Abstract
The benefits of sexual reproduction that outweigh its costs have long puzzled biologists. Increased genetic diversity generated by new allelic combinations, as enhanced by recombination during meiosis, is considered to be a primary benefit of sex. Sex-determining systems have evolved independently on numerous occasions. One of the most familiar is the use of sex chromosomes in vertebrates. Other eukaryotic groups also use sex chromosomes or smaller sex-determining regions within their chromosomes, such as the mating type loci in the fungi. In these organisms, sexual reproduction and its associated meiotic recombination is controlled by regions of the genome that are themselves blocked in recombination. Non-recombining DNA that is essential for recombination presents a paradox. One hypothesis is that sex-determination requires or leads to highly diverse alleles, establishing this block in recombination. A second hypothesis to account for the common occurrence of these types of sex-determining systems is that they combine mechanisms for recombination suppression and reproductive isolation, thereby promoting the evolution of new species. The fungal kingdom represents the ideal eukaryotic lineage to elucidate the functions of non-recombining regions in sex-determination and speciation.
Keywords: Ascomycota, Basidiomycota, chromosomal speciation, homothallism, heterothallism, MAT, Mucormycotina, Mycota, recombination, X/Y
1. A sex paradox: DNA regions that promote recombination do not recombine
The widespread occurrence of sexual reproduction has long served as a mystery for evolutionary biologists. In particular, the expenses associated with sex versus asexual reproduction include a numerical disadvantage in producing gametes, and the possibility that meiotic recombination arranges combinations of alleles that are deleterious or produce a less fit organism. Furthermore, the process of sexual reproduction increases the risk of contracting or spreading infectious diseases. Despite these disadvantages, the prevalence of sex in many eukaryotes indicates that during natural selection the benefits in generating genetic diversity have outweighed the costs.
The differentiation of organisms into separate sexes or mating types is mediated by multiple mechanisms. One that evolved on independent occasions is the use of sex-determining chromosomes or regions (loci) within chromosomes. This is the mechanism with which people are most familiar because it occurs in our own species. Sex chromosomes and mating type loci share a common feature: they originated from normal chromosomes or chromosomal regions and now exhibit high sequence divergence, leading to or caused by blocked recombination during meiosis (Fraser and Heitman, 2005; Marshall Graves, 2006). The trend, while obviously not universal amongst eukaryotes, is striking enough that the DNA differences between sex-determining regions can facilitate their identification. This is illustrated by the recent discovery of the sex-determining mat region in the amoeba Dictyostelium discoideum that used DNA hybridization and bioinformatic analysis to identify highly variable regions within the genome (Bloomfield et al., 2010).
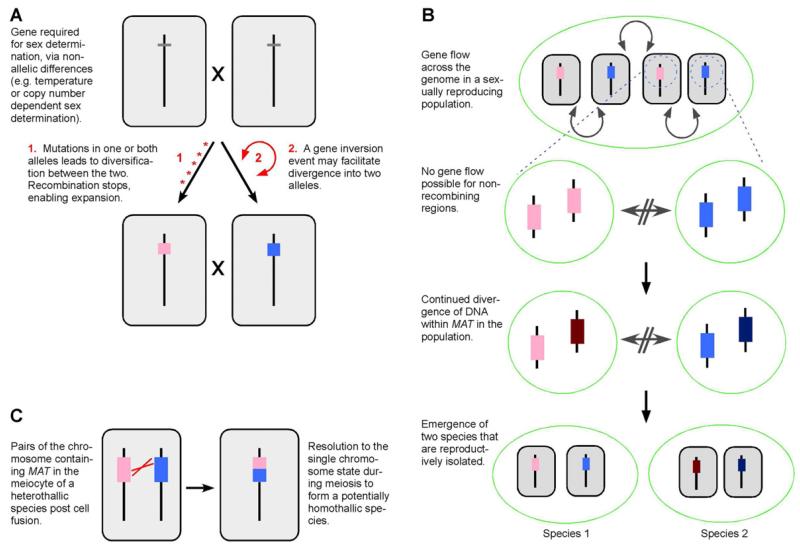
Sex chromosomes and mating type loci represent a paradox from the standpoint of natural selection acting at the level of an individual piece of DNA. Why is it that systems exist that are essential to promote the process of recombination wherein the DNA responsible is unable itself to benefit? One hypothesis is that allele-based sex-determination requires highly diverse alleles of genes. The evolution of such regions would gradually lead to suppression of recombination (Figure 1A). Alternatively, a chromosomal inversion that includes a gene required for sex-determination would prevent recombination and the promote divergence into sex-specific alleles (Figure 1A). A second hypothesis to explain the prevalence of such systems is a possible role in promoting speciation. As such, these “unselfish” genetic regions may contribute to the diversity of eukaryotic species. Interest in the evolution of sex-determination and speciation has focused on larger eukaryotes, especially using Drosophila species. However, insights into this paradox may be best made in members of the fungal kingdom due to the structure of their mating type determining regions and their amenability for experimental biology. This Opinion article outlines the issue of non-recombining DNA and its possible role in speciation, using the fungi as a focal point.
Fig. 1.
A central role for blocking recombination in sex-determination and speciation. A. Conversion of non-allelic sex-determination to allelic determination, through divergence by (1) mutations (* marks) or (2) a chromosomal inversion event (arrows). B. Gradual divergence through mutations leads to sub-populations unable to mate with the original population. C. When rare illegitimate recombination does occur within the MAT locus of heterothallic species, this can lead to the formation of new species that are homothallic. One of several possible recombination and chromosomal rearrangements is illustrated.
2. Sex-determination in fungi is mediated by mating type (MAT) loci: miniature versions of sex chromosomes
Cell type identity in fungi is conferred by mating type loci (MAT), usually small regions of the genome that encode transcription factors and/or pheromones and pheromone receptors (Heitman et al., 2007; Lee et al., 2010). The alleles for each mating type have been termed “idiomorphs” because they lack sequence similarity (Metzenberg and Glass, 1990), although the alleles most likely evolved from a single genetic locus that stopped recombining and then diverged. Mating type specificity in the fungal kingdom is thus controlled by non-recombining regions of their genomes (Lee et al., 2010). In most cases the fungal MAT loci are smaller (<10 kb) relative to sex chromosomes; however, in some species MAT has expanded to larger regions (0.5 Mb) or possibly whole chromosomes (Coelho et al., 2010; Hood, 2002; Lee et al., 1999; Menkis et al., 2008). MAT regions are identical to those in other eukaryotes in promoting recombination without themselves receiving the benefit of recombination.
When MAT evolved in the fungi is an open question. Mating in fungi pre-dates the emergence of many recognizable eukaryotic lineages used to study sex-determination. Despite their soft material and as a consequence very limited record, fossils of fungal mating structures appear identical to those structures made by current day ascomycete and basidiomycete fungi: some of those fossils are 400 million years old (Krings et al., 2011; Taylor et al., 1999). Molecular clock dates for the origin of MAT loci can be inferred by synteny analysis of where MAT resides in the genome of related species. Conserved genes flank the MAT loci in each of the Ascomycota, Basidiomycota and Mucormycotina lineages (Butler et al., 2004; Gryganskyi et al., 2010; James et al., 2004; Lee et al., 2008). This conservation is best-elucidated in the ascomycetes due to the extensive analysis of MAT in these species. Conservation of the APN2 and SLA2 genes adjacent to MAT in members of the Pezizomycotina and Saccharomycotina (Butler et al., 2004; Martin et al., 2010) dates the origin of the ascomycete MAT locus to prior to 400 million years ago, using the most conservative estimate (Berbee and Taylor, 2010). Further, it has been proposed that the presence of an APN2 homolog near the candidate MAT loci of the Microsporidia would indicate a single evolutionary event giving rise to MAT in at least three major lineages of fungi (Martin et al., 2010). This hypothesis requires further investigation by the identification of MAT in other basal fungal lineages. In summary, there is strong evidence that specific MAT loci are ancient and have been maintained, with modifications, over long time frames within fungi.
Although of ancient origin, the alleles of MAT loci house different genes between fungal groups. All known species have a locus that encodes a transcription factor (homeodomain or HMG/α-box type): these genes are essential for sex because strains lose the ability to mate if mutated in most instances (Lee et al., 2010). Transcription factors also control sex-determination in other eukaryotes. For example, human sex chromosomes encode the HMG-domain proteins SRY or SOX3 that were once allelic (Charlesworth et al., 2005; Lahn and Page, 1999; Sutton et al., 2011). The closest parallel to the Eutheria mammals in the fungi are the Mucormycotina sex loci, which have HMG-domain proteins encoded by each allele (Gryganskyi et al., 2010; Idnurm et al., 2008; Lee et al., 2008). Most fungi have two mating types, analogous to the male and female sexes of vertebrates, although in a subset of basidiomycetes the alleles at MAT confer many different mating types.
3. Advantages and disadvantages in suppressing recombination in relation to sex-determination
Interest in the effects of blocked recombination has focused primarily on the evolutionary trajectory of animal and the rarer plant sex chromosomes (Bergero and Charlesworth, 2009; Marshall Graves, 2006). A noted effect of eliminating recombination is the accelerated accumulation of changes in those regions when compared with autosomal regions. For example, the human Y chromosome is estimated to have lost 95% of its original genes, leading to the proposed “extinction” of this chromosome within 14 million years (Aitken and Marshall Graves, 2002; Charlesworth and Charlesworth, 2000; Marshall Graves, 2006). A fungal example exhibiting evidence for rapid degeneration with non-recombining regions is the MAT locus of Neurospora tetrasperma (Whittle and Johannesson, 2011; Whittle et al., 2011b). Thus, sex-determining systems miss the benefits of recombination and are prone to rapid accumulation of potentially detrimental mutations (Rice, 1994).
On the other hand, blocking recombination can be beneficial to link genes encoding proteins with related functions. A human example is the placement of genes for sex-specific functions on the X and Y chromosomes, such as those for sperm production on Y. Among the fungi, the basidiomycete MAT loci provide equivalent examples (Casselton and Olesnicky, 1998). Some species have clusters of genes related to the same mating type in each allele, e.g. Cryptococcus neoformans and C. gattii in which the MATa or MATα specific transcription factors are adjacent to the associated a or α pheromone-encoding genes (Fraser et al., 2004). Alternatively, in the tetrapolar basidiomycetes, i.e. those with two loci controlling mating, blocking recombination enforces separation at one locus of the pheromone receptors from pheromone-producing genes and at a second locus the pairs of transcription factors that must form heterodimers to progress through the sexual cycle (Kämper et al., 1995). Suppression of recombination in MAT may be advantageous; however, such gene linkage is rarely observed beyond the basidiomycetes, while both theory and practical experiments predict a general deterioration of these DNA regions.
4. Can suppression of recombination promote speciation?
Reasons must exist why non-recombining DNA is so prevalent in sex-determining regions. One hypothesis is that this reflects a requirement for “extreme” allelic diversity to establish two different mating types or sexes, thus leading to regions of the genome that are different (Figure 1A). However, other mechanisms of sex-determination, e.g. temperature-dependence, do not require multiple alleles. Another hypothesis is that the block in recombination plays a role in speciation: the prevalence of non-recombining sex-determination systems in eukaryotes would be explained by those lineages that feature such elements becoming more species rich. This hypothesis is explored in more detail during the rest of this article. There are multiple ways to define a species, with these definitions overlapping the forces that generate species (Giraud et al., 2008; Kohn, 2005). The contributions of the underlying mechanisms promoting speciation remain subject to debate (Coyne and Orr, 1998). The topic that recombination suppression mediates speciation has been extensively reviewed (e.g. see Faria and Navarro, 2010; Jackson, 2011; Rieseberg, 2001). Incorporating genes required for sex into chromosomal regions without recombination would have the potential to enhance speciation by mutation, if such mutations altered fertility barriers (Figure 1B).
Blocking recombination is predicted to play a key role in the evolution of new species in the plant kingdom, especially in the angiosperms (Rieseberg, 2001; Soltis and Soltis, 2009). The formation of new allopolyploids includes an intermediate stage of an infertile hybrid, with the infertility attributed to divergence in DNA sequences that prevent proper alignment and recombination of homologous chromosomes. Genome duplication to generate pairs of chromosomes resolves infertility, and generates a species that is no longer able to produce viable progeny when crossed to the original parents due the formation of aneuploid gametes or progeny. Curiously, rapid genetic changes, including chromosomal rearrangements, often follow soon after the development of allopolyploids further restricting their ability to cross with the parental species (Soltis and Soltis, 2009). Thus, recombination blocks are implicated in speciation in one eukaryotic lineage.
But what of the smaller non-recombining parts of genomes that control sex-determination? Plant polyploids illustrate the link between blocked recombination and speciation, but in a manner independent of sex-type determination and on a larger scale that may include the entire genome of each progenitor species. Sex chromosomes and the MAT loci are among the most rapidly diverging regions within eukaryote genomes since the inability of these regions to undergo recombination prevents the removal of deleterious mutations or gene flow between each allele within the population (Figure 1B). Hallmarks of this are phylogenies of MAT-encoded proteins that are not required for mating, that form allele-dependent clades rather than species-dependent clades (Fraser et al., 2004). Many fungi have strains that are divided into one of two mating types or sexes [e.g. A and a; a and α; (+) and (−); MAT1-1 and MAT1-2]; however, this designation does not reflect the natural variation observed when sequencing mating type alleles. At some level of divergence one species with a1, α1, a2 and α2 alleles could become two separate species with a1, α1 and a2, α2 alleles (Figure 1B).
One challenge with a model of speciation led by mutations in MAT is that it requires multiple mutation events. Further, the model also depends upon the MAT locus components conferring reproductive isolation between species. In the simplest system, both a transcription factor and an interacting component such as another transcription factor or a promoter region should be mutated to provide sufficient specificity for successful mating between strains of the new species with each other but not with the progenitor species. Thus, modulating fertility by mutation would require at least two mutation events (Figure 1B). For the fungi, generation of large numbers of progeny from asexual reproduction provides a way of maintaining modified alleles within the population and the probability to achieve two-hit mutation events. The two MAT locus alleles of Botrytris cinerea appear to have originated from two (or more) mutation events from the MAT locus of a homothallic species (Amselem et al., 2011), demonstrating that multiple mutation events can occur within MAT loci to influence speciation.
While meiotic recombination is blocked across the MAT loci, it is worth consideration the possible outcomes of illegitimate recombination events between alleles of a MAT locus, and what role that might play in speciation. These events may happen by chance or be enhanced by factors within MAT itself, such as repetitive elements that are found in some MAT loci that could facilitate non-allelic homologous recombination. In many instances self-fertility (homothallism) requires MAT locus homologs found in the self-incompatible (heterothallic) relatives. This observation suggests these regions are derived from non-allelic homologous recombination or other chromosomal changes. A genome rearrangement that convert what was two alleles of a MAT locus in a heterothallic species into two separate loci or into a single allele at one locus (as illustrated in Figure 1C) is one process to account for the genetic basis of sex-determination in homothallic species (Paoletti et al., 2007; Yun et al., 1999). Thus, a consequence when recombination does occur in a normally non-recombining region is also the evolution of new species.
A final factor for consideration is other recombination-suppressed regions in the genomes of fungi and if they also promote speciation. Suppression of recombination is usually found around centromeres. Probably the only contribution of centromeres to speciation would be if they were incorporated into the MAT loci to generate new alleles (or participated in major chromosomal rearrangments). The unusually large MAT loci of Ustilago hordei and Neurospora tetrasperma may include the centromeres and account in part for their size (Lee et al., 1999; Menkis et al., 2008), while on the other hand the large MAT locus of Cryptococcus neoformans is unlinked to the centromere (Idnurm, 2010). A second genetic mediator, other than MAT loci, that controls the ability of some species of fungi to mate is vegetative incompatibility. Rare examples include pairs of adjacent genes and the suppression of recombination between them (Hall et al., 2010; Micali and Smith, 2006; Powell et al., 2007). It will be interesting to explore the evolution of these types of genetic regions as compared to single vegetative incompatibility genes.
5. Fungi are ideal organisms for experimental biology
Animals and plants present restrictions in understanding how natural selection gave rise to their current sex-determining systems. First, many of the “higher” eukaryotes are obligately sexual for reproduction. As such, selection no longer acts solely on sex-determination. Similar limitations are also present for fungi in which survival of the species depends on sexual reproduction in certain lineages (e.g. the Pucciniomycotina species that cause rust disease).
Further, fungal spores produced during sexual reproduction may serve roles in addition to generating new allelic combinations, such as for dispersal both through space and time or as infectious particles providing access to alternative environmental niches (Zeyl, 2009). Second, the developmental complexity of generating specific sex organs may lead to biased gene content on those chromosomes, by linking genes that promote organ development or gender-specificity onto such regions. Likewise, sex-biased evolutionary forces shape sex chromosome behavior in these organisms. This bias in gene content also occurs in some basidiomycete fungi, as already commented upon. Third, the predominance of diploidy during the life cycle of other eukaryotes is predicted to affect sex chromosome evolution, including skewing the distribution of sexually antagonist genes (Rice, 1984; Zhang et al., 2010). Analysis of the MAT loci of fungi provides a retrospective snap shot of their evolution and, importantly, the ability to test hypotheses about the evolution of non-recombining sex-determination without the issues of highly dimorphic sexes. As noted by others (e.g. see Kohn, 2005), reviews on speciation neglect the fungi despite the fungi being the most species-rich group of organisms on earth (Hawksworth, 2001) and the experimental power of fungi to reveal fundamental aspects about eukaryotic biology.
A few of the many advantages to use fungi for research include the convenience of small organism and genome size, shorter times to complete life cycles, large numbers of progeny, financial cost, and ethical considerations. One primary experimental benefit in working with fungi is species with a suite of different reproductive strategies that can be controlled by environmental conditions or genetically through mutations. The homothallic fungi have sex, but with themselves rather than opposite mating types, i.e. a homothallic strain is self-fertile. Manipulating the mating ability of homothallic strains offers a powerful experimental system to study the effects of outcrossing vs. inbreeding. Haploid, diploid and/or dikaryotic species or life stages are known, and many species reproduce both asexually and sexually, allowing the selective benefits under different reproductive and environmental conditions to be measured (e.g. Goddard et al., 2005; Schoustra et al., 2010; Zeyl and Bell, 1997), something rarely possible in plants or animals.
If speciation is promoted through non-recombining sex-determining parts of genomes, several predictions can be made. One prediction is that reproductive isolation in fungi must be governed by mating type locus components. At present, investigating the role of MAT locus components in fungal speciation is little explored beyond a few groups of species (e.g. see Lu et al., 2011; Yun et al., 1999). A second prediction is that in comparing related lineages, heterothallic species would be more numerous than homothallic species. However, other reasons may account for such an observation, including the disadvantage of inbreeding that is more prevalent than outcrossing in homothallic fungi (Whittle et al., 2011a). These predictions can be explored in future studies. In addition, many research directions for fungi could test the role of blocked recombination in evolution. Three directions are described briefly in the following paragraphs.
The first approach is to elucidate and characterize the sex-determining regions of many fungal species. Those species should represent phylogenetic coverage to encompass the evolutionary diversity within the kingdom, as well as a subset of closely-related lineages. Fungi usually have kilobase MAT loci rather than megabase sex chromosomes of animals. Smaller size provides an opportunity to sequence MAT loci with far greater ease, from more species, and from more strains when compared to animal and plant sex chromosomes with their well-established challenges of size and repetitive nature (Skaletsky et al., 2003). This approach will reveal the trajectory of MAT evolution in extant fungi.
Second is to assess the role of sex-determining regions in speciation based on reproductive isolation. One experiment would be to exchange MAT locus components from different species and examine the fertility of these strains crossed amongst each other and in combinations with the progenitor parents. Swapping of MAT components between species and alleles already suggests that such an experimental strategy is feasible (e.g. Lu et al., 2011; Stanton et al., 2010)]. This approach would test what contribution the MAT locus has in reproductive isolation between species. In particular, if this contribution is minimal then it is necessary to consider other reasons for the prevalence of non-recombining systems in sex-determination.
The third direction is to remake sex chromosomes or MAT loci. Such experiments have been performed in Drosophila (Rice, 1994). The model eukaryote Saccharomyces cerevisiae would be well-suited since it offers the opportunity to use haploid and diploid strains, mutant collections, the ease of constructing yeast artificial chromosomes, and the ability to study selection over time (Delneri et al., 2003; Goddard et al., 2005; Zeyl and Bell, 1997). Experiments with a well-designed set of strains carried over many generations would enable measuring changes in fitness, and its underlying basis by DNA sequence analysis, in a range of genomic and genetic contexts.
6. Concluding comments
One of the unique and yet most perplexing questions in eukaryote evolution is why sexual reproduction, which leads to meiotic recombination, requires DNA regions that lack recombination. These “unselfish” genetic elements attract interest to understand their evolution in diverse species. Multiple or lineage-specific reasons may explain this trend. Here the hypothesis is put forward that coupling non-recombining regions with genes that control sexual reproduction could account for species richness. The underlying paradox and hypothesis is challenging to explore experimentally in animals and plants, but suited for study in the fungi which use mating type loci as their sex-determining genetic systems. Insights from the fungi can be applied to other organisms to establish which factors are common or different during the evolution of groups of eukaryotes. The continuing advances in genomics and experimental techniques in all eukaryotes provides the exciting promise that we will be able to understand this paradox underlying sex-determination, and define its relative contribution to the diversity of species on earth.
Sex-determination is controlled in many eukaryotes by DNA that does not undergo recombination.
A primary advantage of sex is that it facilitates the process of recombination.
Non-recombining regions that promote recombination are a paradox for selection.
Several hypotheses may account for the occurrence of sex chromosomes or MAT loci.
Blocking recombination with sex chromosomes or MAT loci may enhance speciation.
Acknowledgements
I thank Hanna Johannesson, B. Gillian Turgeon and Jianping Xu for comments on the manuscript. I also acknowledge the numerous publications on speciation, MAT loci and sex-determination that are not mentioned in this short article, yet influenced these thoughts. Research in the laboratory is supported by the National Science Foundation (grant MCB-0920581).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aitken RJ, Marshall Graves JA. The future of sex. Nature. 2002;415:963. doi: 10.1038/415963a. [DOI] [PubMed] [Google Scholar]
- Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Plummer KM, Pradier J-M, Quévillion E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett B, Kodira CD, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun M-H, Dickman M. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002230. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee ML, Taylor JW. Dating the molecular clock in fungi - how close are we? Fungal Biol. Rev. 2010;24:1–16. [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Bloomfield G, Skelton J, Ivens A, Tanaka Y, Kay RR. Sex determination in the social amoeba Dictyostelium discoideum. Science. 2010;330:1533–1536. doi: 10.1126/science.1197423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe KH. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Coelho MA, Sampaio JP, Gonçalves P. A deviation from the bipolar-tetrapolar mating paradigm in an early diverged basidiomycete. PLoS Genet. 2010;6:e1001052. doi: 10.1371/journal.pgen.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. The evolutionary genetics of speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998;353:287–305. doi: 10.1098/rstb.1998.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- Faria R, Navarro A. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol. 2010;25:660–669. doi: 10.1016/j.tree.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Heitman J. Chromosomal sex-determining regions in animals, plants and fungi. Curr. Opin. Genet. Dev. 2005;15:645–651. doi: 10.1016/j.gde.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Giraud T, Refrégier G, Le Gac M, de Vienne DM, Hood ME. Speciation in fungi. Fungal Genet. Biol. 2008;45:791–802. doi: 10.1016/j.fgb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Godfray HCJ, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434:636–640. doi: 10.1038/nature03405. [DOI] [PubMed] [Google Scholar]
- Gryganskyi AP, Lee SC, Litvintseva AP, Smith ME, Bonito G, Porter TM, Anishchenko IM, Heitman J, Vilgalys R. Structure, function, and phylogeny of the mating locus in the Rhizopus oryzae complex. PLoS ONE. 2010;5:e15273. doi: 10.1371/journal.pone.0015273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Welch J, Kowbel DJ, Glass NL. Evolution and diversity of a fungal self/nonself recognition locus. PLoS ONE. 2010;5:e14055. doi: 10.1371/journal.pone.0014055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 2001;105:1422–1432. [Google Scholar]
- Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in Fungi: Molecular Determination and Evolutionary Implications. ASM Press; Washington, D.C.: 2007. [Google Scholar]
- Hood ME. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics. 2002;160:457–461. doi: 10.1093/genetics/160.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics. 2010;185:153–163. doi: 10.1534/genetics.109.113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Walton FJ, Floyd A, Heitman J. Identification of the sex genes in an early diverged fungus. Nature. 2008;451:193–196. doi: 10.1038/nature06453. [DOI] [PubMed] [Google Scholar]
- Jackson BC. Recombination-suppression: how many mechanisms for chromosomal speciation? Genetica. 2011;139:393–402. doi: 10.1007/s10709-011-9558-0. [DOI] [PubMed] [Google Scholar]
- James TY, Kües U, Rehner SA, Vilgalys R. Evolution of the gene encoding mitochondrial intermediate peptidase and its cosegregation with the A mating-type locus of mushroom fungi. Fungal Genet. Biol. 2004;41:381–390. doi: 10.1016/j.fgb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Kämper J, Reichmann M, Romeis T, Bölker M, Kahmann R. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- Kohn LM. Mechanisms of fungal speciation. Annu. Rev. Phytopathol. 2005;43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- Krings M, Dotzler N, Galtier J, Taylor TN. Oldest fossil basidiomycete clamp connections. Mycoscience. 2011;52:18–23. [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lee N, Bakkeren G, Wong K, Sherwood JE, Kronstad JW. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. USA. 1999;96:15026–15031. doi: 10.1073/pnas.96.26.15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Corradi N, Byrnes EJ, 3rd, Torres-Martinez S, Dietrich FS, Keeling PJ, Heitman J. Microsporidia evolved from ancestral sexual fungi. Curr. Biol. 2008;18:1675–1679. doi: 10.1016/j.cub.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S-W, Yun S-H, Lee T, Turgeon BG. Altering sexual reproductive mode by interspecific exchange of MAT loci. Fungal Genet. Biol. 2011;48:714–724. doi: 10.1016/j.fgb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Marshall Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Martin T, Lu S-W, van Tilbeurgh H, Ripoll DR, Dixelius C, Turgeon BG, Debuchy R. Tracing the origin of the fungal α1 domain places its ancestor in the HMG-box superfamily: implication for fungal mating-type evolution. PLoS ONE. 2010;5:e15199. doi: 10.1371/journal.pone.0015199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis A, Jacobson DJ, Gustafsson T, Johannesson H. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg RL, Glass NL. Mating type and mating strategies in Neurospora. Bioessays. 1990;12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- Micali CO, Smith ML. A nonself recognition gene complex in Neurospora crassa. Genetics. 2006;173:1991–2004. doi: 10.1534/genetics.106.057562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, Archer DB, Dyer PS. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 2007;17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Powell AJ, Jacobson DJ, Natvig DO. Ancestral polymorphism and linkage disequilibrium at the het-6 region in pseudohomothallic Neurospora tetrasperma. Fungal Genet. Biol. 2007;44:896–904. doi: 10.1016/j.fgb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Degeneration of a nonrecombining chromosome. Science. 1994;263:230–232. doi: 10.1126/science.8284674. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Schoustra S, Rundle HD, Dali R, Kassen R. Fitness-associated sexual reproduction in a filamentous fungus. Curr. Biol. 2010;20:1350–1355. doi: 10.1016/j.cub.2010.05.060. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S-F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang S-P, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Stanton BC, Giles SS, Staudt MW, Kruzel EK, Hull CM. Allelic exchange of pheromones and their receptors reprograms sexual identity in Cryptococcus neoformans. PLoS Genet. 2010;6:e1000860. doi: 10.1371/journal.pgen.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton E, Hughes J, White S, Sekido R, Tan J, Arboleda V, Rogers N, Knower K, Rowley L, Eyre H, Rizzoti K, McAninch D, Goncalves J, Slee J, Turbitt E, Bruno D, Bengtsson H, Harley V, Vilain E, Sinclair A, Lovell-Badge R, Thomas P. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J. Clin. Invest. 2011;121:328–341. doi: 10.1172/JCI42580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TN, Hass H, Kerp H. The oldest fossil ascomycetes. Nature. 1999;399:648. doi: 10.1038/21349. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Johannesson H. Evidence of the accumulation of allele-specific non-synonymous substitutions in the young region of recombination suppression within the mating-type chromosomes of Neurospora tetrasperma. Heredity. 2011;107 doi: 10.1038/hdy.2011.11. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CA, Nygren K, Johannesson H. Consequences of reproductive mode on genome evolution in fungi. Fungal Genet. Biol. 2011a;48:661–667. doi: 10.1016/j.fgb.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Sun Y, Johannesson H. Degeneration in codon usage within the region of suppressed recombination in the mating-type chromosomes of Neurospora tetrasperma. Eukaryot. Cell. 2011b;10:594–603. doi: 10.1128/EC.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S-H, Berbee ML, Yoder OC, Turgeon BG. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. USA. 1999;96:5592–5297. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl C. The role of sex in fungal evolution. Curr. Opin. Microbiol. 2009;12:592–598. doi: 10.1016/j.mib.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Zeyl C, Bell G. The advantage of sex in evolving yeast populations. Nature. 1997;388:465–468. doi: 10.1038/41312. [DOI] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010;8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]