Abstract
The existence of several thousand Salmonella enterica serovar Typhimurium LT2 and LT7 cultures originally collected by M. Demerec and sealed in agar stab vials for 33 to 46 years is a resource for evolutionary and mutational studies. Cultures from 74 of these vials, descendants of cells sealed and stored in nutrient agar stabs several decades ago, were phage typed by the Callow and Felix, Lilleengen, and Anderson systems. Among 53 LT2 archived strains, 16 had the same phage type as the nonarchival sequenced LT2 strain. The other 37 archived cultures differed in phage typing pattern from the sequenced strain. These 37 strains were divided into 10 different phage types. Among the 19 LT7 strains, only one was similar to the parent by phage typing, while 18 were different. These 18 strains fell into eight different phage types. The typing systems were developed to track epidemics from source to consumer, as well as geographic spread. The value of phage typing is dependent upon the stability of the phage type of any given strain throughout the course of the investigation. Thus, the variation over time observed in these archived cultures is particularly surprising. Possible mechanisms for such striking diversity may include loss of prophages, prophage mosaics as a result of recombination events, changes in phage receptor sites on the bacterial cell surface, or mutations in restriction-modification systems.
Decades ago, thousands of Salmonella enterica serovar Typhimurium LT2 and LT7 mutant cultures were collected for inter- and intragenic mapping of the chromosome. The strains were initially isolated by K. Lilleengen (9). In 1950, Lilleengen sent strains to J. Lederberg, and these cultures became the basis of transduction experiments (24). LT2 and LT7 strains were then sent to M. Demerec for development of a serovar Typhimurium genetic map based on recombination between neighboring alleles via phage P22 transduction (3, 4, 17). These strains were stored in sealed agar stabs at room temperature, and most survive today.
Phage typing distinguishes serovar Typhimurium variants based on their susceptibility to a set of phages. The present battery of 209 definitive phage types (DTs) (L. Ward, personal communication) at the Robert Koch Institute has been used for long-term surveillance of serovar Typhimurium (15) and identification of the spread of different, yet closely related, serotypes in the tracking of disease (8), in addition to tracking host-adapted variants within a particular serovar of S. enterica (13). Because the phage type of a given strain has been shown to be stable under standard laboratory conditions, it can be assumed that these strains all had the same phage type when they were introduced into the vials decades ago. The original LT2 strain sent to the former Institute of Experimental Epidemiology (now the Robert Koch Institute) was retyped annually as one of the reference phage type test strains from a Dorset agar (egg yolk) culture, and we found no changes in phage types from 1976 to 1989.
Samples of several LT2 and LT7 archived cultures have now been opened and examined for differences between the genomes of survivors and that of the putative initial parent strain, including differences in phage types. Extensive genetic deviation from parental strains from this archival collection has been reported in terms of carbon utilization, genomic arrangement, and gene expression (5, 10, 20, 21). We now report additional deviations as detected by phage typing.
It is important to note that the value of using the typing schemes in this particular case is based on the relative stability of serovar Typhimurium strains at the phage type level. In diagnostic laboratories, the sources of epidemics have been traced horizontally across continents and vertically from contaminated food and feed to human and animal disease (8, 15, 22, 23). In light of this observed stability, the difference in phage types between these archived samples is particularly striking.
MATERIALS AND METHODS
Selection of strains.
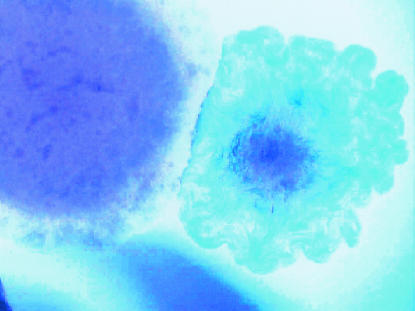
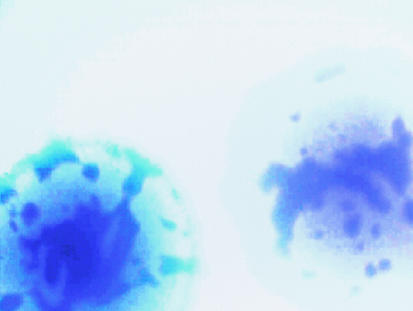
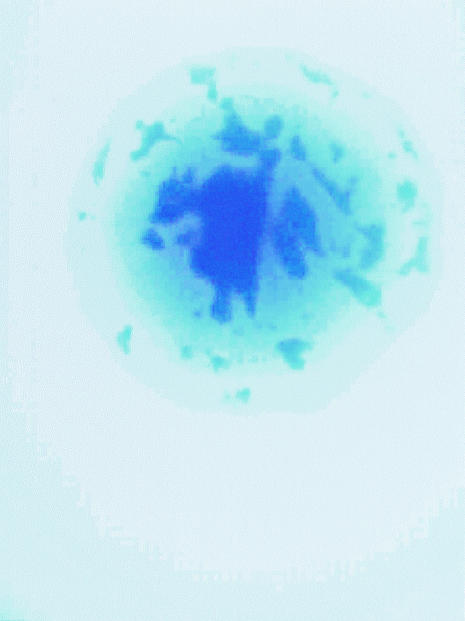
Strains derived from wild-type LT2 and LT7 were used in this study. The most notable difference between LT2 and LT7 is that LT7 has a mutator phenotype resulting in a higher mutation frequency relative to LT2 (7). Strains were revived by introducing 0.5 ml of sterile Luria-Bertani (LB) broth into the archived vial followed by brief agitation and incubation at 37°C for 30 min. The suspension of 500 ml was spread on a plate containing Waterblue glucose agar, developed at the Robert Koch Institute for phage typing in the 1970s (16). Multiple colonies from a single vial were tested to determine whether there was diversity within a single vial. It is possible to distinguish rough colonies and white or blue colonies from strain 6537 (LT7 his-91) (Fig. 1). On some white colonies, deep blue areas could be observed (Fig. 2 and 3). When some of the deep blue area was inoculated into LB broth via a needle, no growth occurred, indicating death. Some colonies grew well, however, and showed an extended lysis spectrum, e.g., DT38 or DT46 (see Table 6).
FIG. 1.
Micrograph of two colonies, one rough and one smooth, from strain 6537, an LT7 his-91 derivative, on Waterblue glucose agar.
FIG. 2.
Micrograph of two colonies from strain 9098, an LT2 his-583 derivative, on Waterblue glucose agar. The dark blue areas, when removed from the colony, did not grow in LB broth.
FIG. 3.
Micrograph of a smooth colony from strain 9098, an LT2 his-583 derivative, on Waterblue glucose agar.
TABLE 6.
Determination of diversity within a single vial
| Total no. of colonies/vial no./ strain derivation (no. of phage types) | Phage type
|
No. of colonies from vial with phage type | |
|---|---|---|---|
| Felix and Callow/Lilleengen | Anderson | ||
| 13/6968/LT2 (3) | 1b/2 | DT4 | 4 |
| UT/ph30 | DT193 | 5 | |
| 01 phage + | UT | 4 | |
| 29/6537/LT7 (5) | 1b/2 | DT4 | 7 |
| 1b vars 2/7 | DT4 | 6 | |
| 1b/7 | DT4 | 11 | |
| 1/1 | DT38 | 3 | |
| 1/1 | DT46 | 2 | |
The observations were made with a Nikon Eclipse E600 microscope and Nikon Plan Fluor 4×/0.13 objective, and photos (Fig. 1 to 3) were taken with a Nikon DXM 1200 digital camera with Lucia G software on DXM 1200, version 4.61.
The archive cultures that were phage typed, and their genotypes, for LT2 and LT7 are listed in Tables 1 and 2, respectively. Cultures were stored in 1-ml nutrient agar stabs in 2-ml glass vials at various times between 33 and 46 years ago. Vials were sealed and were not opened until this study. There was no preconceived pattern of selection among the archive strains for phage typing.
TABLE 1.
Phage typing comparisons between 53 archived LT2 strains and the wild-type sequenced strain 2004 and two other nonarchived strains of serovar Typhimuriuma
| Strain | Allele | Phage type
|
|
|---|---|---|---|
| Felix and Callow/Lilleengen | Anderson DT | ||
| 2004 | Sequenced strain | 1b var. 2/2 | 4 |
| 1596 | hisD2555 | 1b var. 2/2 | 4 |
| 1600 | hisB143 | 1b var. 2/2 | 4 |
| 1601 | hisF144 | 1b var. 2/2 | 4 |
| 1639 | hisD142 | 1b var. 2/2 | 4 |
| 1648 | thyA1566 | 1b var. 2/2 | 4 |
| 1670 | hisD2550 | 1b var. 2/2 | 4 |
| 1671 | hisD2550 | 1b var. 2/2 | 4 |
| 1685 | thyA269 | 1b var. 2/2 | 4 |
| 1693 | thyA274 | 1b var. 2/2 | 4 |
| 1702 | thyA315 | 1b var. 2/2 | 4 |
| 1703 | thyA314 | 1b var. 2/2 | 4 |
| 1737 | thyA324 | 1b var. 2/2 | 4 |
| 1738 | thyA324 | 1b var. 2/2 | 4 |
| 1927 | aro-20 | 1b var. 2/2 | 4 |
| 2011 | thyA314 | 1b var. 2/2 | 4 |
| 2014 | thyA324 | 1b var. 2/2 | 4 |
| 1609 | purD35 | 1b/2 | 4 |
| 1613 | thyA162 | 1b/2 | 4 |
| 1614 | hisD4384 | 1b/2 | 4 |
| 1615 | hisD2543 | 1b/2 | 4 |
| 1626 | thyA173 | 1b/2 | 4 |
| 1632 | hisD2555 | 1b/2 | 4 |
| 1669 | hisD2550 | 1b/2 | 4 |
| 1674 | hisD2550 | 1b/2 | 4 |
| 1681 | thyA270 | 1b/2 | 4 |
| 1688 | thyA272 | 1b/2 | 4 |
| 1706 | thyA316 | 1b/2 | 4 |
| 1747 | hisD2550 | 1b/2 | 4 |
| 1748 | hisD2550 | 1b/2 | 4 |
| 2013 | thyA315 | 1b/2 | 4 |
| 1594 | hisD2555 | 1b/2 | 4 |
| 1598 | hisD141 | 1b var. 2/2 | 4 (+14)b |
| 1684 | thyA273 | 1b/2 | 4 (+14)b |
| 1700 | thyA314 | 1b/2 | 4 (+14)b |
| 1635 | hisD142 | 2b/NCd | RDNC (1)c |
| 1617 | hisD4385 | 2b/NC | RDNC (1) |
| 1657 | hisD2122 | 2b/NC | RDNC (1) |
| 1922 | thyA158 | 2b/NC | RDNC (1) |
| 1636 | hisD142 | 2b/NC | RDNC (1) |
| 2000 | Nonarchivale | 2b/NC | RDNC (2) |
| 2007 | Nonarchivalf | 1a/9 | RDNC (3) |
| 1939 | thyA2422 | 1b/2 | RDNC (4) |
| 1656 | thyA68 | 1/1 | 1 |
| 1666 | hisD2550 | 1/1 | 1 |
| 1652 | hisD2526 | UT/NC | 193 |
| 1672 | hisD2550 | 35/NC | 29 |
| 1692 | thyA275 | Serologically rough 01 + | UT |
| 1695 | thyA272 | Serologically rough 01 + | UT |
| 1704 | thyA316 | Serologically rough 01 + | UT |
| 1935 | thyA2431 | Serologically rough 01 + | UT |
| 1619 | aro-63 | Serologically rough 01 Φ | UT |
| 1687 | thyA273 | Serologically rough 01 Φ | UT |
| 1701 | thyA314 | Serologically rough 01 Φ | UT |
| 1717 | thyA320 | Serologically rough 01 Φ | UT |
| 1745 | thyA329 | Serologically rough 01 Φ | UT |
A total of 11 phage types are represented among the archived strains.
Strain additionally reacts with phage A14.
RDNC, reaction does not conform to a known Anderson phage type but falls in the group given in parentheses.
NC, noncharacteristic of a Lilleengen phage type.
Clinical isolate of serovar Typhimurium.
Non-LT2 strain with Gifsy-1 mutant.
TABLE 2.
Phage typing comparisons between 19 archived LT7 strains and the wild-type strain 1970a
| Strain | Allele | Phage type
|
|
|---|---|---|---|
| Felix and Callow/Lilleengen | Anderson DT | ||
| 1970 | WTb | 1b var. 2/2 | 4 |
| 1999 | met-121 | 1b var. 2/2 | 4 |
| 2020 | met-115 | 1b var. 2/7 | 4 |
| 2038 | met-143 | 1b var. 2/7 | 4 |
| 2040 | met-143 | 1b var. 2/7 | 4 |
| 2041 | ser-85 | 1b var. 2/7 | 4 |
| 2034 | ser-85 | 2b/NCc | RDNC (1)d |
| 1686 | thyA273 | 1/1 | 36 |
| 2022 | met-143 | UT rough 01 +e | UT |
| 1971 | leu-56 | UT rough 01 Φf | UT |
| 2029 | met-121 | Serologically rough 01 + | UT |
| 2036 | met-143 | Serologically rough 01 + | UT |
| 1988 | ser-85 | Serologically rough 01 Φ | UT |
| 1998 | ser-85 | Serologically rough 01 Φ | UT |
| 2024 | met-121 | Serologically rough 01 Φ | UT |
| 2025 | met-143 | Serologically rough 01 Φ | UT |
| 2031 | pro-64 | Serologically rough 01 Φ | UT |
| 2033 | ser-85 | Serologically rough 01 Φ | UT |
| 2043 | ser-92 | Serologically rough 01 Φ | UT |
| 2026 | ser-92 | Serologically rough 01 Φ | 120 |
A total of nine different phage types are represented.
Nonarchival LT7 strain obtained from K. Sanderson.
NC, noncharacteristic of a Lilleengen phage type.
Reaction does not conform to a known Anderson phage type but falls in group 1.
UT by Felix and Callow system but reacts to phage 01.
UT by Felix and Callow system and does not react to phage 01.
Phage typing.
The phage typing was performed by the Felix and Callow, Lilleengen, and Anderson schemes used for detection of Salmonella pathogens, as described previously (2, 13, 14, 16, 18). Briefly, a battery of well-defined phages is introduced to the bacterial strain being tested, and the strain is typed according to which phages cause lysis. Table 3 demonstrates the Anderson phage typing scheme. These schemes are used in routine investigations for epidemiological purposes at the Robert Koch Institute. The Felix-Callow and Anderson systems were developed in England (2), while the Lilleengen scheme was derived in Sweden (9). The wild-type strain LT2 acquired its name because it was initially phage typed by Lilleengen (Lilleengen type 2). This combination of Felix-Callow and Lilleengen schemes has been used at the Robert Koch Institute since 1972.
TABLE 3.
Anderson phage typing schemea
| Phage type | Result with typing phage:
|
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 32 | 35 | 01 | |
| DT36 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| DT1 | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + |
| DT126 | + | + | + | + | + | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + |
| RDNC (3) | − | + | + | + | + | − | + | − | + | + | + | + | + | + | + | − | − | + | + | + | + | + | + | + | + | + | − | + | + | + | + |
| RDNC (4) | − | + | − | + | + | + | − | + | + | + | + | + | − | + | + | − | + | + | + | − | − | + | − | + | − | + | − | + | + | + | + |
| DT4 (+14) | − | − | − | + | + | + | − | − | + | + | + | + | + | + | + | − | − | + | + | − | + | + | − | + | + | + | − | + | + | + | + |
| DT4 | − | − | − | + | + | + | − | − | + | + | + | + | − | + | + | − | − | + | + | − | + | + | − | + | + | + | − | + | + | + | + |
| RDNC (2) | + | + | + | − | − | − | + | − | − | − | − | − | + | − | − | − | − | − | + | − | − | + | + | − | − | + | − | + | + | + | + |
| RDNC (1) | − | − | − | − | − | − | − | − | − | + | − | − | + | + | + | − | − | − | − | − | + | − | − | + | − | + | − | + | − | + | + |
| DT29 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + |
| DT120 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| UT | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
Typing phages in addition to phage 01 are arbitrarily labeled with numbers 1 to 35. + and − indicate that the phage does or does not lyse the strain, respectively. RDNC, reaction does not conform to the phage type.
Additionally, the Anderson scheme was used since it is a widely distributed scheme and is used globally. Thus, it is possible to detect epidemiological associations between food poisoning outbreaks as recently reported (15). Furthermore this technique yields insight into host adaptations to different animal species within a single serovar (13)
The serovar Typhimurium parent LT2 strain carries several prophages, Fels-1, Fels-2, Gifsy-1, and Gifsy-2 and perhaps others (1, 6, 11).
Rough-smooth detection.
Most rough strains agglutinate spontaneously in 0.85% NaCl solution. Such strains are designated as serologically rough strains (Fig. 1). Some rough strains show lysis with phage Felix01, which lyses over 90% of all Salmonella serotypes of subspecies I, II, and IIIb (12). However, some serologically rough strains are negative in phage 01 reactions. All these strains are untypeable (UT) by the Anderson scheme, since the adsorption site of the typing phages is the 012 antigen (19). Two of the typing phages adsorb to proteins. These phages can also lyse rough strains. Thus, we may assign some serologically rough strains to phage type DT120. Furthermore we use a “rough phage” from London (A59/6S/R) and another from Frankfurt/Main (Ffm) that adsorb to a special rough polysaccharide (16). Such strains belong to Anderson phage type DT120 also. Some strains are also labeled “UT rough” (UT by Felix-Callow and Lilleengen) but are lysed by the rough phage (+) since they do not agglutinate spontaneously in 0.85% NaCl solution.
RESULTS
The archival strains tested and their phage types are presented in Table 1 (for LT2 strains) and Table 2 (for LT7 strains). Among 53 LT2 archived strains, there were a total of 11 different phage types. Sixteen samples had the same phage type as the sequenced LT2 strain (1b var. 2/2; DT4 by Felix and Callow/Lilleengen; Anderson), the assumed parent (Table 1). The 37 archived LT2 cultures that differed were divided into 10 phage type patterns. Among 19 LT7 strains, only one was similar to the wild-type LT7 (1b var. 2/2; DT4 by Felix and Callow/Lilleengen; Anderson), while 18 differed (Table 2). The 18 archived LT7 cultures that differed were divided into eight different phage type patterns.
Out of 53 archived LT2 strains, 9 strains were found to be serologically rough, but 12 of the 19 LT7 strains were also serologically rough. While the numbers may be too small to draw conclusions, they would indicate that the presence of a mutator phenotype may be responsible for the increased mutation from smooth to rough. Three LT2 and one LT7 additional strains were UT, although they were serologically smooth. The UT strains could have mutations in membranes or other cell components important for phage attachment or for propagation.
Decades ago when the strains were initially stored, multiple vials were inoculated with Salmonella from a single colony. One reason for establishing replicate vials is that mutants from the collection were frequently sent to colleagues. These replicate archival strains deriving from a single colony are designated as “siblings.” Phage types of three sets of sibling LT2 strains were compared to those of wild-type LT2, and a number of differences were evident, even among sibling strains (Table 4). A similar diversity was found with LT7 sibling strains (Table 5).
TABLE 4.
Phage typing comparisons among three sets of LT2 sibling strainsa
| Allele | Strain | Phage type
|
|
|---|---|---|---|
| Felix and Callow/Lilleengen | Anderson DT | ||
| hisD2550 | 1670 | 1b var. 2/2 | 4 |
| 1671 | 1b var. 2/2 | 4 | |
| 1748 | 1b/2 | 4 | |
| 1669 | 1b/2 | 4 | |
| 1674 | 1b/2 | 4 | |
| 1666 | 1/1 | 1 | |
| 1672 | 35/NCb | 29 | |
| hisD2555 | 1596 | 1b var. 2/2 | 4 |
| 1594 | 1b/2 | 4 | |
| 1632 | 1b/2 | 4 | |
| thyA273 | 1687 | Serologically rough 01 Φc | UT |
| 1684 | 1b/2 | 4 (+14)d | |
| 1686 | 1/1 | 36 | |
Each set of sibling strains was stored from a single isolate nearly 40 years ago.
Noncharacteristic of a Lilleengen phage type.
Does not react with phage 01.
Strain additionally reacts with phage A14.
TABLE 5.
Phage typing comparisons among four sets of LT7 sibling strainsa
| Allele | Strain | Phage type
|
|
|---|---|---|---|
| Felix and Callow/Lilleengen | Anderson DT | ||
| met-121 | 1999 | 1b var. 2/2 | 4 |
| 2029 | Serologically rough 01 +b | UT | |
| 2024 | Serologically rough 01 Φc | UT | |
| met-143 | 2038 | 1b var. 2/7 | 4 |
| 2040 | 1b var. 2/7 | 4 | |
| 2036 | Serologically rough 01 + | UT | |
| 2025 | Serologically rough 01 Φ | UT | |
| 2022 | UT rough 01 + | UT | |
| ser-85 | 2041 | 1b var. 2/7 | 4 |
| 2034 | 2b/NCd | RDNC (1)e | |
| 1988 | Serologically rough 01 Φ | UT | |
| 1998 | Serologically rough 01 Φ | UT | |
| 2033 | Serologically rough 01 Φ | UT | |
| ser-92 | 2043 | Serologically rough 01 Φ | UT |
| 2026 | Serologically rough 01 Φ | 120 | |
Each set of sibling strains was stored from a single isolate nearly 40 years ago.
Reacts with phage 01.
Does not react with phage 01.
NC, noncharacteristic of a Lilleengen phage type.
Reaction does not conform to a known Anderson phage type but falls in group 1.
To support the view that the observed phage type diversity arose in the vials, isolated colonies arising from individual cells within one LT2 vial and one LT7 vial were tested. Thirteen colonies with different microscopic phenotypes on Waterblue glucose agar (white, blue, or white with blue areas [Fig. 1 to 3]) were tested from the LT2 vial, and 29 colonies were tested from the LT7 vial. The 13 colonies from the LT2 vial yielded three different phage types, while the 29 colonies from the LT7 vial yielded five different phage types (Table 6).
It is important to note that, although mutagens were sometimes used in the selection of strains for mapping studies, the auxotrophs in our experiments arose spontaneously, without any mutagen treatment.
DISCUSSION
Phage typing is frequently used to track Salmonella epidemics across the globe (8, 15). Additionally, it is used to identify the initial source of epidemics (22, 23). Epidemiologic information garnered from phage typing is dependent upon the relative stability of any given Salmonella phage type over time. Because diversity of carbon utilization and genomic arrangements has previously been observed in the archived S. enterica serovar Typhimurium strains at the Cancer Research Center, experiments were performed to determine whether the phage type of these strains had also undergone changes compared to that of the wild type. The data indicate that the conditions of storage over decades have a drastic effect on the phage type of these strains. In a variety of both LT2 and LT7 strains, there is a drastic alteration of phage type from that of the parent strain (Tables 1 and 2, respectively).
Regarding a concern that the altered phage types may have existed upon initial inoculation of the stabs decades ago, it was noted that many cultures were initially inoculated in replicate vials from a single colony. Many of these sibling vials remain in the collection and are a resource for testing differences among recovered progeny from a single source. Cells from a number of sets of sibling vials in the collection were tested and exhibited genetic diversity indistinguishable from diversity observed between nonsibling strains. Among sibling sets tested were some of the LT2 and LT7 strains described in this report (Tables 4 and 5, respectively). For example, progeny from sibling vials were previously shown to differ in motility (data not shown) and carbon and nitrogen utilization (21). These observations, coupled with the fact that multiple phage types were found in a single vial, support the view that the scored changes occurred during storage and did not preexist.
Two strains (1656 and 1666) belonging to phage type 1/1 (DT1) could be distinguished in the Anderson scheme. It is possible that these clones have been cured of three or all four of the Gifsy-1, Gifsy-2, Fels-1, and Fels-2 prophages. Strain 1686, typed DT36, may have lost an additional defective prophage. Some colonies were typed from strain 6537 (LT7), which exhibited extensive phage lysis (DT38 and DT46, Table 6). Furthermore it is possible that mutations that occurred during storage may have destroyed the immunity region of the prophages. Note that DT126 phage types and one DT29 phage type are seldom detected in natural isolates. DT193 is frequently observed in isolates from cattle, pigs, and humans. In the future, it would be interesting to isolate and propagate phages from strains with phage types RDNC, DT4 (+14), and UT and to then phage type the lysogens. It will be interesting to further investigate the clones of DT36, DT38, and DT46 in terms of prophages lost and whether phage module exchange occurred.
Several possibilities exist for the cause of such broad diversity among strains over time. A change in phage receptors or lipopolysaccharides on the outer membrane of the cell would not allow the initial phage-host interaction necessary for adsorption. Serovar Typhimurium strain LT2 carries at least four prophages, and loss of one or more of these prophages or possibly the generation of mosaic phages as a result of recombination events could potentially alter the cellular immunity and would thus alter the phage type. Finally, mutations at sites related to restriction-modification systems or in any other bacterial gene that affects phage propagation or release would also have a potential effect on the strains. These possibilities have not yet been tested experimentally in these strains but should be explored in the near future.
The broad diversity in phage type observed among these cultures is intriguing, and further investigation needs to be performed to elucidate possible mechanisms. Phage typing of more strains, coupled with research at the genomic level of the strains identified in this study, will provide insight into the effects of long-term storage on bacterium-phage interactions, identification of closely related strains, and possibly bacterium-host interactions.
Acknowledgments
This research was supported by funds from the Cancer Research Center, Columbia, Mo., and the Robert Koch Institute (Wernigerode, Germany). R.A.H. was supported by the Raymond Freese Memorial Postdoctoral Fellowship Program.
We thank H. Gattermann, V. Trute, S. Kulbe, H. Ragnit, and B. Tannert for skillful technical assistance.
Footnotes
This paper is dedicated to the memory of Philip and Zlata Hartman, pioneers in Salmonella genetics.
REFERENCES
- 1.Affolter, M., C. Parent-Vaugeois, and A. Anderson. 1983. Curing and induction of the Fels 1 and Fels 2 prophages in the Ames mutagen tester strains of Salmonella typhimurium. Mutat. Res. 110:243-262. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, E. S., L. R. Ward, M. J. Saxe, and J. D. de Sa. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (London) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demerec, M., I. Blomstrand, and Z. E. Demerec. 1955. Evidence of complex loci in Salmonella. Proc. Natl. Acad. Sci. USA 41:359-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demerec, M., and P. E. Hartman. 1959. Complex loci in microorganisms. Annu. Rev. Microbiol. 13:377-406. [Google Scholar]
- 5.Edwards, K., I. Linetsky, C. Hueser, and A. Eisenstark. 2001. Genetic variability among archival cultures of Salmonella typhimurium. FEMS Microbiol. Lett. 199:215-219. [DOI] [PubMed] [Google Scholar]
- 6.Figueroa-Bossi, N., S. Uzzau, S. Rubino, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 7.Kirchner, C. E., and M. J. Rudden. 1966. Location of a mutator gene in Salmonella typhimurium by cotransduction. J. Bacteriol. 92:1453-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liesegang, A., D. Davos, J. C. Balzer, W. Rabsch, R. Prager, D. Lightfoot, A. Siitonen, H. Claus, and H. Tschape. 2002. Phage typing and PFGE pattern analysis as tools for epidemiological surveillance of Salmonella enterica serovar Bovismorbificans infections. Epidemiol. Infect. 128:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lilleengen, K. 1948. Typing Salmonella typhimurium by means of bacteriophage. Acta Pathol. Microbiol. Scand. Suppl. 77:11-125. [Google Scholar]
- 10.Liu, G. R., K. Edwards, A. Eisenstark, Y. M. Fu, W. Q. Liu, K. E. Sanderson, R. N. Johnston, and S. L. Liu. 2003. Genomic diversification among archival strains of Salmonella enterica serovar Typhimurium LT7. J. Bacteriol. 185:2131-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 12.Popoff, M. Y. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. WHO Collaborating Centre for Reference and Research on Salmonella, Paris, France.
- 13.Rabsch, W., H. L. Andrews, R. A. Kingsley, R. Prager, H. Tschape, L. G. Adams, and A. J. Baumler. 2002. Salmonella enterica serovar Typhimurium and its host-adapted variants. Infect. Immun. 70:2249-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabsch, W., S. Mirold, W. D. Hardt, and H. Tschape. 2002. The dual role of wild phages for horizontal gene transfer among Salmonella strains. Berl. Muench. Tieraerztl. Wochenschr. 115:355-359. [PubMed] [Google Scholar]
- 15.Rabsch, W., H. Tschape, and A. J. Baumler. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3:237-247. [DOI] [PubMed] [Google Scholar]
- 16.Rische, H. 1973. Infecktionskrankheiten und ihre Erreger. Gustav Fischer Verlag, Jena, Germany.
- 17.Sanderson, K. E., A. Hessel, and B. A. D. Stocker. 1996. Strains of Salmonella typhimurium and other Salmonella species used in genetic analysis, p. 2496-2503. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 18.Schmieger, H. 1999. Molecular survey of the Salmonella phage typing system of Anderson. J. Bacteriol. 181:1630-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinbacher, S., U. Baxa, S. Miller, A. Weintraub, R. Seckler, and R. Huber. 1996. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. USA 93:10584-10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutton, A., R. Buencamino, and A. Eisenstark. 2000. rpoS mutants in archival cultures of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:4375-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tracy, B. S., K. K. Edwards, and A. Eisenstark. 2002. Carbon and nitrogen substrate utilization by archival Salmonella typhimurium LT2 cells. BMC Evol. Biol. 2:14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsen, H. Y., and J. S. Lin. 2001. Analysis of Salmonella enteritidis strains isolated from food-poisoning cases in Taiwan by pulsed field gel electrophoresis, plasmid profile and phage typing. J. Appl. Microbiol. 91:72-79. [DOI] [PubMed] [Google Scholar]
- 23.van Duijkeren, E., W. J. Wannet, M. E. Heck, W. van Pelt, M. M. Sloet van Oldruitenborgh-Oosterbaan, J. A. Smit, and D. J. Houwers. 2002. Serotypes, phage types and antibiotic susceptibilities of Salmonella strains isolated from horses in The Netherlands from 1993 to 2000. Vet. Microbiol. 86:203-212. [DOI] [PubMed] [Google Scholar]
- 24.Zinder, N. D., and J. Lederberg. 1952. Genetic exchange in Salmonella. J. Bacteriol. 64:679-699. [DOI] [PMC free article] [PubMed] [Google Scholar]