs were UV-light cross-linked to the membrane and prehybridized in ULTRAhyb-Oligo buffer (Ambion) for 1 h. DNA probes complementary to the indicated strands were 5′ radio-labeled and added to the hybridization buffer (hybridization for 6 h at 30 °C). Membranes were washed 4 times in 1× SSC with 0.1% SDS at 30 °C and exposed to PhosphorImager screens (GE Healthcare) for 12–48 h. Membranes were stripped by heating in 0.2× SSC containing 0.1% SDS in a microwave twice. Sequences of the oligonucleotide probes are listed in Supplementary Note 3.
Immunoprecipitation
Cell extracts were prepared, evenly split and immunoprecipitated using antibodies to AGO1 (Abcam) or the Flag epitope (Sigma), respectively, as previously described23. RNAs were recovered from the immunoprecipitated samples using TRIzol and used for northern blotting.
Transposon qPCR analysis
Total RNA was extracted from ovaries of 3–5-day-old flies using TRIzol. cDNA was prepared with random primers. qPCR was performed using Maxima SYBR Green/ROX qPCR Master mix (Fermentas). Calculation of steady-state RNA levels was calculated applying the 2−ΔΔCt method25. Rp49 was used for normalization of all samples, and fold enrichments were calculated in comparison to an shRNA knockdown targeting the white gene. Fold changes in steady-state transcript levels and s.d. were calculated from three biological replicates. Primer sequences are available in Supplementary Note 3.
Small RNA libraries
Small RNAs were cloned as described previously19. For this study, the following small RNA libraries from total RNAs were prepared: 19-nucleotide (nt) to 24-nt from S2 cells transfected with shRNA to dlg1 (shRNA-dlg1); 19-nt to 24-nt from S2 cells transfected with shRNA-N; and 19-nt to 24-nt from S2 cells transfected with shRNA-dpp. For each construct, ~4 × 106 S2-NP cells were transfected with 2 μg of Valium20-shRNA construct and 1 μg of pMT-Gal4 plasmid. ShRNA expression in cells was induced by adding 500 μM CuSO4 2 d after transfection. Total RNA was isolated using TRIzol 24 h after induction. Libraries were sequenced in-house using the Illumina GA-II sequencing platform.
Bioinformatic analysis of small RNA libraries
The analysis of small RNA libraries was performed as previously described26. Illumina reads were stripped of the 3′ linker and collapsed, and the resulting small RNA sequences were matched without mismatches to the Drosophila release 5 genome and to the genomes of Drosophila C virus, Flock house virus and Cricket paralysis virus with up to three mismatches. Only reads that met these conditions were analyzed further. For annotations we used a combination of University of California Santa Cruz genome browser, miRBase and Flybase tracks for protein-coding genes, repeats or transposons, noncoding RNAs and microRNAs as well as custom tracks (for shRNAs, synthetic markers, endo-siRNAs from structured loci, miR and miR* strands) with different priorities (annotation priority list is available upon request). For comparison of small RNA counts between libraries, reads were normalized to the same total number after bioinformatic removal of sequences matching to synthetic cloning markers or assumed degradation products of abundant cellular RNAs (rRNAs, snoRNAs and tRNAs). Heatmaps were computed by plotting the abundance and ratio of individual miR, miR* and shRNA strands in each library.
Construction of the shRNA library
An shRNA library representing 83,256 unique synthetic hairpins was synthesized on four custom 22K Agilent microarrays13. The library covered all 14,208 annotated genes (excluding small RNA and noncoding RNA genes) of the Drosophila release 5 genome with up to six shRNAs per gene (14,138 genes were covered by six hairpins, and 14,147 genes were covered by five hairpins). Hairpin constructs were based on the miR-1 backbone and essentially resembled those described above with perfect complementarity between guide and passenger strands. Additional sequence was attached on both ends for PCR amplification. In addition, to eliminate off-target effects only shRNAs that lacked sequence complementarity to annotated microRNA ‘seed’ sequences were considered. DNA pools from microarray chips were amplified13 and cloned into Valium20 and Valium22 destination vectors. Plasmid DNA was transformed, clones were picked (160,000 individual clones per destination vector), and resulting transformants were multiplexed using DNA Sudoku14 at Open Biosystems. Pools were barcoded via PCR, and amplicons were sequenced in-house using the Illumina GA-II sequencing platform. Positive clones were picked into 96-well plates and Sanger sequencing was carried out to validate correct shRNA sequences. Once shRNA clones in Valium20 and Valium22 are available, they will be openly available to the Drosophila community.
Supplementary Material
Acknowledgments
The design and construction of the first shRNAs were supported in part by the Janelia Farm Visitor Program. We thank G. Rubin, C. Zuker and T. Laverty for their interest and support; R. Hardy and C. Zuker for the data presented in Supplementary Table 2; B. Haley for helpful discussion on shRNAs; L. Cooley (Yale University) for the gift of the MTD-Gal4 line; and Z. Xuan for help with library design. S. Zusman and M. Tworoger of Genetic Services, Inc. generated the transgenic lines. R.Z. is supported by the Leukemia and Lymphoma Society. B.C. is supported by a PhD fellowship from the Boehringer Ingelheim Fonds. This work was supported by two US National Institute of General Medical Sciences R01 grants (GM067761 and GM084947) to N.P., an EU FP7 European Research Council starting grant to J.B. and contributions from the US National Institute of Neurological Disorders and Stroke.
Footnotes
Accession codes. Gene Expression Omnibus: GSE27039 (small RNA sequences).
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
J.-Q.N., R.Z. and B.C. carried out major experiments; L.-P.L., L.H., D.Y.-Z., H.-S.S., R.B., M.B. and L.A.P. produced the TRiP lines; P.K. performed the luciferase experiments in ovaries; D.H. and J.B. analyzed the piRNA pathway during oogenesis; and G.J.H. and N.P. supervised the project. R.Z., B.C., J.-Q.N., D.H., J.B., G.J.H. and N.P. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Perrimon N, Ni JQ, Perkins L. Cold Spring Harb Perspect Biol. 2010;2:a003640. doi: 10.1101/cshperspect.a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czech B, Hannon GJ. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haley B, Hendrix D, Trang V, Levine M. Dev Biol. 2008;321:482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CH, et al. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 5.Ni JQ, et al. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrella LN, Smith-Leiker T, Cooley L. Development. 2007;134:703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- 7.Rorth P. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 8.Malone CD, Hannon GJ. Cold Spring Harb Symp Quant Biol. 2009;74:225–234. doi: 10.1101/sqb.2009.74.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkarni MM, et al. Nat Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 10.Birmingham A, et al. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 11.Dietzl G, et al. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 12.Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. BMC Bioinformatics. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleary MA, et al. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 14.Erlich Y, et al. Genome Res. 2009;19:1243–1253. doi: 10.1101/gr.092957.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Doren M, Williamson AL, Lehmann R. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 16.Tracey WD, Jr, et al. Genetics. 2000;154:273–284. doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni JQ, et al. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrimon N, Engstrom L, Mahowald AP. Genetics. 1989;121:333–352. doi: 10.1093/genetics/121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennecke J, et al. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 20.Nishida KM, et al. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 22.Vagin VV, et al. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 23.Czech B, et al. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou R, et al. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Czech B, et al. Mol Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.