Abstract
Campylobacter infections are the most common cause of bacterial enteritis in humans, and nearly 8% of such infections are caused by Campylobacter coli. Most studies have concentrated on Campylobacter jejuni, frequently isolated from intensively farmed poultry and livestock production units, and few studies have examined the spread and relatedness of Campylobacter across a range of geographical and host boundaries. Systematic sampling of a 100-km2 area of mixed farmland in northwest England yielded 88 isolates of C. coli from a range of sample types and locations, and water was heavily represented. Screening for antibiotic resistance revealed a very low prevalence of resistance, while genotyping performed by using three methods (flaA PCR restriction fragment length polymorphism [RFLP], pulsed-field gel electrophoresis [PFGE], and fluorescent amplified fragment length polymorphism [fAFLP]) provided insights into the genomic relatedness of isolates from different locations and hosts. Isolates were classified into 23 flaA groups, 34 PFGE groups, and five major fAFLP clusters. PFGE banding analysis revealed a high level of variability and no clustering by sample type. fAFLP and flaA analyses successfully grouped the isolates by sample type. We report preliminary findings suggesting that there is a strain of C. coli which may have become adapted to survival or persistence in water and that there is a group of mainly water-derived isolates from which unusual flaA PCR fragments were recovered.
Campylobacter spp. are recognized as the most common cause of acute bacterial enteritis in the United Kingdom (16) and in the rest of the industrialized world (10). One recent retrospective Danish study showed that there was a significantly increased risk of death after infection with Campylobacter (14). The most common taxon isolated from clinical samples from patients in the United Kingdom with food poisoning in 2000, the most recent year for which detailed data are available, was Campylobacter jejuni subsp. jejuni (referred to as C. jejuni below) (91.9% of cases), followed by Campylobacter coli (7.9%) (1). Other species, such as Campylobacter lari (0.1%) and Campylobacter fetus (0.04%), had a much lower prevalence, but they are also known to cause infections occasionally.
Estimates of the prevalence of Campylobacter spp. in various groups of animals have varied. Recent studies have suggested that the prevalence in United Kingdom beef and dairy cattle may vary from 53 to 89%, depending on seasonal variation (36), although in one recent study the workers reported that the prevalence in cattle was as low as 23% (26). In lambs, seasonal variation in prevalence from 0 to nearly 92% has also been reported, and isolation methods were a key factor contributing to this variation (35). In broiler chickens, the animal usually associated with Campylobacter, carriage rates ranging from 30 to 100% have been reported, and again the rate depended on the season and the location from which the sample was taken (34).
Several methods of genomically typing these bacteria have been developed. These methods include PCR restriction fragment length polymorphism (PCR-RFLP) analysis based on the flagellin gene, flaA (fla typing), pulsed-field gel electrophoresis (PFGE), and amplified fragment length polymorphism (AFLP) analysis (24, 31, 5). The suitability of these methods for assessing the genotypes of Campylobacter has been reviewed recently (25, 41).
Structured environmental sampling of a 100-km2 area of mainly dairy farmland in Cheshire, United Kingdom, was performed between May and July 2000. The aim of this study was to explore the distribution and diversity of campylobacters in a large-scale farming environment. Feces, soil, and water were sampled systematically, and campylobacters were isolated from the samples by standard microbiological methods. In this study, 1,870 Campylobacter isolates were recovered from a range of sample types. These isolates were characterized by flaA typing, PFGE, and fluorescent AFLP (fAFLP) analysis to compare the levels of discrimination and utility of these typing schemes in epidemiological studies. The study area is important not only because of its role in local and national food production but also because it is an area widely used for recreational purposes (e.g., water sports, walking, and picnicking). Therefore, the presence of pathogenic organisms in this region may be significant in terms of both food and environmental exposure.
Susceptibility to a panel of six antibiotics was also studied as little data relating to levels of antibiotic sensitivity in C. coli from nonclinical (human) isolates or chicken broiler flocks is available. Increasing antimicrobial resistance in Campylobacter is a recognized problem, and quinolone resistance is most common in human isolates of both C. jejuni and C. coli (15, 22, 32, 38).
MATERIALS AND METHODS
Cross-sectional study.
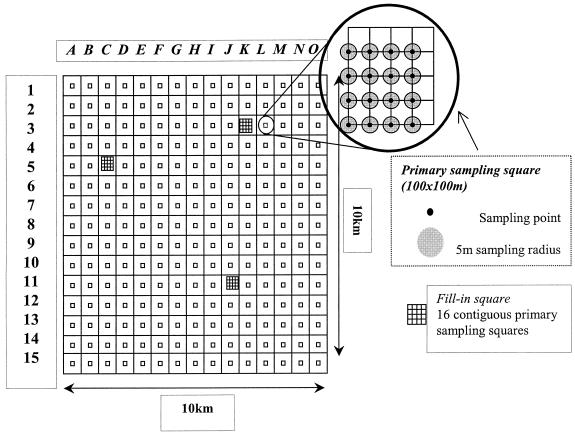
Systematic sampling of feces, soil, and water was carried out in a 100-km2 area of predominantly dairy farmland in Cheshire, United Kingdom. Sampling units were assigned by aligning a 15-by-15 grid over the study region (Fig. 1). Within these sampling units, primary sampling squares that were 100 by 100 m were assigned. Each primary sampling square was divided into four secondary squares and 16 tertiary squares. The bottom left corner of each tertiary sampling square was identified and designated the sampling point. Soil samples were taken from the sampling point, and an area with a radius of 5 m was marked out around this point. Within this area, samples of the cattle feces nearest the sampling point and samples of the most recently voided cattle feces were taken. Water and any other feces (e.g., feces from domestic animals and wildlife) within the primary sampling square were identified and sampled; water samples were taken from different sources, including stream water (running water), ponds, drinking troughs, and standing water in fields (all standing water). Soil samples were pooled in the field by combining the samples within each secondary sampling square; cattle fecal samples were pooled by the same method, but this pooling took place in the laboratory; and fecal samples from sheep and wildlife were pooled in the field at the level of the primary square. Water samples (250 ml) were not pooled and were filtered through a Nalgene 0.45-μm-pore-size nitrocellulose filter (Nalge Nunc International, Rochester, N.Y.).
FIG. 1.
Diagrammatic representation of study area and sampling strategy (not to scale).
Campylobacter spp. were isolated by enrichment (filters were incubated for water samples) in Campylobacter enrichment broth (Lab M, Bury, United Kingdom) for 24 h at 42°C under microaerophilic conditions, followed by plating onto Campylobacter selective agar (Lab M) and incubation for 48 h at 42°C under microaerophilic conditions. Up to three isolates from each sample were purified, subjected to Gram staining and oxidase and catalase testing, assigned a culture collection number, and frozen at −70°C in a cryogenic preservative for later analysis (Microbank; Pro-Lab Diagnostics, Neston, United Kingdom). Campylobacter species were identified by performing PCR with a boiled lysate from a freshly grown culture by using conditions described previously (12, 17); C. coli was identified by using the putative virulence marker ceuE as a target (12). Species identities were confirmed by using a commercial Campylobacter biotyping kit (MAST Diagnostics, Liverpool, United Kingdom). A strain of C. coli isolated from a pig, NCTC 11366 (National Collection of Type Cultures, Central Public Health Laboratory, Colindale, London, United Kingdom), was also included for comparison.
flaA typing.
C. coli strains were typed by using the method of Nachamkin et al. (24) as described on the Campynet web site (http://www.svs.dk/campynet), with the following modifications. In order to obtain cleaner template DNA, the heat lysis method was not used. Instead, DNA was extracted with a Nucleospin tissue kit used according to the manufacturer's instructions (Macherey-Nagel, Duren, Germany). Restriction digestion with DdeI (Roche Diagnostics, Lewes, United Kingdom) was carried out at 37°C overnight, and the product was electrophoresed on a 2% agarose-TAE buffer-ethidium bromide gel for 4 h at 90 V by using a Bio-Rad Sub-Cell GT system (Bio-Rad Laboratories, Hercules, Calif.) (TAE buffer is 40 mM Tris acetate plus 1 mM EDTA [pH 8.3]; Sigma-Aldrich, Poole, United Kingdom).
PFGE.
C. coli strains were typed by using the rapid method of Ribot et al. (31), with the following modifications. Phosphate-buffered saline was used for cell suspension and culture dilution steps, which were carried out in 3-ml preparations in sterile plastic 7-ml pots (bijoux). Agarose plugs were washed once in 3 ml of water and three times in TE buffer (100 mM Tris-HCl, 10 mM EDTA [pH 8.0]; Sigma-Aldrich) for 20 min each at 54°C in sterile plastic bijoux. Prior to restriction with SmaI (Thermo Life Sciences, Basingstoke, United Kingdom), plugs were incubated at 25°C for 20 min, first in 500 μl of 0.1× TE buffer and then in 200 μl of restriction buffer. The gel running conditions were the conditions described below, and the gels were electrophoresed for 16 h with a Bio-Rad CHEF DRIII system at 14°C. The gels were stained in TAE buffer containing 0.5 μg of ethidium bromide ml−1 for 30 min.
Gel visualization.
Both flaA and PFGE gels were visualized with UV illumination by using a Bio-Rad Gel Doc 2000. Images were cropped and the background was filtered as appropriate to achieve maximum resolution of the banding patterns. Gel images were exported as TIFF files for further analysis.
Analysis of banding patterns.
Both flaA and PFGE banding patterns were analyzed by using the Molecular Analyst software (Bio-Rad Laboratories). Gels were normalized by alignment with the appropriate size standard lanes; the size standards used were a 100-bp ladder for flaA (AbGene, Epsom, United Kingdom) and a concatemer of bacteriophage λ genomes for PFGE (New England Biolabs Inc., Hitchin, United Kingdom). Matching and construction of a dendrogram of the banding patterns by the unweighted pair group method with averages were performed by using the Dice coefficient with a 2% tolerance window.
fAFLP analysis.
The fAFLP analysis was carried out with a subset of 47 C. coli isolates representing all pooled samples from which C. coli strains were isolated and all flaA and PFGE types. The method of Duim et al. (5) was used throughout, except that the selectively amplified products were analyzed by using an ABI Prism 377 automated sequencer (Applied Biosystems). C. jejuni NCTC strain 81116 was included to check for reproducibility between gels. Data were collected by using the GeneScan software and were analyzed by using the BioNumerics package (Applied Maths, Kortrijk, Belgium) with the Pearson correlation coefficient and the unweighted pair group method with averages with a 2% tolerance window.
Determination of antibiotic resistance.
Eighty-one C. coli isolates were screened for resistance to a panel of six antibiotics by the disk diffusion method as described on the British Society for Antimicrobial Chemotherapy (BSAC) web site (http://www.bsac.org.uk) (see Table 2); control strain NCTC 11366, laboratory strain LS002 (serotype HS28), and erythromycin-resistant control strain NCTC 11437 were also included. The antibiotics used were chosen as representatives of the various classes of antimicrobial agents in common use and included nalidixic acid, ciprofloxacin, erythromycin, ampicillin, augmentin (co-amoxyclav), and trimethoprim. The MIC was determined for any isolate determined to be resistant by disk diffusion by using the E-test gradient strip method as described by the manufacturer (AB Biodisk, Dalvagen, Sweden). No MICs were determined for trimethoprim as Campylobacter strains are inherently resistant to this antibiotic; this was confirmed for the C. coli strains in this study by the disk diffusion method (there were no inhibition zones). MICs were also determined for other selected isolates (data not shown).
TABLE 2.
Antibiotic resistance screening of C. colia
| Antibiotic | Disc content (μg) | Zone size interpretation (mm)
|
MIC classification (μg ml−1)
|
No. of isolates resistant | MIC (μg ml−1) | ||
|---|---|---|---|---|---|---|---|
| Resistant | Susceptible | Resistant | Susceptible | ||||
| Nalidixic acidb | 30 | ≤15 | ≥16 | NAc | NA | 0 | |
| Ciprofloxacin | 1 | ≤17 | ≥18 | ≥4 | ≤2 | 0 | |
| Erythromycin | 5 | ≤19 | ≥20 | ≥2 | ≤1 | 6 | 2 (3), 3 (2), >256 (1) |
| Ampicillinc | 10 | ≤17 | ≥18 | ≥16 | ≤8 | 0 | |
| Augmentinc | 30 | ≤17 | ≥18 | ≥16 | ≤8 | 0 | |
| Trimethoprim | 2.5 | ≤14 | ≥20 | ≥4 | ≤0.5 | 81 | |
Unless otherwise indicated, interpretations were based on tentative BSAC values for Campylobacter. The MIC is shown only for resistant isolates; the number of isolates for each MIC is indicated in parentheses. Eighty-one C. coli isolates were screened in this manner. Control strain NCTC 11366, negative control strain LS002, and erythromycin-resistant control strain NCTC 11437 were also included.
No interpretation for nalidixic acid MIC was available, so only zone sizes are indicated. NA, not applicable.
No BSAC Campylobacter values are available for ampicillin and augmentin, so the data are based on BSAC values for the Enterobacteriaceae.
The MIC and disk diffusion zone size breakpoints for antibiotic resistance remain nonstandardized for Campylobacter. The values used for ciprofloxacin and erythromycin were taken from BSAC web site, where they are listed as tentative for Campylobacter. There are no specific Campylobacter BSAC MIC data for ampicillin, augmentin, and trimethoprim, so interpretations of the data for these antibiotics were based on the values listed for the Enterobacteriaceae. Interpretations of the data for nalidixic acid were based on Campylobacter values for antibiotic disk zone sizes, as no MIC data are available for either Campylobacter or the Enterobacteriaceae.
RESULTS
Bacterial isolates.
Eighty-eight (4.7%) of the Campylobacter strains isolated were identified as C. coli by PCR and a commercial biotyping kit (Table 1); isolates of C. jejuni, C. lari, C. hyointestinalis, and unidentified Campylobacter sp. were also characterized.
TABLE 1.
Relative proportions of C. coli isolates by sample type
| Sample type | No. of isolates | % of total |
|---|---|---|
| Avian | 3 | 3.41 |
| Cattle | 20 | 22.72 |
| Wildlife (rabbit) | 1 | 1.14 |
| Sheep | 14 | 15.91 |
| Soil | 1 | 1.14 |
| Water (all types) | 49 | 55.68 |
| Total | 88 | 100 |
Recovery of isolates from frozen cultures.
Five isolates (1155, 1156, 1157, 1163, and 2622) were recovered from frozen samples only once, permitting extraction of DNA for flaA and fAFLP analyses. Subsequent recovery attempts were not successful. Two other isolates (1164 and 1147) could be recovered only for PFGE analysis. None of these isolates could be recovered to test for antibiotic resistance.
flaA PCR-RFLP analysis.
Most C. coli DNA extracts yielded the expected fragment size (1,719 bp) after amplification with the flaA primers. Thirteen isolates yielded fragments that were approximately 100 bp larger than expected, and one isolate produced a fragment that was approximately 200 bp smaller than expected. Of these 14 atypical flaA products, 12 were from isolates derived from water samples (representing five samples), and the other 2 were derived from a single cattle sample (Fig. 2).
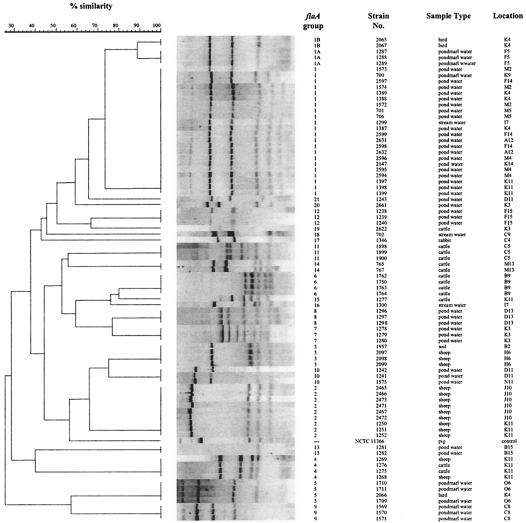
FIG. 2.
Comparison of C. coli flaA banding patterns. Of the 88 C. coli strains isolated, 81 could be typed by this method. The assigned flaA groups are indicated, as are the strain designations, the types of samples from which the strains were isolated, and the primary sampling squares. The dendrogram shows the results of a phylogenetic comparison. Control strain NCTC 11366 was included, but it was not placed in an flaA group.
A total of 81 of the 88 isolates from 36 pooled samples were successfully genotyped by this method, and flaA groups were assigned on the basis of the phylogenetic dendrogram derived from the banding pattern analysis (Fig. 2). A total of 23 flaA genotypes were assigned; the largest group, group 1, comprised 22 of the 81 isolates typed, all of which were isolated from water samples (referred to as the water group). flaA groups 1A and 1B were closely related to group 1 but differed in the number and position of the bands below 220 bp. The 22 isolates in group 1 represented 12 pooled samples. However, two pooled samples, one from a bird (isolates 2065 and 2066) and one from cattle (isolates 1275 and 1277), plus two water samples (isolates 1241 and 1243 and isolates 1299 and 1300) contained two flaA genotypes. The isolates from which flaA fragments of unusual sizes were recovered did not cluster together by this method, nor did they cluster with any other isolates.
PFGE.
A total of 80 of the 88 isolates from 36 samples were genotyped by the PFGE method, and 34 PFGE types were assigned by using the same criteria that were used for the flaA analysis (Fig. 3). One isolate, 1387, could not be typed by PFGE despite successful recovery from a frozen culture; the reason for this repeated failure is unknown. Apart from the multisample groups which exhibited identical PFGE banding patterns, very few pairs of isolates had patterns with less than four band differences. One group of isolates, from sheep samples in square K11, shared five bands of the same size, yet even within this closely defined population there were considerable differences in the banding patterns (types 2, 20, 21, and 23). These observations highlight the very high level of genotypic diversity among a relatively small population; more PFGE types than flaA groups were observed.
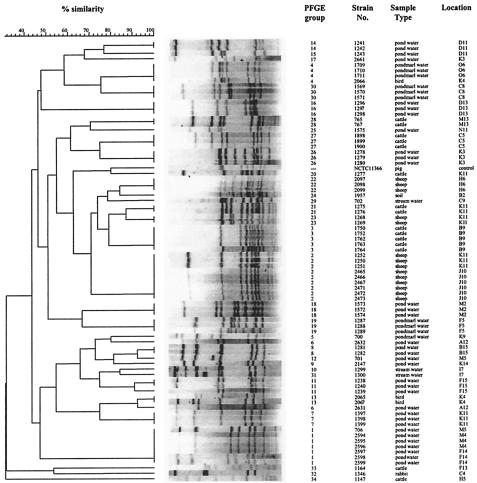
FIG. 3.
Comparison of C. coli PFGE banding patterns. Of the 88 C. coli strains isolated, 80 could be typed by this method. The assigned PFGE groups are indicated, as are the strain designations, the types of samples from which the strains were isolated, and the primary sampling squares. The dendrogram shows the results of a phylogenetic comparison. Control strain NCTC 11366 was included, but it was not placed in a PFGE group.
The largest group was type 2, which comprised nine isolates from three pooled sheep samples (Fig. 2). Type 1 contained seven isolates from three water samples; four water samples contained two different PFGE types, two of which were associated with different flaA types. Overall, isolates could be divided into three main clusters. Cluster 1 contained most of the water isolates, including all isolates belonging to flaA group 1, plus one bird isolate (2065) which belonged to flaA group 1B. Cluster 2 contained all the sheep isolates, all but one of the water isolates not in cluster 1, the single soil isolate, the other bird isolate (2066), and most of the cattle isolates. Four isolates fell outside these two clusters; three of the isolates (one water isolate, the only rabbit isolate, and one cattle isolate which could not be flaA typed) were in a small cluster (cluster 3), and the other isolate (1147 from cattle, not typed by the flaA method) was on its own.
The isolates belonging to flaA group 1 (the water group) could be discriminated by PFGE analysis (i.e., the members of this group exhibited multiple PFGE types).
fAFLP analysis.
A total of 47 of the 88 isolates, representing 39 samples, were successfully genotyped by fAFLP analysis; these isolates included four isolates (from two samples) which could not be typed by either of the other methods. In the resultant dendrogram the isolates occurred in five broad clusters, which exhibited equivalent levels of within-cluster similarity in the case of clusters 2 to 5 (Fig. 4). Cluster 1 comprised three samples with very low levels of similarity to each other (<30%) and to its nearest neighbor (<20%). Due to the highly discriminatory nature of the fAFLP method, no two isolates exhibited identical banding patterns; the most closely related isolates exhibited a level of similarity of >95%. It is generally accepted that fAFLP banding patterns with levels of similarity of ≥95% are indistinguishable, an assertion which has been demonstrated by replicate pattern analysis (28); fAFLP types were assigned by using this cutoff value (Fig. 4). fAFLP cluster groups were assigned on the basis of the largest clusters that the isolates fell into. In the case of clusters 2 to 5, subclustering was also observed (similarity, ≥60%).
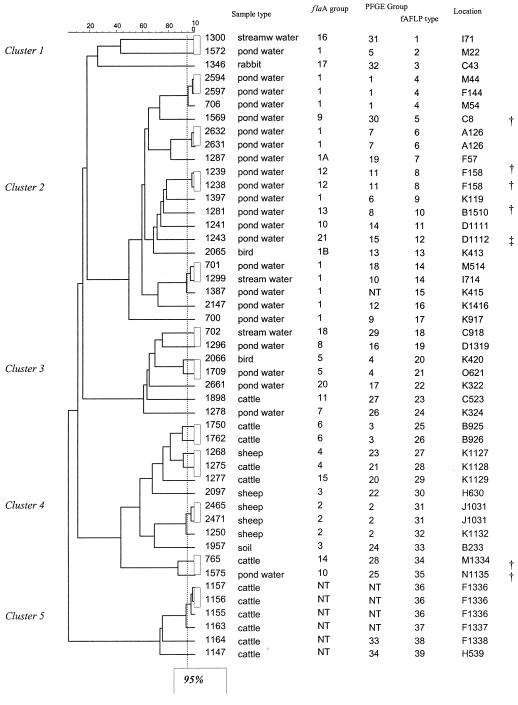
FIG. 4.
fAFLP dendrogram for C. coli isolates and a comparison with the flaA and PFGE groups. The 95% similarity cutoff for the fAFLP method is indicated by the dotted line. Forty-seven C. coli isolates representing every combination of sample type, flaA group, and PFGE group were analyzed by this method; four isolates which could not be typed by either method were also included. An internal control was included to ensure gel-to-gel reproducibility (results not shown). NT, not typeable. The daggers indicate isolates that produced unusually large flaA products, while the double dagger indicates an isolate that produced an unusually small flaA product.
Clustering by sample type was observed by this method. Water isolates were grouped in clusters 1, 2, and 3, and cattle and sheep were grouped in clusters 4 and 5. Most water isolates belonging to flaA group 1 were found in cluster 2; the only exception was isolate 1572. All of the other water isolates except isolate 1575 (cluster 4) were in cluster 3. The rabbit isolate (1346) was in cluster 1. This cluster also contained two water isolates, one of which (1572) was also in flaA group 1 but was genetically distinct from the other members of this group. The two bird isolates also clustered with the water isolates, as did a single cattle isolate. Sheep isolates were found only in cluster 4, which also contained isolates from cattle, soil, and water. Cluster 5 contained only cattle isolates, all of which were untypeable by the flaA method and/or by PFGE.
Antibiotic resistance.
A total of 6 of 81 isolates tested exhibited resistance to erythromycin; these six isolates were sensitive to all other antibiotics tested except trimethoprim. All other isolates were sensitive to all of the antibiotics except trimethoprim, to which all of the isolates were resistant (Table 2). The MIC of erythromycin for one isolate (1957) was >256 μg ml−1, the MIC of erythromycin for two isolates (1275 and 1276) was 3 μg ml−1, and the MIC of erythromycin for three isolates (2097, 2098, and 2099) was 2 μg ml−1. The inhibition zone sizes for control strain NCTC 11366 were as follows (MICs are indicated in parentheses): nalidixic acid, 14.5 mm (6 μg ml−1); ciprofloxacin, 19 mm (0.5 μg ml−1); erythromycin, 8 mm (4 μg ml−1); ampicillin, 11.5 mm (8 μg ml−1); augmentin, 27.5 mm (2 μg ml−1); and trimethoprim, 0 mm. Both the zone size and MIC data suggest that this strain is resistant to erythromycin, and the zone size suggests that it is marginally resistant to nalidixic acid (BSAC Campylobacter tentative breakpoints). The zone size also suggested that it is resistant to ampicillin, but this suggestion is not supported by the MIC data (BSAC Enterobacteriaceae breakpoints). The inhibition zone sizes for laboratory control strain LS002 were as follows (MICs are indicated in parentheses); nalidixic acid, 24.5 mm (0.5 μg ml−1); ciprofloxacin, 22 mm (0.008 μg ml−1); erythromycin, 20 mm (0.5 μg ml−1); ampicillin, 16 mm (0.25 μg ml−1); augmentin, 27.5 mm (0.125 μg ml−1); and trimethoprim, 0 mm. These results confirm the sensitivity of strain LS002 to all of the antibiotics tested except trimethoprim. Erythromycin-resistant control strain NCTC 11437 had no inhibition zone and an MIC of >256 μg ml−1 when erythromycin was used; these data are clearly consistent with erythromycin resistance (BSAC Campylobacter tentative breakpoints).
DISCUSSION
Comparison of different methods of genotyping.
The three different genotyping methods (flaA PCR-RFLP analysis, PFGE, and fAFLP analysis) provided different levels of discrimination of isolates. This was consistent with previous studies (8, 25). For example, flaA group 1 (the water group) could be subdivided into nine genotypes on the basis of PFGE data. Based on the number of genotypes observed, the degree of discrimination obtained by these methods increased from flaA typing (the least discriminatory method, producing 23 groups) to PFGE (34 groups) and fAFLP analysis (39 types, based on a 95% cutoff value). Although five of the additional fAFLP types corresponded to isolates which could not be typed by PFGE (although one isolate, isolate 1387, was typed by flaA), in three cases (isolates 2066 and 1709, isolates 1750 and 1762, and isolates 2471 and 1250) fAFLP analysis discriminated between isolates which were identical in both flaA and PFGE analyses at the 95% cutoff value.
The other multisample flaA groups (groups 2, 3, 4, 5, 6, and 10) could also be examined in terms of PFGE and fAFLP. With the exception of groups 2, 5, and 6, the isolates could all be discriminated from each other on the basis of PFGE data; groups 2, 5, and 6 could be discriminated only by fAFLP profiles. When flaA was used as a marker for genetic relatedness, the comparisons were between populations of flagellin genes and not between whole genomes. Hence, isolates falling into the same flaA group had identical or very closely related flaA genes but may have differed significantly in the rest of the genome. It has been reported that the flaA and flaB genes exhibit concerted evolution (19), and this may help explain why a relatively small number of flaA types were observed in what is clearly a genetically diverse population. A factor that also affects the long-term stability of flaA types is the reported evidence of recombination between flaA genes of different strains of C. jejuni and also recombination between flaA and its close homologue flaB (13). It has been suggested that one way to overcome some of these shortcomings is to genotype by using PCR-RFLP for both flaA and flaB (29, 41).
The ability of these typing methods to examine relatedness between isolates is crucial for epidemiological studies. Generally, grouping by flaA analysis and grouping by fAFLP analysis were consistent; both methods grouped the isolates by sample type. By comparison, PFGE analysis generated many different types with less clear grouping by sample type and thus appeared to provide a less useful method for examining strain-host associations or epidemiological relationships. These observations support the convention that at least two typing methods are necessary for Campylobacter relatedness studies, but they also highlight the importance of selecting the correct methods according to the requirements of the study.
The reasons for the differences in banding pattern consistency may mainly involve the nonclonal nature of the organisms. The chromosomes of Campylobacter species are known to be subject to genetic instability (27, 40), and this affects the three typing methods differently. As flaA is specifically concerned with motility, associated selective pressures such as colonization potential may be applied to the flaA region but not the rest of the chromosome. As fAFLP analysis relies on sampling a random portion of the bacterial chromosome, it is relatively insensitive to chromosomal instability (41). The polymorphisms observed by AFLP analysis are due to mutations in the restriction sites, mutations in the sequences adjacent to the restriction sites and complementary to the selective primer extensions, and insertions, deletions, or rearrangements within the amplified fragments. PFGE utilizes a rare cutting enzyme that typically produces 6 to 15 fragments. Hence, gross chromosomal rearrangements can dramatically alter the pattern of bands observed. As so few bands are produced compared with the number produced by the fAFLP method, this effect is exaggerated when profiles are compared. This observation may explain the case of flaA group 4 (isolates 1268 and 1275), which had two PFGE profiles but only one fAFLP subcluster. A possible explanation for this is that chromosomal instability has led to deletion of a piece of DNA containing an SmaI restriction site, resulting in the loss during PFGE analysis of a fragment of one size and the gain of two fragments of different sizes.
Comparisons by sample type and location.
The methods which we used can also be used to search for evidence of a relationship between isolates derived from the same sample type or sampling square. flaA group 1 contained only isolates from water samples, and flaA group 2 and PFGE type 1 contained only isolates from sheep feces from adjacent squares J10 and K11. Relatively few C. coli strains were isolated from cattle feces, and few of these strains proved to be typeable by any method other than the fAFLP method; PFGE could be performed with only around 50% of the cattle isolates as the other isolates could not be recovered from frozen samples following the initial culturing to generate template DNA. The reason for the failure of flaA typing with these cattle isolates is unclear, although development of improved primers may help. The success of the fAFLP analysis confirms the quality of the template DNA used. When they could be compared by the flaA and PFGE methods, cattle isolates appeared to have a distinct group of genotypes. On most occasions when isolates from different sampling squares belonged to identical flaA groups, the groups could be resolved as distinct groups by PFGE and fAFLP analysis. With the exception of isolates from squares J10 and K11 and the bird isolates, isolates from different types of samples rarely had the same genotype.
In general, C. coli isolates appeared to cluster according to the type of sample from which they were isolated by flaA typing and fAFLP analysis but not by PFGE. It is difficult to make comparisons with other studies regarding these grouping characteristics as previous studies have tended to be related to specific outbreaks or host animals and very few studies have been carried out with C. coli. Additionally, when two methods have been used (usually flaA analysis and PFGE), the outcome is often recorded as a combined genotype. This approach was specifically avoided in this study, in part because three methods rather than two were used but also because combined grouping may mask detail evident in results from the separate methods. There have been limited reports that C. jejuni exhibits clustering by sample type by fAFLP analysis (4) and by flaA typing (23) but not by PFGE. The lack of such reports is probably because very few isolates from multiple sample types have been examined in very few studies.
The J10 and K11 sampling squares were diagonally adjacent in the study area. C. coli strains were isolated from six sheep samples and one water sample from these two squares. The water isolate belonged to flaA group 1. The sheep isolates belonged to three flaA groups, two of which (groups 4 and 15) were unique to square K11 and the other of which (group 2) was found in squares J10 and K11; the common flaA group could not be resolved by either PFGE or fAFLP analysis, and the data are consistent with the hypothesis that the isolates are the same strain.
Water isolates.
The majority of water isolates were clustered by the fAFLP analysis and had a common flaA type. A group of three isolates, from squares F14, M4, and M5, were closely related as determined by all three typing methods. The squares were not connected by any water course, although M4 and M5 were adjacent squares. Wildlife, particularly birds, may be important in the dissemination of this organism. The possibility that gulls transmit Campylobacter to humans via water was proposed as early as 1983 (9), and various species of Campylobacter have been isolated from birds of numerous types, including wildfowl (7) and sparrows (2). The only other sample that shared a similar flaA and fAFLP profile with a water sample was a bird sample from square K4. This is consistent with the hypothesis that the organism is transmitted via bird feces. An alternative explanation is that a clonal strain of C. coli persists across a wide geographical area.
We observed the presence of a large flaA group specific to water samples from unconnected sites across a broad geographical area, although not all water isolates belonged to this group. It is possible that in the group 1 isolates, the flagellin gene is associated with survival and/or persistence in water, a medium in which Campylobacter is normally thought to be present only as a fecal contaminant. It may be that the group 1 strains represent organisms adapted to survival in the water, whereas strains in other flaA groups represent contaminants from fecal runoff from the surrounding pasture. However, no identical flaA patterns were obtained from other sources in the same squares.
The presence of C. coli in samples of environmental water has been reported previously (33, 39), and there have been a small number of reports linking cases of human disease to water supplies (11, 21). It has also been reported that a laboratory strain of C. coli is less well adapted to survival in water than C. jejuni or C. lari is (37). The present study supports the view that C. coli is widely distributed in environmental water and further suggests the possibility that there is a strain or strains associated with this medium, whose existence would have significant ramifications for exposure risks associated with environmental water sources.
Atypical flagellin genes.
The presence of atypically large flaA genes in C. coli isolated from samples of river water has been reported by Linton et al. (18). We believe that this is the first report of the presence of this larger fragment in an isolate from cattle feces and of a smaller flagellin gene associated with water sources. Linton et al. (18) suggested that the additional material in the flagellin gene was in the hypervariable region of flaA (HV1) which encodes a surface-exposed area of the protein associated with serospecificity of the flagellar filament (30). Sequence analysis is required to determine the nature and locations of the additional and deleted DNA in the isolates described here. It has also been suggested that genetic material may be exchanged between species (13, 20), and although in vivo exchange remains uncharacterized, it is reasonable to assume that if such exchange occurs, then DNA from the flaA gene could be transferred in this manner. Indeed, comparisons of the flaA profiles from this study with examples of human clinical C. jejuni isolates revealed indistinguishable fla types, suggesting fla gene homology (data not shown).
Antibiotic resistance.
The resistance profile of strain NCTC 11366 was surprising, as there have been no reports concerning the resistance of this organism in the various international culture collections where this strain is held. It is possible that subculturing has led to acquisition of this profile or that screening for the antibiotics which we used has not been performed previously. Strain LS002 was, however, confirmed to be sensitive to all of the antibiotics tested and was regarded as the negative control strain.
Screening with a panel of six antibiotics demonstrated that there was a very low prevalence of resistance among the C. coli isolates. Six samples yielded resistant isolates, all of which were resistant to erythromycin. These findings are quite surprising as there have been reports of multiple-antibiotic resistance in C. coli (3, 38). One of the isolates, isolate 1957 from soil, exhibited the highest MIC (>256 μg ml−1). Interestingly, isolates 1275 and 1276, which were obtained from the same pooled cattle sample and had identical flaA and PFGE patterns, were resistant, whereas isolate 1277 (which had different flaA and PFGE patterns) from the same sample was not resistant, confirming that it was a different strain. The mechanism of resistance has not been determined yet, and this represents the next area for investigation.
Conclusions.
In this study we demonstrated the diversity and variability of an environmental population of C. coli in the United Kingdom. We observed low levels of antibiotic resistance, high genetic diversity, and the appearance of a possible water-adapted strain. We concluded that C. coli is widespread and may have adapted to particular niches in animal populations and the environment. Such findings have implications for human exposure through both food and environmental pathways.
Acknowledgments
This work was supported by the Department for Environment, Food and Rural Affairs (DEFRA).
REFERENCES
- 1.CDSC. 2000. Sentinel surveillance of Campylobacter in England and Wales. Commun. Dis. Rep. Wkly. 10:169, 172. [PubMed] [Google Scholar]
- 2.Chuma, T., S. Hashimoto, and K. Okamoto. 2000. Detection of thermophilic Campylobacter from sparrows by multiplex PCR: the role of sparrows as a source of contamination of broilers with Campylobacter. J. Vet. Med. Sci. 62:1291-1295. [DOI] [PubMed] [Google Scholar]
- 3.Chuma, T., T. Ikeda, T. Maeda, H. Niwa, and K. Okamoto. 2001. Antimicrobial susceptibilities of Campylobacter strains isolated from broilers in the southern part of Japan from 1995 to 1999. J. Vet. Med. Sci. 63:1027-1029. [DOI] [PubMed] [Google Scholar]
- 4.Desai, M., J. M. J. Logan, J. A. Frost, and J. Stanley. 2001. Genome sequence-based fluorescent amplified length polymorphism of Campylobacter jejuni, its relationship to serotyping, and its implications for epidemiological analysis. J. Clin. Microbiol. 39:3823-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duim, B., T. M. Wassenaar, A. Rigter, and J. A. Wagenaar. 1999. High resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 65:2369-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberhart-Philips, J., N. Walker, N. Garrett, D. Ball, D. Sinclair, W. Rainger, and M. Bates. 1997. Campylobacteriosis in New Zealand: results of a case-control study. J. Epidemiol. Community Health 51:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fallacara, D. M., C. M. Monahan, T. Y. Morishita, and R. F. Wack. 2001. Fecal shedding and antimicrobial susceptibility of selected bacterial pathogens and a survey of intestinal parasites in free-living waterfowl. Avian Dis. 45:128-135. [PubMed] [Google Scholar]
- 8.Fitzgerald, C., K. Stanley, S. Andrew, and K. Jones. 2001. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl. Environ. Microbiol. 67:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fricker, C. R., R. W. A. Girdwood, and D. Munro. 1983. A comparison of procedures for the isolation of campylobacters from seagull faeces. J. Hyg. 91:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialised nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology Press, Washington, D. C.
- 11.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, K. R. Neal, and the Campylobacter Sentinel Scheme Collaborators. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez, I., K. A. Grant, P. T. Richardson, S. F. Park, and M. D. Collins. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helms, M., P. Vastrup, P. Gerner-Smidt, and K. Mølbak. 2003. Short and long term mortality associated with foodborne bacterial gastrointestinal infections: registry based study. Br. Med. J. 326:357-361. [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, L. B., and F. M. Aarestrup. 2001. Macrolide resistance in Campylobacter coli of animal origin in Denmark. Antimicrob. Agents Chemother. 45:371-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 17.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 18.Linton, D., A. Hurtado, A. J. Lawson, J. P. Clewey, H. Chart, and J. Stanley. 1999. Campylobacter coli strains with enlarged flagellin genes isolated from river water. Res. Microbiol. 150:247-25515. [DOI] [PubMed] [Google Scholar]
- 19.Meinersmann, R. J., and K. L. Hiett. 2000. Concerted evolution of duplicate fla genes in Campylobacter. Microbiology 146:2283-2290. [DOI] [PubMed] [Google Scholar]
- 20.Meinersmann, R. J., C. M. Patton, G. M. Evins, I. K. Wachsmuth, and P. I. Fields. 2002. Genetic diversity and relationships of Campylobacter species and subspecies. Int. J. Syst. Bacteriol. Evol. Microbiol. 52:1789-1797. [DOI] [PubMed] [Google Scholar]
- 21.Melby, K. K., J. G. Svendby, T. Eggebø, L. A. Holmen, B. M. Andersen, L. Lind, E. Sjøgren, and B. Kaijser. 2000. Outbreak of Campylobacter infection in a subartic community. Eur. J. Microbiol. Infect. Dis. 19:542-544. [DOI] [PubMed] [Google Scholar]
- 22.Mirelis, B., T. Llovet, C. Muñoz, F. Navarro, and G. Prats. 1999. Resistance of Campylobacter species to antimicrobial agents. Eur. J. Clin. Microbiol. Infect. Dis. 18:312. [DOI] [PubMed] [Google Scholar]
- 23.Nachamkin, I., K. Bohachick, and C. M. Patton. 1993. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nachamkin, I., H. Ung, and C. M. Patton. 1996. Analysis of HL and O serotypes of Campylobacter strains by the flagellin gene typing system. J. Clin. Microbiol. 34:277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen, E. M., J. Engberg, V. Fussing, L. Petersen, C.-H. Brogren, and S. L. W. On. 2000. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J. Clin. Microbiol. 38:3800-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen, E. M. 2002. Occurrence and strain diversity of thermophilic campylobacters in cattle of different age groups in dairy herds. Lett. Appl. Microbiol. 35:85-89. [DOI] [PubMed] [Google Scholar]
- 27.On, S. L. W. 1998. In vitro genotypic variation of Campylobacter coli documented by pulsed-field gel electrophoretic DNA profiling: implications for epidemiological studies. FEMS Microbiol. Lett. 165:341-346. [DOI] [PubMed] [Google Scholar]
- 28.On, S. L. W., and C. S. Harrington. 2000. Identification of taxonomic and epidemiological relationships among Campylobacter species by numerical analysis of AFLP profiles. FEMS Microbiol. Lett. 193:161-169. [DOI] [PubMed] [Google Scholar]
- 29.Petersen, L., and D. G. Newell. 2001. The ability of fla-typing schemes to discriminate between strains of Campylobacter jejuni. J. Appl. Microbiol. 91:217-224. [DOI] [PubMed] [Google Scholar]
- 30.Power, M. E., P. Guerry, W. D. McCubbin, C. M. Kay, and T. J. Trust. 1994. Structural and antigenic characteristics of Campylobacter coli FlaA flagellin. J. Bacteriol. 176:3303-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sáenz, Y., M. Zarazaga, M. Lantero, M. José Gastañares, F. Baquero, and C. Torres. 2000. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997-1998. Antimicrob. Agents Chemother. 44:267-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sails, A. D., F. J. Bolton, A. J. Fox, D. R. A. Wareing, and D. L. A. Greenway. 2002. Detection of Campylobacter jejuni and Campylobacter coli in environmental waters by PCR enzyme-linked immunosorbent assay. Appl. Environ. Microbiol. 68:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley, K. N., J. S. Wallace, J. E. Currie, P. J. Diggle, and K. Jones. 1997. Seasonality of thermophilic Campylobacter populations in chickens. J. Appl. Microbiol. 82:219-224. [PubMed] [Google Scholar]
- 35.Stanley, K. N., J. S. Wallace, J. E. Currie, P. J. Diggle, and K. Jones. 1998. Seasonal variation of thermophilic campylobacters in lambs at slaughter. J. Appl. Microbiol. 84:1111-1116. [DOI] [PubMed] [Google Scholar]
- 36.Stanley, K. N., J. S. Wallace, J. E. Currie, P. J. Diggle, and K. Jones. 1998. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85:472-480. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, C., D. J. Hill, and M. Mabey. 1999. Evaluation of the effect of temperature and nutrients on the survival of Campylobacter spp. in water microcosms. J. Appl. Microbiol. 86:1024-1032. [DOI] [PubMed] [Google Scholar]
- 38.Thwaites, R. T., and J. A. Frost. 1999. Drug resistance in Campylobacter jejuni, C. coli and C. lari isolated from humans in north west England and Wales, 1997. J. Clin. Pathol. 52:812-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waage, S. A., T. Vardund, V. Lund, and G. Kapperud. 1999. Detection of small numbers of Campylobacter jejuni and Campylobacter coli cells in environmental water, sewage, and food samples by a seminested PCR assay. Appl. Environ. Microbiol. 65:1636-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]