Abstract
Three mutations on the penicillin acylase surface (increasing the number of Lys in a defined area) were performed. They did not alter the enzyme's stability and kinetic properties; however, after immobilization on glyoxyl-agarose, the mutant enzyme showed improved stability under all tested conditions (e.g., pH 2.5 at 4°C, pH 5 at 60°C, pH 7 at 55°C, or 60% dimethylformamide), with stabilization factors ranging from 4 to 11 compared with the native enzyme immobilized on glyoxyl-agarose.
A new strategy to improve enzyme stability that consists of using site-directed mutagenesis not to directly improve enzyme activity or stability but to increase the multipoint covalent attachment is proposed in this study. Thus, by using glyoxyl-agarose supports (3, 8) and by increasing the lysine content of the lysine-rich regions of the enzyme surface, we intend to achieve a more intense multipoint attachment of the penicillin G acylase (PGA) to the support, which contributes to improving its overall rigidity and, therefore, its stability against any inactivating agent (2). The advantage of this strategy compared to strategies directed at stabilizing the structure of the soluble enzyme via protein engineering (13) is that, in contrast to the high unpredictability of the amino acid changes required to enhance enzyme stability in solution, it provides an easy way to determine the amino acid changes that could be introduced in the protein surface to improve enzyme immobilization without altering the enzyme's properties. Moreover, whereas changes that stabilize enzyme structure might provide only limited stability under specific conditions, the overall rigidification achieved by this approach should render an enzyme stable under any adverse conditions (e.g., high temperature, organic solvents, or extreme pH).
Molecular dynamics of PGA enzyme structure.
Models of the different mutants were built on the basis of the crystal structure of PGA (9). Amino acid changes were introduced by using the O graphic program (10) from a database of more common conformers (12). Models were energy minimized by a slow-cooling protocol by simulated annealing (5) starting at 2,500 K as implemented in XPLOR (4).
DNA manipulation and protein purification.
The pac gene was amplified by PCR with the primers TOPO5′ (5′-ATGAAAAATAGAAATCGTATGA-3′) and TOPO3′ (5′-TACAACCTCCCGACCAATGAAA-3′), and Escherichia coli ATCC 11105 DNA (6) was used as a template. The PCR product was cloned into the vector T101/D-TOPO (Invitrogen) and the resulting plasmid pOAF was expressed in E. coli BL21(DE3). Site-directed mutagenesis of the pac gene was performed in two steps by a PCR megaprimer method (1, 15) with TOPO5′ and TOPO3′ as external primers. The mutagenic primers MUTb13-5′ (5′-CGGCAAAAGCAAAGCCCAGAAAGCGAAAGCAATCATGG-3′) and MUTb13-3′ (5′-CCATGATTGCTTTCGCTTTCTGGGCTTTGCTTTTGCCG-3′) wereused for constructing the first pac mutant (Asp13Lys). With this mutant used as a template, the triple pac mutant containing the additional mutations Glu272Lys and Arg276Lys was constructed with the primers MUTb272-276-5′ (5′-GAGATCGACCGACTGCTTAAACAAAAGCCAAAATTAACTGCTGATCAGCATG-3′) and MUTb272-276-3′ (5′-CATGCCTGATCAGCAGTTAATTTTGGCT TTTGTTTAAGCAGTCGGTCGATCTCC-3′). The triple pac mutant was cloned into plasmid pT101/D-TOPO, and the resulting plasmid pOAF3mut was expressed in E. coli BL21(DE3). Transformants were incubated at 22°C in Luria-Bertani medium containing ampicillin (0.1 mg/ml) and 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (14), and cells were broken by sonication. The wild-type and mutant PGA enzymes were purified by fast-performance liquid chromatography with a Q-Sepharose high-performance column (Pharmacia) and a gradient of 0 to 100 mM NaCl in 50 mM Tris-HCl, pH 8.5.
Studies on the activity and stability of PGA-immobilized derivatives.
The stability of PGA-immobilized derivatives is expressed as the initial rate of inactivation of the different enzyme preparations. The initial inactivation rate, defined as Ki, is the percentage of enzyme activity lost per hour under defined experimental conditions. This value was calculated based on the time when the activity loss reached 20%. The stabilization factor was calculated as the ratio between the initial rate of inactivation of the reference and that of the studied derivative. Experiments were carried out in triplicate, and standard errors were never over 2%. Activity was determined by using 6-nitro-3-phenylacetamidebenzoic acid (5 mM) at pH 7.5 and 25°C (11) or penicillin G (10 mM) at pH 8 and 25°C.
Determination of suitable mutations.
We decided to focus our attention on a region located just opposite an active center very rich in Lys. To avoid altering the properties of the enzymes, two types of amino acid replacements were analyzed: (i) conservative mutations such as replacing arginine by lysine or (ii) nonconservative mutations such as replacing acidic residues (glutamic or aspartic acids) with lysine by using groups not involved in salt bridges. Moreover, the rationale for proposing the substitution of acidic amino acids in PGA was derived from a previous study in which it was demonstrated that all external glutamic and aspartic residues of PGA could be modified with ethylenediamine (a much more dramatic modification) without having a significant effect on PGA activity (7). In accordance with this strategy, we located in this region three residues (Aspβ13, Gluβ272, and Argβ276) that were suitable for our purpose.
When both wild-type PGA and mutant PGA structures were compared, no significant folding differences were found, even in the regions close to the mutations.
Effect of mutations on the characteristics of the soluble enzyme.
The specific activities and Km of both pure enzymes were very similar (respectively, 50 ± 2 U/mg and 25 ± 2 μM for the native PGA and 46 ± 2 U/mg and 28 ± 2 μM for the mutant enzyme). The thermal stability of both enzymes at different pH values (from pH 5 to pH 10) was also comparable.
Properties of native and mutant PGA enzymes immobilized on CNBr-Sepharose or on glyoxyl-agarose.
Both the wild-type and mutant PGA enzymes were immobilized on CNBr-Sepharose (as a reference). Stability (Fig. 1) and kinetic constants of both enzymes remained parallel after this immobilization.
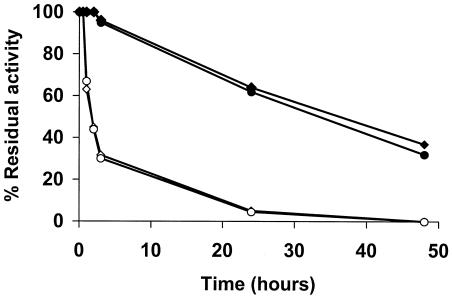
FIG. 1.
Thermal stability of CNBr-Sepharose derivatives of wild-type and mutant PGA enzymes. Thermal stability was determined by assaying the residual activity of the CNBr-Sepharose derivatives of wild-type PGA (◊ and ⧫) and mutant PGA (○ and •) after incubation at 50°C in 50 mM sodium phosphate buffer, pH 7.0 (⧫ and •), or at 55°C in 50 mM sodium acetate buffer, pH 5.0 (◊ and ○).
Immobilization on glyoxyl-agarose occurred in less than 10 min and allowed enzyme activity to be preserved. With this support, the immobilized mutant enzyme was always more stable than the immobilized wild-type PGA under various inactivation conditions: at different pH values (e.g., at pH 5 and 60°C) (Fig. 2) or with organic solvents (Table 1). Bearing in mind that the native immobilized enzyme was 10,000-fold more stable than the soluble enzyme (8), we can assume that the new mutant PGA derivative showed a half-life that was 100,000-fold higher than that of the soluble wild-type enzyme. Both immobilized enzymes presented similar activity and pH profiles (Fig. 3).
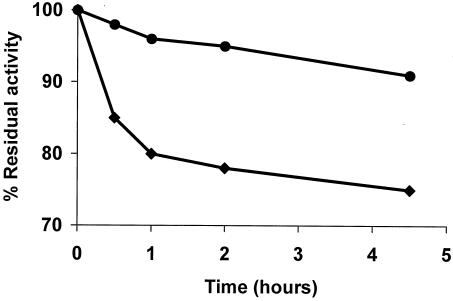
FIG. 2.
Thermal stability of glyoxyl-agarose derivatives of wild-type and mutant PGA enzymes. Thermal stability was determined by assaying the residual activity of the glyoxyl-agarose derivatives of wild-type PGA (⧫) and mutant PGA (•) after incubation at 60°C in 50 mM sodium acetate buffer, pH 5.0.
TABLE 1.
Inactivation constants (Ki) for wild-type PGA and mutant PGA glyoxyl derivatives under different inactivating conditions
| Conditions |
Kia
|
Stabilization factor | |
|---|---|---|---|
| PGA | Mutant PGA | ||
| pH 5, 60°C | 0.22 | 0.02 | 11 |
| pH 7, 50°C | 0.15 | 0.04 | 4 |
| pH 5, 60% DMF, 4°C | 0.26 | 0.12 | 2 |
| pH 2.5, 4°C | 0.39 | 0.05 | 8 |
Values are Ki h−1.
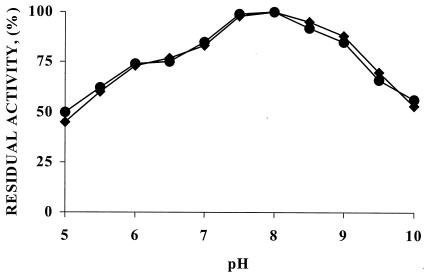
FIG. 3.
Activity and pH profile of glyoxyl-agarose derivatives of wild-type (⧫) and mutant (•) PGA enzymes.
To summarize, we have demonstrated the feasibility of a novel strategy that involves the use of site-directed mutagenesis to improve the stabilization of enzymes via multipoint covalent attachment, instead of creating mutations to stabilize the soluble enzyme. To increase the number of attachment sites, we have proposed two types of amino acid changes on the enzyme surface, i.e., replacing either arginine or acidic residues by lysine. These changes have been found to cause a negligible effect on the activity and stability of soluble enzymes. The success of this approach is closely related to the use of a suitable immobilization system; based on the results, glyoxyl supports are revealed as a very suitable immobilization system. By using this combined approach, the rigidification of the enzyme structure after immobilization was significantly increased, hindering the conformational changes induced by any inactivating agents. This strategy could be of general applicability to other enzymes.
Acknowledgments
This work has been funded by the Spanish CICYT (grants BIO2000-0747-C05-02 and BIO2001-2259). A Ph.D. fellowship from CAM for O.A. is gratefully recognized.
REFERENCES
- 1.Bi, W., and P. J. Stambrook. 1998. Site-directed mutagenesis by combined chain reaction. Anal. Biochem. 256:137-140. [DOI] [PubMed] [Google Scholar]
- 2.Blanco, R. M., J. J. Calvete, and J. M. Guisán. 1989. Immobilization-stabilization of enzymes: variables that control the intensity of the trypsin (amine)-agarose (aldehyde) multipoint attachment. Enzyme Microb. Technol. 11:353-359. [Google Scholar]
- 3.Blanco, R. M., and J. M. Guisán. 1989. Stabilization of enzymes by multipoint covalent attachment to agarose-aldehyde gels. Borohydride reduction of trypsin-agarose derivatives. Enzyme Microb. Technol. 11:360-366. [Google Scholar]
- 4.Brünger, A. T. 1992. A system for X-ray crystallography and NMR: XPLOR version 3.1.C. Yale University, New Haven, Conn.
- 5.Brünger, A. T., A. Krukowski, and J. Erickson. 1990. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr. Sect. A 46:585-593. [DOI] [PubMed] [Google Scholar]
- 6.Burkholder, R. P. 1951. Determination of vitamin B12 with a mutant strain of Escherichia coli. Science 114:459-460. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Lafuente, R., C. M. Rosell, V. Rodríguez, and J. M. Guisán. 1995. Strategies for enzyme stabilization by intramolecular crosslinking with bifunctional reagents. Enzyme Microb. Technol. 17:17-523. [Google Scholar]
- 8.Guisán, J. M. 1988. Aldehyde-agarose gels as activated supports for immobilization-stabilization of enzymes. Enzyme Microb. Technol. 10:375-382. [Google Scholar]
- 9.Hunt, P. D., S. P. Tolley, R. J. Ward, C. P. Hill, and G. G. Dobson. 1990. Expression, purification and crystallization of penicillin G acylase from Escherichia coli ATCC 11105. Protein Eng. 3:635-639. [DOI] [PubMed] [Google Scholar]
- 10.Jones, T. A., J. Y. Zon, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. B 47:110-119. [DOI] [PubMed] [Google Scholar]
- 11.Kutzbach, C., and E. Ravenbush. 1974. Preparation and general properties of crystalline penicillin acylase from Escherichia coli ATCC 11105. Hoppe- Seyler’s Z. Physiol. Chem. 355:45-53. [DOI] [PubMed] [Google Scholar]
- 12.Ponder, J., and F. Richards. 1987. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J. Mol. Biol. 193:775-779. [DOI] [PubMed] [Google Scholar]
- 13.Rodríguez, E., Z. A. Wood, P. A. Karplus, and G. X. Lei. 2000. Site-directed mutagenesis improves catalytic efficiency and thermostability of Escherichia coli pH 2.5 acid phosphatase/phytase expressed in Pichia pastoris. Arch. Biochem. Biophys. 382:105-112. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Sarkar, G., and S. S. Sommers. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 8:404-407. [PubMed] [Google Scholar]