Abstract
The occurrence, structure, and mobility of Tn1546-like elements were studied in environmental vancomycin-resistant enterococci (VRE) isolated from municipal sewage, activated sludge, pharmaceutical waste derived from antibiotic production, seawater, blue mussels, and soil. Of 200 presumptive VRE isolates tested, 71 (35%) harbored vanA. Pulsed-field gel electrophoresis analysis allowed the detection of 26 subtypes, which were identified as Enterococcus faecium (n = 13), E. casseliflavus (n = 6), E. mundtii (n = 3), E. faecalis (n = 3), and E. durans (n = 1) by phenotypic tests and 16S ribosomal DNA sequencing. Long PCR-restriction fragment length polymorphism (L-PCR-RFLP) analysis of Tn1546-like elements and PCR analysis of internal regions revealed the presence of seven groups among the 29 strains studied. The most common group (group 1) corresponded to the structure of Tn1546 in the prototype strain E. faecium BM4147. Two novel L-PCR-RFLP patterns (groups 3 and 4) were found for E. casseliflavus strains. Indistinguishable Tn1546-like elements occurred in VRE strains belonging to different species or originating from different sources. Interspecies plasmid-mediated transfer of vancomycin resistance to E. faecium BM4105 was demonstrated for E. faecalis, E. mundtii, and E. durans. This study indicates that VRE, including species other than E. faecium and E. faecalis, are widespread in nature and in environments that are not exposed to vancomycin selection and not heavily contaminated with feces, such as seawater, blue mussels, and nonagricultural soil. Tn1546-like elements can readily transfer between enterococci of different species and ecological origins, therefore raising questions about the origin of these transposable elements and their possible transfer between environmental and clinical settings.
Glycopeptide antibiotic resistance of enterococci is considered a global threat to public health. This is because glycopeptide antibiotics, namely vancomycin and teicoplanin, are the drugs of choice and often the last option for the treatment of hospital infections caused by multiresistant gram-positive bacteria (29). The transferable nature of the gene clusters encoding high-level glycopeptide resistance in enterococci (i.e., vanA and vanB) has caused concern in the scientific community due to the possible emergence of vancomycin resistance in highly pathogenic bacteria such as Staphylococcus aureus, which was indeed reported for the first time in the United States in 2002 (17). At present, six gene clusters conferring glycopeptide resistance have been sequenced from enterococci (vanA to vanG) (19), with vanA being the most commonly encountered in clinical enterococci in Europe (2). The occurrence of VRE of the VanA type has been reported for sewage treatment plants in Denmark, Germany, Spain, Sweden, and the United States (4, 6, 7, 10, 22).
The structure of Tn1546, the transposable element associated with VanA glycopeptide resistance, has been widely studied with vancomycin-resistant enterococci (VRE) isolated from humans and animals in different geographical areas (8, 9, 16, 20, 21, 26, 27). However, information on the diversity of Tn1546-like elements in environmental enterococci is scarce and limited to the study of a few Enterococcus faecium isolates from sewage (21, 25, 27). Furthermore, very little information is available on interspecies transfer of such transposable elements.
This study was conceived to provide information on the occurrence, structure, and mobility of Tn1546-like elements in enterococci from different environmental samples. The diversity of Tn1546-like elements was investigated by using previously described techniques, such as long PCR-restriction fragment length polymorphism (L-PCR-RFLP), PCR analysis of internal regions, and restriction analysis of vanX. Filter mating experiments were performed to assess the mobility of Tn1546-like elements between enterococci of different species and ecological origins.
MATERIALS AND METHODS
Sample types and collection.
The sample types analyzed included sludge derived from vancomycin production, waste effluent derived from vancomycin and polymyxin B production, raw sewage, activated sludge, treated sewage, seawater, marine sediment, blue mussels, and soil. Samples were collected twice at each sampling site between October 2001 and August 2002. Sludge from vancomycin production originated from storage tanks at a Danish pharmaceutical plant, where such sludge was stored prior to transportation to a sewage treatment plant. Waste effluent from vancomycin and polymyxin B production was collected at the same pharmaceutical plant. Sewage was collected at four different sites situated upstream and downstream of the discharge points of the two waste effluents. Twenty-four-hour flow proportional samples of incoming raw sewage and outgoing treated sewage were collected at the sewage treatment plant receiving waste derived from vancomycin production (Lynetten, Denmark) and at a control plant not receiving such waste (Damhusaaen, Denmark). At Lynetten, raw sewage was collected at the south inlet, receiving vancomycin waste, and at the north inlet, which was not exposed to waste from vancomycin production. Activated sludge samples were collected from different biological tanks within each treatment plant and bulked together to obtain composite samples. Professional divers collected composite samples of surface marine water, sediment, and blue mussels at a distance between 10 and 150 m from the outlet of Lynetten. Core samples of agricultural soil previously treated with pig manure and nonagricultural soil which had not received animal manure over the last 30 years were collected at the research station of the Danish Institute of Agricultural Science in Flakkebjerg.
Isolation of presumptive VRE.
Presumptive VRE were isolated by direct plating on Slanetz-Bartley agar (SBA; Oxoid) supplemented with 20 μg of vancomycin/ml. For each sample, two to five colonies with different colony morphologies were subcultured on SBA with vancomycin, and the obtained pure cultures were stored in the freezer at −80°C prior to phenotypic and genotypic characterization. Membrane filtration of volumes of 0.1 and 1 liter of seawater and treated sewage was used to detect low numbers of VRE. For all sample types, presumptive VRE isolates were also obtained by selective enrichment in azide dextrose broth (ADB; Merck) supplemented with 20 μg of vancomycin/ml. After 2 to 3 days of incubation at 37°C, enrichment cultures were inoculated on SBA, and representative colonies with different morphologies were selected for further analysis. Ten-gram composite samples of marine sediment and blue mussels were mixed with 90 ml of physiological saline in a stomacher before inoculation onto SBA and enrichment in ADB. Soil composite samples were treated with Chelex-100 (Bio-Rad) and polyethylene glycol 6000 (Mallinckrodt Baker, Inc.) for soil dispersion and concentration of bacterial cells (24) before culturing. Four strains that were previously isolated from vancomycin sludge originating from the same pharmaceutical plant (4) were included in the study (see Table 2).
TABLE 2.
Environmental VRE in which the structure and mobility of Tn1546-like elements were analyzed
| Strain | Source | Species | PFGE type | Resistance patterna | Transfer of vancomycin resistance
|
|
|---|---|---|---|---|---|---|
| Approximate frequency | Plasmid detection | |||||
| 3b | Vancomycin sludge | E. faecium | A1 | Van | 1 × 10−4 | Yes |
| 69b | Vancomycin sludge | E. faecium | A2 | Van | Not tested | |
| 174b | Vancomycin sludge | E. faecium | A3 | Van | Not tested | |
| WE3 | Vancomycin sludge | E. faecium | A4 | Van | Not tested | |
| W5 | Vancomycin sludge | E. faecium | B1 | Van, Pen | 2 × 10−5 | Yes |
| W2 | Vancomycin sludge | E. faecium | B2 | Van, Pen | Not tested | |
| LB 7 | Activated sludge | E. faecium | B3 | Van, Pen | Not tested | |
| B2-S2 | Raw sewage | E. faecium | B4 | Van, Pen | Not tested | |
| ØME1 | Blue mussels | E. faecium | C | Van, Pen, Ery | 1 × 10−4 | Yesc |
| 68b | Vancomycin sludge | E. mundtii | D1 | Van, Ery | 7 × 10−3 | Yes |
| C2-W3 | Vancomycin effluent | E. mundtii | D2 | Van, Ery | Not tested | |
| B3-W8 | Raw sewage | E. mundtii | E | Van | 8 × 10−4 | Yesc |
| C1-S4 | Polymyxin effluent | E. faecium | F | Van | No transfer | |
| C1-S6 | Polymyxin effluent | E. faecium | G | Van | 3 × 10−5 | Yes |
| C1-W5 | Polymyxin effluent | E. durans | H | Van, Ery | 2 × 10−3 | Yes |
| C1-S2 | Polymyxin effluent | E. casseliflavus | I1 | Van, Pen | No transfer | |
| C2-W4 | Vancomycin effluent | E. casseliflavus | I2 | Van | No transfer | |
| A2/1-W1 | Raw sewage | E. casseliflavus | I3 | Van | Not tested | |
| A2/1-S1 | Raw sewage | E. casseliflavus | I4 | Van | No transfer | |
| B2-W5 | Raw sewage | E. casseliflavus | I5 | Van | No transfer | |
| C2-W2 | Vancomycin effluent | E. casseliflavus | K | Van | No transfer | |
| LSIE1 | Raw sewage | E. faecalis | J | Van, Pen | No transfer | |
| LSIE6 | Raw sewage | E. faecalis | L1 | Van | 5 × 10−6 | No |
| A2/2-S1 | Raw sewage | E. faecalis | L2 | Van | 1 × 10−7 | No |
| E2-F | Nonagricultural soil | E. faecium | M | Van | 6 × 10−5 | Yesc |
| E4-F | Agricultural soil | E. faecium | N | Van | No transfer | |
| C2-W1 | Vancomycin effluent | E. durans | —d | Van | 2 × 10−3 | Yes |
| B2-S5 | Raw sewage | E. mundtii | —d | Van, Ery | 2 × 10−4 | Yesc |
| E3-F | Agricultural soil | E. mundtii | —d | Van, Ery | 3 × 10−4 | No |
Van, vancomycin; Ery, erythromycin; Pen, penicillin.
Isolated in a previous study (4).
The plasmid was detected in the transconjugant but not in the donor strain.
—, not typeable by PFGE.
PCR detection of vanA.
All presumptive VRE isolates were analyzed by PCR for the occurrence of vanA (4). This screening allowed selection of VRE of the VanA type and excluded intrinsically resistant bacteria (e.g., lactic acid bacteria).
PFGE.
VanA-positive isolates were typed by pulsed-field gel electrophoresis (PFGE) according to the rapid protocol described by Turabelidze et al. (23), which includes bacterial lysis with lysozyme (2.5 mg/ml) and mutanolysin (1.250 U/ml) and DNA restriction digestion with SmaI (30 U for 2 h at 30°C). Strains differing by three or fewer bands were grouped into the same PFGE type (A, B, C, etc.) and were subdivided into different PFGE subtypes (A1, A2, A3, etc.) based on single-band differences. Cluster analysis was done with Gel Compar (Applied Maths, Kortrijk, Belgium), using the unweighted pair group method. Levels of similarity between PFGE patterns were calculated based on the Dice coefficient, with a position tolerance of 2%.
Identification and antimicrobial susceptibility testing.
Strains showing distinct PFGE patterns were identified at the species level by phenotypic tests and 16S ribosomal DNA (rDNA) sequencing. Phenotypic identification was done according to the scheme described by Manero and Blanch (11) with biochemical tests using the following reagents: arabinose, arginine, α-galactosidase, mannitol, methyl-α-d-glucopyranoside, ribose, sorbose, and pyrrolidonyl aminopeptidase. Carbohydrate fermentation tests were performed in a basal medium, with bromthymol blue as an acid-base indicator (1). Pyrrolidonyl aminopeptidase and α-galactosidase activities were evaluated by using the diagnostic kits of Rosco Diagnostica (Taastrup, Denmark). Antimicrobial susceptibility testing was done by the disk method described by the Swedish Reference Group for Antibiotics (14). The following six antimicrobial disks (Oxoid) were included: penicillin, ampicillin, chloramphenicol, ciprofloxacin, erythromycin, and tetracycline.
DNA templates for the amplification of 16S rDNA and further PCR analysis were prepared by using a High Pure PCR template preparation kit (Roche Diagnostics Corporation). PCR amplification of 16S rDNA was done by a PCR using the forward primer AGA GTT TGA TYM TGG CT (positions 9 to 25) and the reverse primer TAC GGY TAC CTT TGT TAC GAC T (positions 1493 to 1513) (17). DNA sequencing was done on an ABI 377 sequencer with DNA Technology A/S (Aarhus, Denmark). The sequences were analyzed by BLAST for homologies with sequences in the GenBank database.
Molecular analysis of Tn1546-like elements.
The structures of Tn1546-like elements were analyzed by L-PCR-RFLP with strains representative of distinct PFGE subtypes as well as strains that could not be typed by PFGE. Long-range PCR amplification of Tn1546-like elements was performed with an Expand long-template PCR system (Boehringer-Mannheim) using a single primer targeting the inverted repeat sequence of Tn1546 (5′-GGA AAA TGC GGA TTT ACA ACG CTA AG-3′) (28). Restriction analysis of Tn1546 PCR amplicons was done with ClaI, HpaI, and BamHI. PCR amplification of the proximal and distal regions of ORF1, vanS-vanH, vanX, vanY-vanZ, and vanZ was performed according to the method of Simonsen et al. (21). PCR amplicons of vanX were digested with DdeI for detection of a point mutation at position 8234 (8). Tn1546-like elements that could be differentiated by L-PCR-RFLP or PCR analysis of internal regions were assigned to different groups (group 1, 2, 3, etc.). Letters were used for the designation of subgroups of Tn1546-like elements showing the same L-PCR-RFLP patterns and PCR products, but differing in the vanX point mutation (e.g., 1a and 1b).
Filter mating experiments.
The mobility of Tn1546-like elements was studied by the filter mating method, with 21 selected strains representing different species and PFGE types as donors. A rifampin- and fusidic acid-resistant mutant of E. faecium BM4105 was used as a recipient. Briefly, 0.5-ml samples of donor and recipient overnight cultures were mixed, and 0.1 ml of the mixture was plated onto a cellulose nitrate filter with a 0.45-μm pore size (Millipore) placed on brain heart infusion (BHI; Difco) agar. After overnight incubation at 37°C, filters were vortexed in 5 ml of physiological saline. Serial dilutions were then plated on BHI agar containing 20 μg of vancomycin/ml, 50 μg of rifampin/ml, and 10 μg of fusidic acid/ml for the enumeration of transconjugants and on BHI agar containing rifampin and fusidic acid only for the enumeration of the recipient strain. Overnight cultures of donor strains were inoculated onto control plates containing rifampin and fusidic acid.
Plasmid profiling and hybridization.
Two presumptive transconjugant colonies were picked up for each mating pair showing transfer of vancomycin resistance, and their plasmid profiles were determined by a hot alkaline method (5) for evaluation of the possible acquisition of plasmids. Plasmid profiles were transferred onto nylon membranes by Southern blotting and were hybridized with a digoxigenin-labeled probe made from the vanA amplicon by using a random primed DNA labeling kit (Roche). Hybridization was performed according to the DIG System User's Guide for Filter Hybridization (Boehringer Mannheim).
RESULTS
Sludge derived from vancomycin production contained high numbers of presumptive VRE (∼109 CFU/ml), and most isolates obtained from this sample type (94%) contained vanA. Numbers of presumptive VRE in sewage ranged between 10 and 104 CFU/ml depending on the sampling time and site. Although numbers of presumptive VRE were similar in sewage and activated sludge from the two sewage treatment plants, vanA was only detected among isolates from Lynetten, the plant receiving sludge from vancomycin production (Table 1). VRE of the VanA type also occurred in waste effluent from vancomycin and polymyxin B production, sewage collected upstream and downstream of the pharmaceutical plant, seawater, blue mussels, and agricultural and nonagricultural soil, but could not be detected in treated sewage and marine sediments (Table 1). Due to low numbers (<10 CFU/ml), the detection of VRE in blue mussels and soil was not possible by direct plating on SBA and required the use of selective enrichment in ADB containing vancomycin.
TABLE 1.
Counts of presumptive VRE and occurrence of vanA-positive isolates in different environmental samples
| Sample type | Source | Total no. (CFU/ml or g [%]) of presumptive VREa |
vanA PCR
|
|
|---|---|---|---|---|
| No. of tested isolates | No. (%) of positive isolates | |||
| Vancomycin sludge | Pharmaceutical plant | 109 (>100) | 16 | 15 (94) |
| Vancomycin effluent | Pharmaceutical plant | 103 (<1-100) | 11 | 9 (82) |
| Polymyxin effluent | Pharmaceutical plant | <10-103 (11-28) | 11 | 9 (82) |
| Raw sewage | Lynetten south inlet (exposed to vancomycin waste) | 103-104 (≤1) | 18 | 3 (17) |
| Raw sewage | Lynetten north inlet (not exposed to vancomycin waste) | 102-104 (≤1) | 16 | 0 |
| Raw sewage | Damhusaaen (not exposed to vancomycin waste) | 103 (≤1) | 15 | 0 |
| Raw sewage | Pharmaceutical plant (upstream) | <10-103 (<1-87) | 13 | 9 (69) |
| Raw sewage | Pharmaceutical plant (downstream) | 102-104 (<1-45) | 27 | 12 (44) |
| Activated sludge | Lynetten (exposed to vancomycin waste) | 103-104 (1-16) | 14 | 7 (50) |
| Activated sludge | Damhusaaen (not exposed to vancomycin waste) | 103 (≤1) | 14 | 0 |
| Treated sewage | Lynetten | 10−1-10 (1) | 14 | 0 |
| Treated sewage | Damhusaaen | 10−1 (1) | 12 | 0 |
| Seawater | Lynetten outlet | ≤10−2 (2) | 8 | 2 (25) |
| Sediment | Lynetten outlet | <10 ND | 3 | 0 |
| Blue mussels | Lynetten outlet | <10 ND | 4 | 2 (50) |
| Soil | Agricultural field (exposed to animal manure) | <10 ND | 2 | 2 (100) |
| Soil | Nonagricultural field (not exposed to animal manure) | <10 ND | 2 | 2 (100) |
Ranges of total numbers obtained from two bacteriological counts are reported for each sample type. For vancomycin sludge, numbers of colonies were higher on plates with vancomycin than on plates without vancomycin, therefore resulting in VRE percentages above 100%. ND, VRE were not detected by direct plating on SBA with vancomycin.
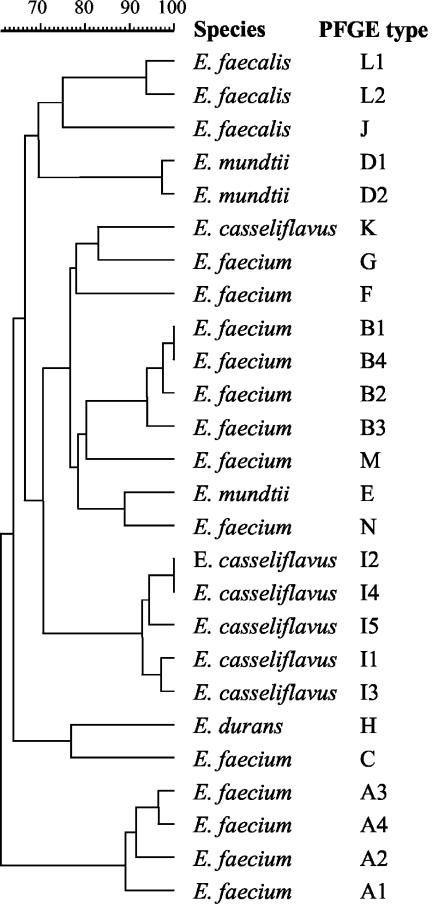
Of 200 presumptive VRE isolates tested, 71 (35%) harbored vanA. vanA-positive isolates were differentiated into 14 types (A to N) and 26 subtypes by PFGE analysis (Fig. 1). Three VRE strains showed no bands after repeated PFGE and were included in further analyses (Table 2). PFGE subtypes A4 and B1 were found in both activated sludge from the Lynetten plant and sludge derived from vancomycin production (Fig. 2). VRE isolates from seawater and blue mussels collected in proximity of the sewage outlet from Lynetten showed a different PFGE type (C). PFGE types F and G were prevalent among VRE isolates from the two antibiotic waste effluents and sewage collected upstream and downstream of the discharge points of such effluents. The distribution of PFGE types in VRE isolates from different sites is shown in Table 3.
FIG. 1.
Dendrogram of 26 environmental VRE typed by PFGE. Levels of similarity were determined based on the Dice coefficient by using the unweighted pair group method and a position tolerance of 2%.
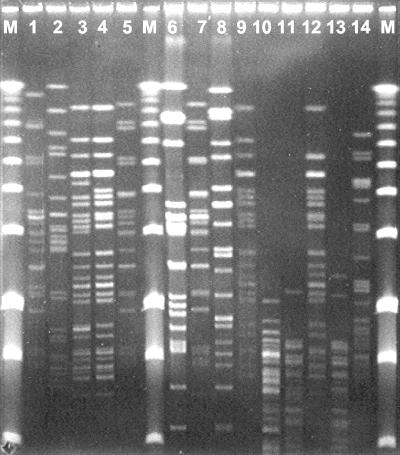
FIG. 2.
Representative PFGE patterns among VRE isolated from sludge derived from the industrial production of vancomycin (lanes 1 to 5), from the sewage treatment plant receiving such sludge (lanes 6 to 13), and from blue mussels collected in the proximity of the sewage effluent (lane 14). Lanes M, molecular weight marker; lane 1, E. faecium strain 3 (PFGE type A1); lane 2, E. mundtii strain 68 (PFGE type D); lane 3, E. faecium strain W2 (PFGE type B2); lane 4, E. faecium strain W5 (PFGE type B1); lane 5, E. faecium strain WE3 (PFGE type A4); lane 6, E. faecalis strain LSIE1 (PFGE type J); lane 7, E. faecium strain LSI5 (PFGE type A4); lane 8, E. faecalis strain LSIE6 (PFGE type L1); lane 9, E. faecium strain LB1 (PFGE type B1); lane 12, E. faecium strain LB7 (PFGE type B3); lane 14, E. faecium strain ΦME1 (PFGE type C); lanes 10, 11, and 13, VanA-negative strains of Pediococcus acidilattici.
TABLE 3.
Distribution of PFGE types in different environmental samplesa
| Sample type | Source | No. of isolates of PFGE type
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
E. faecium
|
E. durans
|
E. casseliflavus
|
Otherb | ||||||
| A | B | C | F | G | H | I | |||
| Vancomycin sludge | Pharmaceutical plant | 1 | 14 | 0 | 0 | 0 | 0 | 0 | 0 |
| Vancomycin effluent | Pharmaceutical plant | 0 | 0 | 0 | 0 | 1 | 3 | 2 | 3 (3) |
| Polymyxin effluent | Pharmaceutical plant | 0 | 0 | 0 | 1 | 4 | 3 | 1 | 0 |
| Raw sewage | Lynetten plant | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (2) |
| Raw sewage | Pharmaceutical plant (upstream) | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 5 (2) |
| Raw sewage | Pharmaceutical plant (downstream) | 2 | 2 | 0 | 0 | 0 | 3 | 2 | 3 (2) |
| Activated sludge | Lynetten plant | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| Seawater | Lynetten outlet | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Blue mussels | Lynetten outlet | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Soil | Flakkebjerg | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (3) |
PFGE patterns differing by three or fewer bands were included in the same PFGE type.
Numbers of isolates belonging to other PFGE types. The numbers of distinct PFGE types are shown in parentheses.
There was accordance between phenotypic and 16S rDNA-based identification, with biochemical tests confirming the most likely species indicated by 16S rDNA sequences. The strains selected based on PFGE results were identified as E. faecium (n = 13), E. casseliflavus (n = 6), E. mundtii (n = 5), E. faecalis (n = 3), and E. durans (n = 2). All strains were generally susceptible to antimicrobials other than glycopeptides, except for penicillin and/or erythromycin (Table 2).
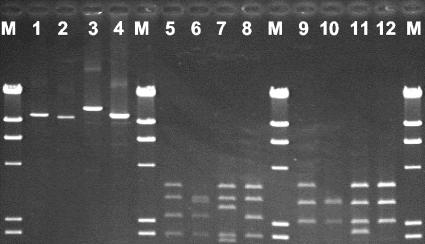
L-PCR-RFLP of Tn1546-like elements and PCR analysis of internal regions revealed the presence of seven groups among the 29 strains tested (Table 4). Twelve strains of five enterococcal species contained the same structure of Tn1546 as the prototype strain E. faecium BM4147 (group 1). Nine strains representative of four species showed the same L-PCR-RFLP pattern accompanied with mutations in the proximal ORF1 primer binding region, as indicated by the failure of PCR amplification of such a region. Different L-PCR-RFLP patterns (groups 3 and 4) were found for two E. casseliflavus strains, one of which was characterized by an insertion of approximately 12,000 bp in the vanY-vanZ region. The remaining groups (5, 6, and 7) could not by typed by L-PCR-RFLP and were differentiated by PCR amplification of the proximal and distal regions of ORF1 (Table 4). Amplicons and restriction profiles of Tn1546-like elements that could be typed by L-PCR-RFLP are shown in Fig. 3. The point mutation G→T in vanX at position 8234 was present in E. faecium and E. mundtii strains from different environmental sources and was not associated with a particular group of Tn1546-like elements (Table 4). The mutation was present in all strains isolated from sludge derived from vancomycin production, including different species (E. faecium and E. mundtii), PFGE types (A, B, and D), and groups of Tn1546-like elements (1, 2, and 5).
TABLE 4.
Distribution and structure of distinct groups of Tn1546-like elements in environmental VRE
| Group | Species (PFGE subtypes) | Source of isolation | Tn1546 PCR-RFLPa
|
vanX RFLPb | PCR of polymorphic regionsa
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| L-PCR | RFLP pattern | ORF1 11c | ORF1 21c | VanSH | VanYZ | VanZ | ||||
| 1a | E. faecium (A) | Vancomycin sludge, sewage | + | 1 | T | + | + | + | + | + |
| 1b | E. faecium (M, N), E. faecalis (J, L), E. casseliflavus (I5, K), E. durans (untypeable) | Vancomycin effluent, sewage, soil | + | 1 | G | + | + | + | + | + |
| 2a | E. faecium (B) | Vancomycin sludge, sewage | + | 1 | T | − | + | + | + | + |
| 2b | E. faecium (F), E. mundtii (E), E. casseliflavus (I3), E. durans (H) | Polymyxin effluent, sewage | + | 1 | G | − | + | + | + | + |
| 3 | E. casseliflavus (I2) | Vancomycin effluent | + | 2 | G | − | + | + | + | + |
| 4 | E. casseliflavus (I4) | Sewage | 12,000 | 3 | G | − | + | + | 2,500 | + |
| 5 | E. faecium (C), E. mundtii (D) | Blue mussels, vancomycin sludge, vancomycin effluent | − | T | − | + | + | + | + | |
| 6 | E. mundtii (not typeable) | Sewage, soil | − | T | + | + | + | + | + | |
| 7 | E. faecium (G) | Polymyxin effluent | − | T | − | − | + | + | + | |
+, presence of a PCR product of the expected size in Tn1546; −, no PCR product was detected. The detection of PCR products of other sizes than expected is indicated by the approximate size (bp) of these products.
Detection of the point mutation G→T in vanX at position 8,234.
FIG. 3.
Long-range PCR amplicons and RFLP profiles of Tn1546-like elements from environmental VRE. Lanes M, phage λ digested with HindIII; lanes 1 to 4, long-range PCR amplicons of E. faecium 3 (lane 1), E. casseliflavus C2-W4 (lane 2), E. casseliflavus A2/1-S1 (lane 3), and E. faecium BM4147 (lane 4); lanes 5 to 8, RFLP profiles of the strains shown in lanes 1 to 4 following digestion with ClaI; lanes 9 to 12, RFLP profiles of the strains shown in lanes 1 to 4 following digestion with HpaI and BamHI.
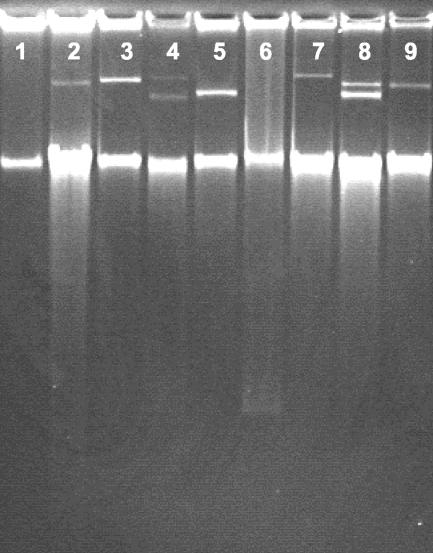
Interspecies transfer of vancomycin resistance to E. faecium BM4105 was demonstrated for E. mundtii, E. faecalis, and E. durans, but not for E. casseliflavus (Table 2). Of 21 donor strains tested, 13 strains were able to transfer vancomycin resistance, with frequencies varying between 10−3 and 10−7 transconjungants per recipient (Table 2). The transfer of vancomycin resistance was generally mediated by plasmids (Table 2). After repeated plasmid profiling, plasmid DNA could not be detected in the transconjugants obtained from three donor strains. In three cases, the presence of plasmids was demonstrated for the transconjugants, but not for the corresponding donor strains (see example in Fig. 4). All plasmids detected in transconjugants hybridized with the vanA probe (data not shown).
FIG. 4.
Examples of plasmid profiles in transconjugants obtained after filter mating of environmental VRE with the recipient strain E. faecium BM4105. In some cases, the plasmid acquired by the transconjugant could not be detected for the corresponding donor strain (e.g., lanes 6 and 7). The intense lower bands in the gel are chromosomal DNA. Lane 1, recipient E. faecium BM4105; lane 2, E. faecium strain 3 isolated from vancomycin sludge; lane 3, transconjugant obtained from strain 3; lane 4, E. faecium strain W5 isolated from vancomycin sludge; lane 5, transconjugant obtained from strain W5; lane 6, E. faecium strain ΦME1 isolated from blue mussels; lane 7, transconjugant obtained from strain ΦME1; lane 8, E. mundtii strain 68 isolated from vancomycin sludge; lane 9, transconjugant obtained from strain 68.
DISCUSSION
According to the results of our investigation, VRE of the VanA type are widespread in nature, including habitats that have not been exposed to vancomycin selection and are not heavily contaminated with feces. We report for the first time the occurrence of vanA in soil, seawater, and blue mussels. Strains isolated from soil harbored Tn1546-like elements whose structures were indistinguishable from the structure of Tn1546 in the prototype strain E. faecium BM4147 (group 1), which corresponds to group A according to the scheme proposed by Palepou et al. (16). The source of VRE in soil, especially in nonagricultural soil, remains unknown. Nonagricultural soil was reported not to have been exposed to any type of organic fertilizer in the last 30 years. Agricultural soil was treated with pig manure 6 months before sampling. However, pig manure was unlikely to be the source of VRE isolated from both agricultural and nonagricultural soil, as the VRE isolates lacked the point mutation in vanX that is usually present in VRE isolates from pigs in Denmark (8).
Tn1546-like elements were found in five different enterococcal species, namely E. faecium, E. faecalis, E. mundtii, E. casseliflavus, and E. durans. Some strains had mutations in the ORF1 primer binding region. Mutations in this region could explain the failure of L-PCR amplification observed with some strains, as previously described for VRE of human and animal origin (16, 21). Based on a comparison with the schematic representation described by Palepou et al. (16), group 2 in this study corresponds to group F, whereas groups 3 and 4 found in E. casseliflavus strains do not correspond to any of the groups described by these authors. Structural rearrangements that were previously described for the vanS-vanH and vanZ regions were not found among environmental VRE. The structure of Tn1546-like elements in environmental VRE was not associated with species and sample type, since the same structure was observed for strains of different species and ecological origins (Tables 2 and 4). This suggests that interspecies horizontal transfer of Tn1546-like elements is likely to occur in nature. Indeed, vancomycin resistance was readily transferred under laboratory conditions from E. faecalis, E. mundtii, and E. durans to E. faecium. On the contrary, Tn1546-like elements could not be mobilized from E. casseliflavus to E. faecium, indicating that horizontal transfer between these two species is a less likely event.
The recovery of VRE of the VanA type from natural habitats raises questions about the ecology of enterococci. It is generally assumed that enterococci are fecal bacteria with an excellent ability for adaptation to adverse environmental conditions. However, enterococci could also be environmental bacteria that subsequently adapted to the intestinal tracts of humans and animals. Recent studies have demonstrated that enterococci, including species that are usually associated with human and animal feces, such as E. faecium and E. faecalis, are normal inhabitants of the phyllosphere of grasses (13, 15). The ecology of enterococci has important implications in relation to the origins of glycopeptide resistance. Various authors have suggested that gene clusters encoding high-level glycopeptide resistance (i.e., vanA and vanB) could originate from self-protecting genes in antibiotic-producing soil bacteria (3, 12, 18). Thus, environmental enterococci could have played an important role in the evolution of glycopeptide resistance by acting as intermediate hosts in the transfer of resistance genes between soil bacteria and enterococci living in warm-blooded animals. This question may be answered by further investigations on the occurrence and diversity of glycopeptide resistance genes in soil bacterial communities, including both enterococcal and nonenterococcal species.
A previous study by Guardabassi et al. (4) showed epidemiological evidence of the dissemination of VRE by the disposal of vancomycin sludge originating from the pharmaceutical plant investigated in this study. The present study confirms the hypothesis that the disposal of such sludge is an important source for the occurrence of VRE in the recipient sewage treatment plant. Although the tanks where the sludge was stored at the pharmaceutical plant were periodically washed and disinfected in the period between the previous and the present study, vancomycin sludge contained 1,000-fold higher concentrations of VRE than was reported in the previous study (106 CFU/ml), indicating that disinfection of the storage tanks was unable to prevent sludge contamination with VRE. A likely source for VRE was fecal contamination, e.g., through connecting hoses from trucks collecting the sludge, as suggested by the observed presence of different strains of Enterobacteriaceae in the sludge (data not shown). As in the previous study, identical PFGE patterns were observed among VRE isolates from vancomycin sludge and those from the sewage treatment plant receiving such sludge (Fig. 2). Furthermore, the association between the occurrence of VRE at the sewage treatment plant and the disposal of vancomycin sludge was confirmed by the lack of detection of vanA isolates at control sites that were not exposed to such waste (Table 1).
VRE of the VanA type also occurred in the two waste effluents from vancomycin and polymyxin B production. The high variability in the total and relative numbers of VRE in both antibiotic waste effluents and their recipient sewers (Table 1) suggests that the occurrence of VRE at these sites was influenced by the production cycles at the pharmaceutical plant. The discharge of antibiotic waste effluent from the pharmaceutical plant did not seem to contribute to the occurrence of VRE at the Lynetten sewage treatment plant, since the PFGE types occurring in the two waste effluents differed from those found at the plant (Table 3). The occurrence of VRE in seawater and blue mussels from the proximity of the treated sewage effluent of Lynetten was not associated with the disposal of vancomycin waste, as VRE isolates from these samples had different PFGE profiles (Table 3 and Fig. 2) and a distinct group of Tn1546-like elements (Table 4). However, the presence of VRE in this marine environment could be enhanced by vancomycin residues occurring in the sewage effluent from the Lynetten plant, where large amounts of vancomycin sludge with high concentrations of active vancomycin (0.1 to 1 mg/ml) are disposed (4). More research is necessary in this field to assess the impact of waste from the industrial production of antibiotics on the spread of antibiotic resistance and to determine whether specific regulations on the disposal of such waste are needed.
A comparison of the numbers of presumptive VRE at the two sewage treatment plants was of limited value without the support of genotypic analysis (Table 1). Counts of presumptive VRE in sewage provide little useful information about the actual occurrence of VRE due to the presence of intrinsically resistant bacteria (e.g., lactic acid bacteria) that are able to grow on media that are selective for enterococci. When presumptive VRE were counted in sludge from vancomycin production, the numbers of colonies on SBA with vancomycin were higher than those observed on SBA without vancomycin (Table 1). This result reflects a situation in which bacteria present in the sludge are exposed to high concentrations of vancomycin (4) and are therefore all resistant to vancomycin. The higher numbers of colonies on SBA containing vancomycin could be due to better growth of these bacteria in the presence of vancomycin. Selective isolation on media containing vancomycin is necessary for the detection of VRE in environmental samples because VRE usually represent a small proportion of the total enterococcal population (Table 1). For certain sample types, i.e., soil and blue mussels, VRE could only be detected by selective enrichment. Accordingly, the nonuse of selective procedures of isolation could explain the absence of previously reported VRE in samples of environmental origin (13, 15).
In conclusion, this investigation demonstrates that Tn1546-like elements occur in environmental enterococci belonging to different species and living in different ecological niches. The structures of Tn1546-like elements in environmental VRE are generally indistinguishable from those occurring in human and animal VRE, indicating a possible flow of such transposable elements through different enterococcal species and populations. The mobility of Tn1546-like elements in the environment, associated with the remarkable ability of enterococci to adapt to adverse environmental conditions, could explain the rapid spread of VRE observed in the last 2 decades.
Acknowledgments
This work was supported by the Danish Agricultural and Veterinary Research Council (project no. 23-01-0170).
We thank Kirstina Marie Holm for excellent technical assistance, Kim Rindel (Lynettefælleskabet I/S) and André Koefoed (Koebenhavns Miljoekontrol) for providing samples of sewage and pharmaceutical waste, and Mogens Greve (Danish Institute of Agricultural Science) for making possible the collection of soil samples at the research station in Flakkebjerg, Denmark.
REFERENCES
- 1.Angen, O., B. Aalbaek, E. Falsen, J. E. Olsen, and M. Bisgaard. 1997. Relationships among strains classified with the ruminant Pasteurella haemolytica-complex using quantitative evaluation of phenotypic data. Zentbl. Bakteriol. 285:459-479. [DOI] [PubMed] [Google Scholar]
- 2.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gholizadeh, Y., and P. Courvalin. 2000. Acquired and intrinsic glycopeptide resistance in enterococci. Int. J. Antimicrob. Agents 16:S11-S17. [DOI] [PubMed] [Google Scholar]
- 4.Guardabassi, L., P. T. Bronnum, R. Dano, A. Forslund, and A. Dalsgaard. 2002. Dissemination of vancomycin-resistant enterococci harboring vanA through disposal of waste derived from industrial production of vancomycin. Microb. Drug Resist. 8:401-406. [DOI] [PubMed] [Google Scholar]
- 5.Guardabassi, L., L. Dijkshoorn, J. M. Collard, J. E. Olsen, and A. Dalsgaard. 2000. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 49:929-936. [DOI] [PubMed] [Google Scholar]
- 6.Harwood, V. J., M. Brownell, W. Perusek, and J. E. Whitlock. 2001. Vancomycin-resistant Enterococcus spp. isolated from wastewater and chicken feces in the United States. Appl. Environ. Microbiol. 67:4930-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iversen, A., I. Kuhn, A. Franklin, and R. Mollby. 2002. High prevalence of vancomycin-resistant enterococci in Swedish sewage. Appl. Environ. Microbiol. 68:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen, L. B. 1998. Differences in the occurrence of two base pair variants of Tn1546 from vancomycin-resistant enterococci from humans, pigs, and poultry. Antimicrob. Agents Chemother. 42:2463-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen, L. B., P. Ahrens, L. Dons, R. N. Jones, A. M. Hammerum, and F. M. Aarestrup. 1998. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J. Clin. Microbiol. 36:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klare, I., H. Heier, H. Claus, and W. Witte. 1993. Environmental strains of Enterococcus faecium with inducible high-level resistance to glycopeptides. FEMS Microbiol. Lett. 106:23-30. [DOI] [PubMed] [Google Scholar]
- 11.Manero, A., and A. R. Blanch. 1999. Identification of Enterococcus spp. with a biochemical key. Appl. Environ. Microbiol. 65:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall, C. G., I. A. D. Lessard, I. S. Park, and G. D. Wright. 1998. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42:2215-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller, T., A. Ulrich, E. M. Ott, and M. Muller. 2001. Identification of plant-associated enterococci. J. Appl. Microbiol. 91:268-278. [DOI] [PubMed] [Google Scholar]
- 14.Olsson-Liljequist, B., P. Larsson, M. Walder, and H. Miorner. 1997. Antimicrobial susceptibility testing in Sweden. III. Methodology for susceptibility testing. Scand. J. Infect. Dis. 105:13-23. [PubMed] [Google Scholar]
- 15.Ott, E. M., T. Muller, M. Muller, C. M. A. P. Franz, A. Ulrich, M. Gabel, and W. Seyfarth. 2001. Population dynamics and antagonistic potential of enterococci colonizing the phyllosphere of grasses. J. Appl. Microbiol. 91:54-66. [DOI] [PubMed] [Google Scholar]
- 16.Palepou, M. F. I., A. M. A. Adebiyi, C. H. Tremlett, L. B. Jensen, and N. Woodford. 1998. Molecular analysis of diverse elements mediating VanA glycopeptide resistance in enterococci. J. Antimicrob. Chemother. 42:605-612. [DOI] [PubMed] [Google Scholar]
- 17.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of campylobacters, Wolinella, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic-acid sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 18.Patel, R. 2000. Enterococcal-type glycopeptide resistance genes in non-enterococcal organisms. FEMS Microbiol. Lett. 185:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Pootoolal, J., J. Neu, and G. D. Wright. 2002. Glycopeptide antibiotic resistance. Annu. Rev. Pharmacol. Toxicol. 42:381-408. [DOI] [PubMed] [Google Scholar]
- 20.Robredo, B., C. Torres, K. V. Singh, and B. E. Murray. 2000. Molecular analysis of Tn1546 in vanA-containing Enterococcus spp. isolated from humans and poultry. Antimicrob. Agents Chemother. 44:2588-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonsen, G. S., M. R. M. Myhre, K. H. Dahl, O. Olsvik, and A. Sundsfjord. 2000. Typeability of Tn1546-like elements in vancomycin-resistant enterococci using long-range PCRs and specific analysis of polymorphic regions. Microb. Drug Resist. 6:49-57. [DOI] [PubMed] [Google Scholar]
- 22.Torres, C., J. A. Reguera, M. J. Sanmartin, J. C. Perezdiaz, and F. Baquero. 1994. VanA-mediated vancomycin-resistant Enterococcus spp. in sewage. J. Antimicrob. Chemother. 33:553-561. [DOI] [PubMed] [Google Scholar]
- 23.Turabelidze, D., M. Kotetishvili, A. Kreger, J. G. Morris, and A. Sulakvelidze. 2000. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. J. Clin. Microbiol. 38:4242-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellington, E. M. H., P. Marsh, J. E. M. Watts, and J. Burden. 1997. Indirect approaches for studying soil microorganisms based on cell extraction and culturing, p. 311-329. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, N.Y.
- 25.Werner, G., I. Klare, and W. Witte. 1997. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol. Lett. 155:55-61. [DOI] [PubMed] [Google Scholar]
- 26.Willems, R. J. L., J. Top, N. van den Braak, A. van Belkum, D. J. Mevius, G. Hendriks, M. Santen-Verheuvel, and J. D. A. van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodford, N., A. M. A. Adebiyi, M. F. I. Palepou, and B. D. Cookson. 1998. Diversity of VanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob. Agents Chemother. 42:502-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodford, N., A. P. Watson, and P. R. Chadwick. 1997. Investigation by long PCR of the genetic elements mediating VaNa glycopeptide resistance in enterococci from uncooked meat in South Manchester. Adv. Exp. Med. Biol. 418:409-412. [DOI] [PubMed] [Google Scholar]
- 29.Ziglam, H. M., and R. G. Finch. 2001. Limitations of presently available glycopeptides in the treatment of gram-positive infection. Clin. Microbiol. Infect. 7:53-65. [DOI] [PubMed] [Google Scholar]