Abstract
Integrons are horizontal gene transfer (HGT) systems containing elements necessary for site-specific recombination and expression of foreign DNA. The overall phylogenetic distribution of integrons and range of genes that can be transferred by integrons are unknown. This report contains an exploration of integrons in an environmental microbial community and an investigation of integron evolution. First, using culture-independent techniques, we explored the diversity of integrons and integron-transferred genes in heavy-metal-contaminated mine tailings. Using degenerate primers, we amplified integron integrase genes from the tailings. We discovered 14 previously undescribed integrase genes, including six novel gene lineages. In addition, we found 11 novel gene cassettes in this sample. One of the gene cassettes that we sequenced is similar to a gene that codes for a step in a pathway for nitroaromatic catabolism, a group of compounds associated with mining activity. This suggests that integrons may be important for gene transfer in response to selective pressures other than the presence of antibiotics. We also investigated the evolution of integrons by statistically comparing the phylogenies of 16S rRNA and integrase genes from the same organisms, using sequences from GenBank and various sequencing projects. We found significant differences between the organismal (16S rRNA) and integrase trees, and we suggest that these differences may be due to HGT.
Horizontal gene transfer (HGT) is the flow of genetic information across species boundaries. The importance of HGT was recognized in the 1960s with the first documentations of antibiotic resistance gene transfer (1, 27). More recently, HGT has been implicated in the dispersal of genes underlying complex cellular processes, including photosynthesis (23), nitrogen fixation (51), carbon fixation (54), sulfate reduction (15), and pathogenicity (16, 19). In addition, genomic studies support the importance of HGT in shaping entire microbial genomes (29, 37). Formerly considered a special case in the generation of variation, evidence is mounting that HGT is a significant force driving microbial evolution (38). This study describes the diversity of and genes transferred by one HGT system, integrons, in a heavy-metal-contaminated environment and provides a discussion of integron evolution.
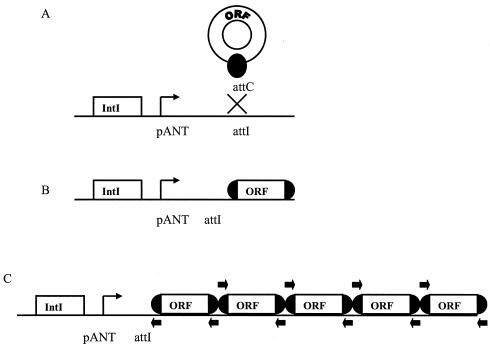
Integrons are HGT systems containing elements necessary for site-specific recombination and expression of foreign DNA (17, 45). Integrons consist of two parts: (i) the stationary integron platform, including the integrase gene (intI), a strong promoter (pant), and a recombination site (attI); and (ii) the mobile gene cassettes, which are promoterless open reading frames (ORFs) with a recombination site (attC) (Fig. 1A). IntI catalyzes a site-specific recombination event between the attI and attC sites and integrates or excises gene cassettes. Gene cassettes that are integrated into the integron platform are expressed from the promoter pant (Fig. 1B).
FIG. 1.
(A) Cartoon showing the stationary integron platform containing the intI gene, attI recombination site, and the pant promoter and the mobile gene cassette containing an ORF and attC site (shaded circle). The recombination site is marked with an X. (B) MR integron with one gene cassette. (C) SI with several gene cassettes and homogenous attC sites. Gene cassette primer sites are indicated with arrows.
Work on integrons began with the discovery of multiresistance (MR) integrons that usually contain one or two gene cassettes encoding antibiotic resistance (30) (Fig. 1B). MR integrons have been identified in many types of pathogenic Proteobacteria (31) and in the firmicute Corynebacterium glutamicum (34). MR integrons are commonly associated with mobile elements, including plasmids and transposons (6, 34, 41, 53).
Recently, genome sequence analysis of Vibrio cholerae led to the discovery of a superintegron (SI) (5, 32). SIs are found on bacterial chromosomes and consist of an array of gene cassettes adjacent to the integrase gene. They differ from MR integrons because they contain many gene cassettes, the activities of the gene cassettes are not limited to antibiotic resistance, and the attC sites associated with the arrays are homogenous (for reviews, see references 5, 32, and 44 to 46) (Fig. 1C). SIs are also found in other species of Vibrio (8) and in an assortment of Pseudomonas species (20, 55). Genome sequencing projects have also uncovered SI intI-like genes in Gamma-, Delta-, and Betaproteobacteria and in the spirochete Treponema denticola (45).
Integrons are found in several major lineages of bacteria, and the view of integron diversity is expanding. There are currently 32 unique integron integrase genes available in GenBank and various sequencing projects, an eightfold increase in the past 3 years. However, the culture-based methods and mostly pathogenic Proteobacteria used for the study of integrons leave two central questions unanswered: (i) what is the phylogenetic distribution of these elements? and (ii) what types of genes can be transferred by integrons?
Soil microbial communities contain phylogenetically diverse organisms living in an array of niches and present an ideal environment to investigate these questions. Molecular techniques, typified by 16S rRNA gene analysis, have revolutionized environmental microbiology and provide the tools necessary to explore microbial communities without the biases associated with cultivation (3, 39). Recently Nield et al. (35) developed degenerate primers for integrons and identified three new classes of integrons from soil microbial communities. The same group (49) also designed primers targeted to the attC recombination sites of gene cassettes. These primers amplify genes flanked by two recombination sites, an arrangement present in any multicassette integron (Fig. 1C). Their study revealed gene cassette diversity in environmental samples. However, most sequences from that study did not share significant sequence similarity with known genes, and the functions of these gene cassettes are unknown.
In this study, we used molecular techniques to examine the diversity of integrons in heavy-metal-contaminated mine tailings. We chose this environment because it is under intense selection pressure for traits other than antibiotic resistance. We used both published primers and newly designed primers to uncover 14 new integron integrase genes and 11 new gene cassettes from this environment. One of the gene cassettes that we discovered is similar to a gene that codes for a step in a pathway for nitroaromatic catabolism. We also discuss the evolution of chromosomally associated SIs by statistically comparing the phylogenies of 16S rRNA and SI integrase gene trees from the same organisms, using sequences available from GenBank and sequencing projects. We found significant differences between the organismal (16S rRNA) and integrase trees, and we suggest that these differences may be due to HGT.
MATERIALS AND METHODS
Sampling procedure.
Samples were collected from the tailings (pH 7) of an abandoned gold mine near Nederland, Colo. (elevation, 2,512 m). Surface (1 to 5 cm) tailings were gathered into sterile 50-ml conical tubes, placed on ice for transport, and stored at −80°C prior to DNA extraction. To measure metal concentrations in the tailings, samples were digested according to U.S. Environmental Protection Agency (EPA) method 3050B and analyzed using atomic emission spectrometry according to EPA method 6010A at the University of Colorado Department of Geology Central Analysis Laboratory.
DNA extraction and clone libraries.
DNA was extracted from the tailings using a modification of the protocol described by Zhou et al. (56). Five grams of soil was added to 10 ml of buffer (100 mM Tris-HCl [pH 8.0], 100 mM EDTA [pH 8.3], 100 mM phosphate buffer [pH 8.0], 1.5 M NaCl, 1% cetyltrimethylammonium bromide), 50 μl of proteinase K (20 mg/ml), 60 μl of lysozyme (100 mg/ml), and 9 μl of RNase (10 mg/ml). Samples were incubated at 37°C with shaking at 80 rpm for 30 min. A 1.5-ml volume of 20% sodium dodecyl sulfate was added, and the tubes were gently agitated and then incubated at 65°C for 2 h. Samples were centrifuged at 3,850 × g for 10 min. The supernatant was removed and extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1). DNA was precipitated with 0.6 volumes of isopropanol and washed with 1 ml of 70% ethanol. Four separate DNA extractions were pooled and purified over Sepharose 4B (Sigma, St. Louis, Mo.) packed columns as described by Jackson et al. (24).
Approximately 30 ng of DNA was amplified with a variety of primer sets (Table 1). 16S rRNA genes were amplified with 27f and 1492r (28), integrase genes were amplified with int1.F/R and intlld F/R, and gene cassettes were amplified with HS286 and HS287. The reaction conditions consisted of a 400 nM (27f/1492r and int1.F/R) or 4 μM (intltdF/intltdR and HS286/HS287) concentration of each primer, a 200 μM concentration of each deoxynucleoside triphosphate, and 1.25 U of Taq DNA polymerase (Promega, Madison, Wis.) in Taq DNA polymerase buffer containing MgCl2 (Promega). After an initial denaturation step at 94°C for 1 min, 35 cycles of 94°C for 1 min, 58°C for 30 s, and 72°C for 2.5 min with a terminal 10-min extension at 72°C were performed. PCR products from the 16S rRNA and integrase gene amplifications were gel purified and ligated into the vector TOPO 2.1 (Invitrogen, Carlsbad, Calif.) and transformed into Escherichia coli cells following the manufacturer's instructions. Gene cassette PCR products were ligated into the pGEM 2.1 vector (Promega) and transformed into E. coli following the manufacturer's instructions. For each cloning reaction, 96 colonies were selected for plasmid extraction.
TABLE 1.
Primers used in this study
| Primer | Target | Sequence (degree of degeneracy) | Reference |
|---|---|---|---|
| 27f | 16S rRNA gene | AGAGTTTGATCMTGGCTCAG (2) | 28 |
| 1492r | 16S rRNA gene | TACGGYTACCTTGTTACGACTT (2) | 28 |
| int1.F | Integron integrases | GGGTCAAGGATCTGGATTTCG | 31 |
| int1.R | Integron integrases | ACATGCGTGTAAATCATCGTCG | 31 |
| intltdF | Integron integrase | CTNYTNTAYGGNWCNGG (2,048) | This study |
| intltdR | Integron integrase | TCYTGNACNGWNCKDATRTC (3,072) | This study |
| HS286 | Gene cassettes | GGGATCCTCSGCTKGARCGAMTTGTTAGVC (48) | 35 |
| HS287 | Gene cassettes | GGGATCCGCSGCTKANCTCVRRCGTTAGSC (384) | 49 |
| M13F | Cloned inserts | GTAAAACGACGGCCAG | Invitrogen, Promega |
| M13R | Cloned inserts | CAGGAAACAGCTATGAC | Invitrogen, Promega |
| T7 | Cloned inserts | TAATACGACTCACTATAGGG | Invitrogen, Promega |
| 534r | 16S rRNA gene | ATTACCGCGGCTGCTGG | 33 |
| 1100r | 16S rRNA gene | GGGTTGCGCTCGTTG | 28 |
| R1113 | 16S rRNA gene | GGGTTGCGCYCGTT (2) | This study |
DNA sequencing.
Inserted sequences were PCR amplified using the primers M13F and M13R and cleaned using exonuclease I and shrimp alkaline phosphatase (New England Biolabs, Beverly, Mass.) or QIAquick PCR purification columns (Qiagen, Valencia, Calif.). The 16S rRNA genes were sequenced with the primers 534r (33), 1100r or R1113 (TM7 clade), and 27f. The integrase genes and gene cassettes were sequenced using the T7 promoter primer. Integrase genes and some 16S rRNA sequencing reactions were performed and sequencing products were run at the MCD Biology Sequencing Facility, University of Colorado. Gene cassettes and other 16S rRNA genes were sequenced using the BigDye terminator cycle sequencing kit (version 3.0; Applied Biosystems, Foster City, Calif.) following the manufacturer's directions. These sequencing products were analyzed at the Iowa State University DNA Sequencing Facility.
Sequence and phylogenetic analysis.
Sequences were edited in Sequencher 4.1 (Gene Codes Co., Ann Arbor, Mich.) and subjected to BLAST (2) or BLASTX searches for protein sequences. 16S rRNA gene sequences were subjected to chimera check in RDP (9) and aligned in an ARB database (http://www.arb-home.de/). Closely related sequences from the ARB database and from BLAST searches were used as reference taxa for phylogenetic analyses, and two archaeal sequences from the ARB database were used as an outgroup. We selected putative integron integrase genes by screening for the presence of the integron integrase-specific insertion (36) and aligned these sequences with other integron integrase proteins and the XerC outgroup (13) in ClustalX.
Alignments were subjected to Bayesian phylogenetic analysis as implemented in MRBAYES (21). Separate analyses were performed for all data combined (environmental genes and published gene sequences collected from GenBank and various sequencing projects) and for reduced taxa data sets for which both intI and 16S rRNA genes were available from the same organisms. For the IntI amino acid data, Bayesian analysis employed the Jones model of sequence evolution (26). For the 16S rRNA data, we used the GTR+ gamma model of evolution. For the IntI analyses, 250,000 generations were run and trees were sampled every 100 generations. For the 16S rRNA analyses, 1,000,000 generations were run and trees were sampled every 100 generations. Burn-in values were determined by plotting the likelihood scores against generation number and retaining trees for which stationarity was evident. In addition, all alignments were subjected to phylogenetic analyses in PAUP* (version 4; D. L. Swofford, Sinauer Associates, Sunderland, Mass.) using both the maximum parsimony optimality criterion and the neighbor-joining tree-building algorithm. Maximum parsimony and neighbor-joining phylogenetic inferences were subject to bootstrap analyses with 1,000 replicates. Finally, all phylogenies were tested with collections of outgroup sequences to confirm the robustness of these estimates.
We examined the relationship between the organismal and SI integrase trees by comparing inferred phylogenies of SI integrase and 16S rRNA genes from the same organisms, using sequences from GenBank and various sequencing projects. IntI genes are available for two subspecies (BAM and Q) of Pseudomonas stutzeri and for two subspecies (badrii and campestris) of Xanthomonas campestris; however, individual 16S rRNA genes are not available for these organisms. P. stutzeri integrase genes are nearly identical (<0.05% difference in amino acid sequence), and so we arbitrarily selected one integrase gene, Q, to represent this lineage. X. campestris genes are also nearly identical, and we selected the subspecies campestris integrase gene to represent this lineage because there was more sequence information available for this gene. A test of concordance between 16S rRNA gene and IntI protein trees was accomplished using the Shimodaira-Hasegawa test (48) and the Wilcoxon signed-rank test (52). These tests compare the likelihood and parsimony, respectively, scores of alternative trees using both the 16S rRNA gene and integrase data. Finally, the extent that the two genes recorded the same evolutionary history was estimated by determining agreement subtrees (implemented in PAUP* [version 4; Sinauer Associates]).
For the gene cassette analysis, sequences were determined to be cassettes if (i) they possessed the eight invariant residues of attC sites (50), (ii) the ends were flanked by two putative IntI-like simple sites including complementary 1R and 1L recombination sequences (51), and (iii) the recombination sites flanked an ORF of greater than 80 amino acids. For some sequences, the stop codon was derived from the 1L sequence. Putative translated sequences were subjected to pBLAST searches, and matches were considered significant if the e value was <0.001.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the gene cassettes are AY271679 to AY271689. The accession numbers for the 16S rRNA gene sequences are AF337861 to AF337888 and AY274120 to AY274164. The accession numbers for the integrase gene sequences are AY283623 to AY283638.
RESULTS
Heavy metals and bacterial communities in gold mine tailings.
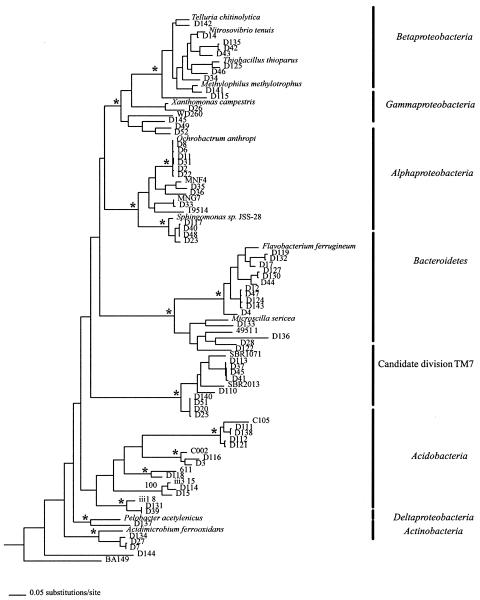
The tailings were assayed for concentrations of a variety of metals, and these results are presented in Table 2. There were high levels of barium, copper, and zinc at the site. The levels of copper and zinc were 66 and 45 times higher, respectively, in these tailings than in a nearby pristine soil (47). We successfully sequenced 67 16S rRNA genes from this environmental sample (Fig. 2) and obtained only 40 unique (>2% different) sequences. Using the ChaoI and ACE estimators of diversity (10) as calculated in EstimateS (5th ed.; R. K. Colwell, http://viceroy.eeb.uconn.edu/estimates), this corresponded to 40% of the predicted total number of 16S rRNA gene sequences in this environmental sample. We sequenced a significant number of 16S rRNA gene repeats, with six sequences that were identical to Ochrobactrum anthropi, four Sphingomonas-like sequences, four identical sequences from within the Bacteroidetes, and four identical sequences from within the candidate division TM7.
TABLE 2.
Metal concentrations in the tailings used for this study
| Metal | Concn (ppm) |
|---|---|
| Arsenic | 5.1 |
| Barium | 414 |
| Cadmium | 6.3 |
| Cobalt | 20.7 |
| Copper | 1,200 |
| Lead | 8.1 |
| Manganese | 5.8 |
| Molybdenum | 61.5 |
| Selenium | 3.5 |
| Zinc | 766 |
FIG. 2.
A 50% majority rule consensus tree of 16S rRNA genes derived from Bayesian phylogenetics. An asterisk indicates a node with a Bayesian posterior probability of >0.95, maximum parsimony bootstrap support of >80, and neighbor-joining bootstrap support of >80. The sequences from this work are the D series. The tree is rooted with Methanosarcina acetivorans (M59137) and Natronobacterium chahannaoensis (AJ004806). Branch lengths are drawn proportional to the amount of evolution based on uncorrected genetic distances. Accession numbers are as follows: Telluria chitinolytica, X65590; Nitrosovibrio tenuis, M96405; Thiobacillus thioparus, M79426; Methylophilus methylotrophus, L15475; X. campestris ATTC, 339113; WD260, AJ292673; O. anthropi, D63837; MNF4, AF292996; MNG7, AF292997; 19514, AF097791; Sphingomonas sp. strain JSS-28, AF031240; Flavobacterium ferrugineum, M28237; Microscilla sericea, M58794; 49511, AF097805; SBR1071, AF268996; SBR2013, AF269000; C105, AF013530; C002, AF013515; 611, Y11629; ii3_15, Z95725; iii1_8, Z95729; Pelobacter acetylenicus, X70955; Acidimicrobium ferrooxidans, U75647; BA149, AF323777.
Diversity of integron integrase genes.
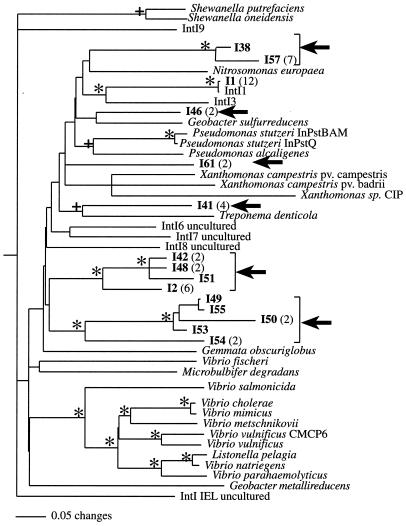
We used int1.F/R (31) and the newly designed intltdF/R (Table 1) primers to amplify integron integrase genes from the mine tailing sample. We sequenced 96 clones and selected 46 sequences that contained the IntI patch, a 16-amino-acid insertion that is specific to integron integrases (36). We sequenced many identical integrase genes and obtained only 15 unique sequences (>2% different from other sequences in our library). Twelve of these genes were obtained using the int1.F/R primer set, and three were obtained with the intltdF/R primers. Using the ChaoI and ACE estimators of diversity (10) as calculated in EstimateS (5th ed., http://viceroy.eeb.uconn.edu/estimates), this corresponded to 82% of the predicted total number of integron integrase gene sequences in this environmental sample. The alignment of these putative IntI proteins showed a great diversity that is typical of integron integrases; in 145 amino acids of total sequence, only 25 amino acids were conserved across all sequences. Phylogenetic analysis of the alignment of our sequences with published sequences revealed that all of our IntI proteins formed a clade with known integron integrase sequences exclusive of the XerC recombinase outgroup (Fig. 3). We recovered 12 sequences (I1) that were nearly exact matches to class 1 integrases (IntI1) (Fig. 3), confirming the specificity of our primers. Notably, we discovered 14 previously undescribed integron integrase genes. We recovered six novel (>10% sequence difference from known integrase genes) gene lineages (Fig. 3). We discovered genes with phylogenetic affinity to IntI proteins from T. denticola (I41) and Nitrosomonas europaea (I38, I56, and I57), although in both cases the extent of sequence divergence was large (>20% difference in amino acid sequence). Given the immense diversity of bacteria in soil and the few described integrase gene sequences, it is not surprising that most of what we found was novel.
FIG. 3.
A 50% majority rule consensus tree of IntI proteins derived from Bayesian phylogenetics. The sequences from this work are the I series, and numbers in parentheses denote the number of identical sequences obtained in this study. The tree was rooted using XerC from E. coli (P22885) and Salmonella enterica serovar Typhimurium (AAF33443) (14). Branch lengths were drawn proportional to the amount of evolution based on uncorrected genetic distances. An asterisk indicates a Bayesian posterior probability of >0.95, maximum parsimony bootstrap support of >80, and neighbor-joining bootstrap support of >80; + indicates a Bayesian posterior probability of >0.95 and either maximum parsimony bootstrap support of >90 or neighbor-joining bootstrap support of >90. Novel lineages are indicated with arrows. Accession numbers (when available) and sequencing project homepages are as follows: S. putrefaciens, AAK01408; S. oneidensis, MR-1 (The Institute for Genomic Research [TIGR] website [http://www.tigr.org]); IntI9, AAK95987; N. europaea (DOE Joint Genome Initiative website [http://www.jgi.doe.gov]); IntI1, AAM89398; IntI3, AAO32355; G. sulfurreducens (TIGR website); P. stutzeri BAM, AAN16071; P. stutzeri Q, AAN16061; P. alcaligenes, AAK73287; X. campestris pv. campestris, AAK07444; X. campestris pv. badrii, AAK07443; Xanthomonas sp. strain CIP, AAK07447; T. denticola (TIGR website); IntI6, AAK00307; IntI7, AAK00305; IntI8, AAK00304; Gemmata obscuriglobus (TIGR website); V. fischeri, AAK02079; Microbulbifer degradans (DOE website); V. salmonicida, CAC35342; V. cholerae, NP_232687; V. mimicus, AAD55407; V. metschnikovii, AAK02074; V. vulnificus CMCP6, AAO10775; V. vulnificus, AAN33109; Listonella pelagia, AAK02082; Vibrio natriegens, AAO38263; V. parahaemolyticus, AAK02076; G. metallireducens (Environmental Biotechnology Center, University of Massachusetts website [http://zdna.micro.umass.edu/]); IntI_IEL, AAN16072.
SI phylogeny.
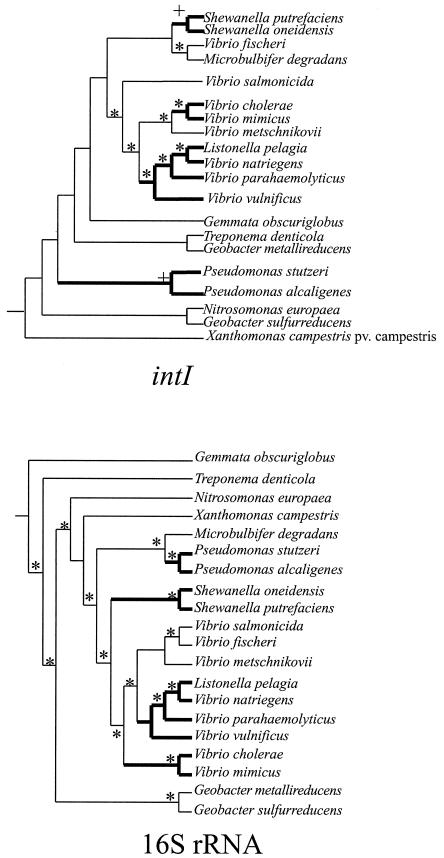
The IntI phylogeny for the chromosomally associated SIs presented in Fig. 3 did not match organismal phylogenies; for instance, Vibrio fischeri clustered with Microbulbifer degradans rather than within Vibrio. We further examined the relationship between the organismal and SI integrase trees by comparing inferred phylogenies of SI integrase and 16S rRNA genes from the same organisms, using sequences from GenBank and various sequencing projects (Fig. 4). We used statistical tests to assess phylogenetic concordance of these two gene trees. The Shimodaira-Hasegawa test (48) compares the maximum likelihood scores of the two trees, and the Wilcoxon signed-rank test (52) compares the parsimony scores of two trees. When we employed these tests using the 16S rRNA gene and integrase data, all trees differed significantly (P < 0.0001). Therefore, we reject the hypothesis that the two genes record the same history.
FIG. 4.
Gene trees for the SI integrase and 16S rRNA genes. Sequences were obtained from GenBank and various sequencing projects. Trees are majority rule consensus trees determined using Bayesian analysis of the amino acid (IntI) and DNA (16S rRNA) alignments. An asterisk indicates a Bayesian posterior probability of >0.95, maximum parsimony bootstrap support of >80, and neighbor-joining bootstrap support of >80. A + indicates a Bayesian posterior probability of >0.95 and either maximum parsimony bootstrap support of >90 or neighbor-joining bootstrap support of >90. Thick lines indicate shared cospeciation events for the 16S rRNA gene and integrase trees. The 16S rRNA gene tree was rooted using Methanococcus jannaschii (L77117). The tree was rooted using XerC from E. coli (P22885) and S. enterica serovar Typhimurium (AAF33443) (14). Accession numbers for integrase genes are presented in the legend to Fig. 3. 16S rRNA gene accession numbers were as follows: G. obscuriglobus, X54522; T. denticola, M71236; N. europaea, AF037106; X. campestris, X95917; Microbulbifer degradans, AF055269; P. stutzeri, U65012; P. alcaligenes, D84006; S. oneidensis, AF039055; S. putrefaciens, X81623; V. salmonicida, X70643; V. fischeri, X70640; V. metschnikovii, X74712; Listonella pelagia, X74722; V. natriegens, X74714; V. parahaemolyticus, M59161; V. vulnificus, X76334; V. cholerae, X76337; V. mimicus, X74713; G. metallireducens, L07834; G. sulfurreducens, U13928.
Coding capacity of gene cassettes.
We used primers targeted to the attC recombination sites (Fig. 1) to amplify gene cassettes from the mine tailings. The physical arrangement of gene cassettes and the closest pBLAST matches for these ORFs are listed in Table 3. We sequenced 11 unique gene cassettes that contained complementary 1R and 1L recombination sequences and ORFs that were greater than 80 amino acids. Our gene cassette library was dominated by two sequences, HMIC11 (49% of sequences) and HMIC12 (33% of sequences). For both of these sequences, several repeats were completely conserved within the coding frame but had mutated, noncomplementary recombination sequences.
TABLE 3.
Structures of gene cassettes from heavy-metal-contaminated mine tailingsa
| Cassette (sequence no.) | Space 1b (bp) | ORF (aa) | Space 2c (bp) | BLAST match (e value) | Organism | Comment |
|---|---|---|---|---|---|---|
| HMIC5 (1) | 24 | 82 | −4 | CAD18309 (e-19) | R. solanacearum | Associated with urease operon |
| HMIC7 (1) | 31 | 131 | −4 | AAK73292 (7e-23) | P. alcaligenes | SI cassette Ypar6 |
| HMIC8 (1) | 3 | 126 | −4 | None | ||
| HMIC9 (2) | 7 | 112 | 54 | None | ||
| HMIC10 (1) | 4 | 93 | 4 | None | ||
| HMIC11 (20) | 15 | 134 | 7 | NP_768178 (0.006) | B. japonicum | Unknown |
| HMIC12 (17) | 20 | 120 | 10 | None | ||
| HMIC15 (4) | 25 | 143 | −4 | NP_337685 (2e-9) | M. tuberculosis | Hydroxylaminobenzene mutase |
| HMIC16 (1) | 12 | 136 | 9 | NP_711470 (1e-11) | Leptospira interrogans serovar lai str | Unknown |
| HMIC18 (1) | 3 | 131 | −4 | None | ||
| HMIC20 (1) | 47 | 103 | 34 | None |
Gene cassette arrangement and significant BLAST matches. All gene cassettes had a TTAGGC 1R sequence and a GCCTAA 2R sequence.
Space between 1R sequence and start codon.
Space between stop codon and 1L sequence.
As with other studies of gene cassettes from environmental samples (35, 49) and from SIs (20, 46, 55), few of these genes share significant sequence similarity with genes of known function. No gene cassettes were related to genes known to be involved in heavy metal resistance. The only cassette that was similar to a gene with a known function was HMIC15, which was highly similar (72% over 54 amino acids) to a hydroxylaminobenzene mutase gene from Mycobacterium tuberculosis. Additionally, HMIC7 was 60% similar to Ypar6, a gene cassette from the Pseudomonas alcaligenes SI (55). Finally, HMIC5 was 78% similar to a hypothetical conserved protein that is associated with the urease operon, ORF3, and is of unknown function.
DISCUSSION
The recent explosion in the known diversity of integron integrase genes is due to both sequencing projects and environmental molecular analyses. Whole-genome sequencing projects have revealed the presence of intI genes in Beta-, Gamma-, and Deltaproteobacteria, a spirochete, and a planctomycete. Our environmental survey, and other published work (35, 49), implies that we are only beginning to appreciate the diversity of integrons. Given the importance of HGT for bacterial evolution and the potential negative effects (i.e., the spread of antibiotic resistance) and beneficial features (i.e., the potential for bioremediation of contaminated environments), it is imperative that we gain a better understanding of the role of the integron in nature.
The bacterial community in a metal-contaminated site.
The bacterial divisions represented in the mine tailings (Fig. 2), Proteobacteria, Bacteroidetes, Acidobacteria, Actinobacteria, and candidate division TM7, are common in other, noncontaminated and contaminated soil surveys (7, 12, 22). We found no evidence for the existence of clades unique to the contaminated site. However, we did find a significant number of sequence repeats: of the 67 rRNA genes sequenced, only 40 were unique. This lack of diversity has been reported from other contaminated soils (7, 43). There were several sequence repeats related to O. anthropi, a sequence from the genus Sphingomonas, a sequence that falls in the candidate division TM7, and a sequence that falls in the Bacteroidetes. Organisms from the Sphingomonas genus are common in contaminated soils (42, 43). O. anthropi is known for the ability to utilize a variety of nitrogen sources, including d and l amino acids (4), urea- formaldehyde (25), and methylammonium (14). This result may suggest the presence of unusual nitrogen sources in these tailings. The other two highly repeated sequences have no known cultured close relatives and, therefore, the significance of their presence in this sample is unknown.
Diversity of integrons in a metal-contaminated site.
In this study we discovered 14 new integron integrase genes (Fig. 3). Nield et al. (35) used a similar molecular approach and identified three new integrase genes from several environments. Although our primers amplify only 444 nucleotides of intI genes and, unlike the primers used by Nield et al. (35), they do not amplify associated gene cassettes, the primer sets used here amplified a more diverse array of genes.
Currently, there are 32 unique integron integrase genes available in GenBank and various sequencing projects, and 22 of these genes are found exclusively in the Gammaproteobacteria. Our 16S rRNA gene library (Fig. 2) included only one sequence that was closely related to an organism known to harbor an integron (X. campestris) and only included four Gammaproteobacteria-like sequences. This may suggest that some of the 14 new integrase genes that we sequenced (Fig. 3) are from organisms previously not known to contain integrons. Although all IntI sequences from our library form a clade with known integron integrases that is exclusive of the XerC outgroup, the relationships between these integrase genes are largely unresolved (Fig. 3). Therefore, it is difficult to speculate on the phylogenetic origins of these integrase genes. More integrase genes from known organisms need to be sequenced and the evolution of integrase genes (see below) needs to be better understood before we can make inferences about the phylogenies of these previously undescribed genes.
Evolution of SI integrase genes.
We further examined the relationship between the organismal and SI integrase trees by comparing inferred phylogenies of SI integrase and 16S rRNA genes from the same organisms, using sequences from GenBank and various sequencing projects (Fig. 4). We found some shared speciation events between the IntI and 16S rRNA gene trees (Fig. 4), including support for the clustering of P. stutzeri and P. alcaligenes, support for the clustering of Shewanella oneidensis and Shewanella putrefaciens, and support for the Vibrio integrase clade (excluding V. fischeri). However, a statistical comparison of trees derived from 16S rRNA and SI integrase genes from the same taxa allowed us to reject the hypothesis that SI integrases track organismal phylogeny. Both HGT and the sampling of paralogous genes could explain the lack of concordance between the two gene trees.
We cannot exclude that other, paralogous intI gene families exist. Indeed, there is support for gene duplication and translocation to plasmids in Vibrio vulnificus (Fig. 3). However, we note that this gene duplication is likely to be a recent event, because the sequences are nearly identical and are therefore unlikely to explain the lack of concordance between the integrase and 16S rRNA gene trees. In addition, some of the integrase gene sequences we used are from entire genome sequences (e.g., N. europaea, V. cholerae), and there were no paralogous integrase genes found. Other integrase genes, however, are from partial genome sequences or pure-culture studies that may have missed paralogous integrase genes.
HGT of integrase genes could also explain the differences between the integrase and 16S rRNA gene tree. For example, V. fischeri is “out of place” on the integrase gene tree, clustering with M. degradans and falling outside the well-supported Vibrio clade (Fig. 4). IntI genes are likely to have been present in the ancestor of the entire Vibrio clade, because these genes largely mirror the organismal (16S rRNA gene) tree (Fig. 4). Therefore, it is possible that the ancestor of V. fischeri lost its Vibrio-type integrase gene and inherited a divergent integrase gene by HGT. The phylogeny of Geobacter sulfurreducens and Geobacter metallireducens integrase genes is also different from the organismal relationships. Comparison of 16S rRNA genes from these two organisms indicates that they have approximately the same number of pairwise differences as Vibrio parahaemolyticus and Vibrio metschnikovii yet, unlike V. parahaemolyticus and V. metschnikovii, their integrase genes do not cluster together (Fig. 4). It is possible that integrase genes are evolving more quickly in the Geobacter lineage. However, HGT could also explain the difference between the Geobacter 16S rRNA genes and integrase gene trees. For example, an integrase gene could have been present in the ancestor of the two Geobacter species, and this gene could have been lost in one lineage and another, more distant integrase gene gained. Alternatively, both lineages could have gained different integrase genes after their divergence. More Geobacter lineages should be sampled for integrase genes to test these two hypotheses.
Rowe-Magnus et al. (45) suggested that SIs were present in the ancestor of the Proteobacteria. If we assume that X. campestris marks the most basal lineage of Gammaproteobacteria sampled (as suggested by the 16S rRNA tree), then its basal placement on the IntI tree may indicate that SIs were present in the ancestor of the Gammaproteobacteria (45). We surveyed for the presence of integrons in the published, completed genome sequences available in GenBank. Surprisingly, we failed to detect integron integrase genes in many Gammaproteobacteria, even in organisms with close relatives that contain integrons. For example, neither P. aeruginosa nor Pseudomonas putida has intI genes, yet other species of Pseudomonas are known to contain integrons (20, 55). Furthermore, we only found intI genes in 5 of the 25 sequenced Gammaproteobacteria. Assuming an ancient origin, the apparent lack of integrons in many Gammaproteobacteria suggests that integrons have been lost numerous times in diverging lineages. The implications of this inference remain to be explored.
Selection pressures and gene cassettes.
Unlike Stokes et al. (49), we found many repeats in our gene cassette library. Although this could be due to PCR bias or lower diversity of the microbial community, it could also be a reflection of selection and could imply that these genes are important in this environment. Further support for the importance of these repeated gene cassettes came from examination of the associated recombination sites. In many cases, the ORF was entirely conserved while the recombination sites were not complementary, preventing them from being recognized by the integrase enzyme. If these gene cassettes were essential for survival, this would select for the loss of these recombination sites to prevent gene excision.
Most of the gene cassettes that we sequenced were related to genes of unknown function (Table 3). For each sequence that had a significant pBLAST hit, we searched the DNA adjacent to the matches, looking for integrase genes or integron recombination sites. With the exception of HMIC7, which is related to an SI cassette, we found no indication that these genes were also located on integrons. We sequenced four repeats of a cassette related to a gene of known function, hydroxylaminobenzene mutase. Homologues of this gene have been found on other mobile gene elements, including a plasmid in P. putida (40) and a transposon in Pseudomonas pseudoalcaligenes JS45 (11, 18), and our results suggest that it is also mobilized by integrons. Hydroxylaminobenzene mutase catalyzes a reduction reaction in the catabolism of nitroaromatics (40). Nitroaromatics are xenobiotic compounds that are used as explosives and solvents, both of which are used in gold extraction. The ability to catabolize nitroaromatics may be a metabolic advantage, or it may be important in detoxification. This suggests that integrons may be important for gene transfer in response to selective pressures other than the presence of antibiotics.
Acknowledgments
This work was supported by the National Science Foundation under grants MCB-0084223 and IBN-9817164.
We thank Kevin Shiley for help with construction and sequencing of the 16S rRNA gene library and Hatch Stokes for the sequence of primer HS287. We also thank members of the Schmidt and Martin labs and three anonymous reviewers for insightful comments on the manuscript.
REFERENCES
- 1.Alberti, S., V. Ortali, and E. J. Salas. 1965. On the transduction of certain metabolic characters in staphylococci. Ann. 1st Super Sanita. 1:61-66. [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in-situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano, Y., M. Umezaki, Y. F. Li, S. Tsubota, and T. L. Lubbehusen. 2001. Isolation of microorganisms which utilize acidic D-amino acid oligomers. J. Mol. Catalysis B 12:53-59. [Google Scholar]
- 5.Barker, A., C. A. Clark, and P. A. Manning. 1994. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J. Bacteriol. 176:5450-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissonnette, L., and P. H. Roy. 1992. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J. Bacteriol. 174:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 8.Clark, C. A., L. Purins, P. Kaewrakon, T. Focareta, and P. A. Manning. 2000. The Vibrio cholerae O1 chromosomal integron. Microbiology 146:2605-2612. [DOI] [PubMed] [Google Scholar]
- 9.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colwell, R. K., and J. A. Coddington. 1994. Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. London B 345:101-118. [DOI] [PubMed] [Google Scholar]
- 11.Davis, J. K., G. C. Paoli, Z. Q. He, L. J. Nadeau, C. C. Somerville, and J. C. Spain. 2000. Sequence analysis and initial characterization of two isozymes of hydroxylaminobenzene mutase from Pseudomonas pseudoalcaligenes JS45. Appl. Environ. Microbiol. 66:2965-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewen, H., H. Kaltwasser, and T. Jahns. 2000. Ammonium and methylammonium uptake in a fertilizer-degrading strain of Ochrobactrum anthropi. Antonie Leeuwenhoek 77:263-270. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich, M. W. 2002. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J. Bacteriol. 184:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamieldien, J., A. Ptitsyn, and W. Hide. 2002. Eukaryotic genes in Mycobacterium tuberculosis could have a role in pathogenesis and immunomodulation. Trends Genet. 18:5-8. [DOI] [PubMed] [Google Scholar]
- 17.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 18.He, Z. Q., L. J. Nadeau, and J. C. Spain. 2000. Characterization of hydroxylaminobenzene mutase from pNBZ139 cloned from Pseudomonas pseudoalcaligenes JS45. A highly associated SDS-stable enzyme catalyzing an intramolecular transfer of hydroxy groups. Eur. J. Biochem. 267:1110-1116. [DOI] [PubMed] [Google Scholar]
- 19.Hentschel, U., and J. Hacker. 2001. Pathogenicity islands: the tip of the iceberg. Microbes Infect. 3:545-548. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, A. J., M. P. Holley, A. Mahon, B. Nield, M. Gillings, and H. W. Stokes. 2003. Recombination activity of a distinctive integron-gene cassette system associated with Pseudomonas stutzeri populations in soil. J. Bacteriol. 185:918-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 22.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igarashi, N., J. Harada, S. Nagashima, K. Matsuura, K. Shimada, and K. V. P. Nagashima. 2001. Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J. Mol. Evol. 52:333-341. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, C. R., J. P. Harper, D. Willoughby, E. E. Roden, and P. F. Churchill. 1997. A simple, efficient method for the separation of humic substances and DNA from environmental samples. Appl. Environ. Microbiol. 63:4993-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahns, T., R. Schepp, C. Siersdorfer, and H. Kaltwasser. 1998. Microbial urea-formaldehyde degradation involves a new enzyme, methylenediurease. Acta Biol. Hung. 49:449-454. [PubMed] [Google Scholar]
- 26.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 27.Klimov, K. V., T. S. M. Levina, and T. N. Kushneva. 1965. Determination of the sensitivity of dysenterial bacteria to tetracycline antibiotics using the agar diffusion method. Antibiotiki 10:544-546. [PubMed] [Google Scholar]
- 28.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., West Sussex, England.
- 29.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 33.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nesvera, J., J. Hochmannova, and M. Patek. 1998. An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol. Lett. 169:391-395. [DOI] [PubMed] [Google Scholar]
- 35.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. H. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 36.Nunes-Duby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochman, H., and I. B. Jones. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 39.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 40.Park, H. S., and H. S. Kim. 2000. Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J. Bacteriol. 182:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radehaus, P. M., and S. K. Schmidt. 1992. Characterization of a novel Pseudomonas sp. that mineralizes high concentrations of pentachlorophenol. Appl. Environ. Microbiol. 58:2879-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roling, W. F. M., M. G. Milner, D. M. Jones, K. Lee, F. Daniel, R. J. P. Swannell, and I. M. Head. 2002. Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl. Environ. Microbiol. 68:5537-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowe-Magnus, D. A., A. M. Guerout, and D. Mazel. 1999. Super-integrons. Res. Microbiol. 150:641-651. [DOI] [PubMed] [Google Scholar]
- 45.Rowe-Magnus, D. A., A. M. Guerout, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. USA 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowe-Magnus, D. A., and D. Mazel. 2001. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 4:565-569. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, S. K., and M. J. Gier. 1990. Coexisting bacterial populations responsible for multiphasic mineralization kinetics in soil. Appl. Environ. Microbiol. 56:2692-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 49.Stokes, H. W., A. J. Holmes, B. S. Nield, M. P. Holley, K. M. H. Nevalainen, B. C. Mabbutt, and M. R. Gillings. 2001. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 67:5240-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stokes, H. W., D. B. O’Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 51.Sullivan, J. T., S. D. Brown, R. R. Yocum, and C. W. Ronson. 2001. The bio operon on the acquired symbiosis island of Mesorhizobium sp. strain R7A includes a novel gene involved in pimeloyl-CoA synthesis. Microbiology 147:1315-1322. [DOI] [PubMed] [Google Scholar]
- 52.Templeton, A. R. 1983. Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution 37:221-244. [DOI] [PubMed] [Google Scholar]
- 53.Tribuddharat, C., and M. Fennewald. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Utaker, J. B., K. Andersen, A. Aakra, B. Moen, and I. F. Nes. 2002. Phylogeny and functional expression of ribulose 1,5-bisphosphate carboxylase/oxygenase from the autotrophic ammonia-oxidizing bacterium Nitrosospira sp. isolate 40KI. J. Bacteriol. 184:468-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaisvila, R., R. D. Morgan, J. Posfai, and E. A. Raleigh. 2001. Discovery and distribution of super-integrons among pseudomonads. Mol. Microbiol. 42:587-601. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, J. Z., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]