Abstract
A two-step strategy for disaccharide modulation using vancomycin as a model is reported. The strategy relies upon a glycosyltransferase-catalyzed ‘reverse’ reaction to enable the facile attachment of an alkoxyamine-bearing sugar to the vancomycin core. Neoglycosylation of the corresponding aglycon led to a novel set of vancomycin 1,6-disaccharide variants. While the in vitro antibacterial properties of corresponding vancomycin 1,6-disaccharide analogs were equipotent to the parent antibiotic, the chemoenzymatic method presented is expected to be broadly applicable.
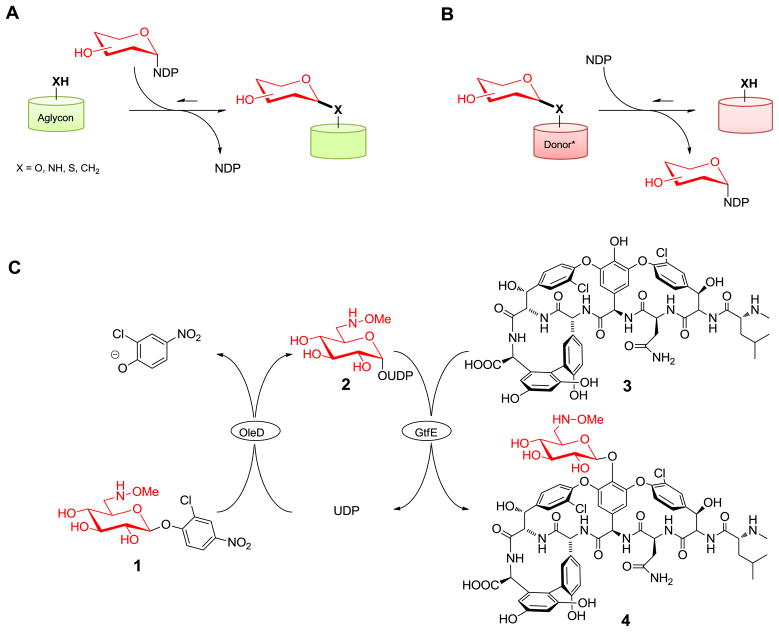
Leloir (sugar nucleotide–dependent) glycosyltransferases (GTs) are ubiquitous in nature where they serve to catalyze the transfer of monosaccharide to a wide array of acceptors including nucleic acids, polysaccharides, proteins, lipids, carbohydrates and medicinally relevant secondary metabolites (Figure 1A).1 A growing appreciation for the reversibility of GT-catalyzed reactions (Figure 1B) has led to new GT-catalyzed methods for the exchange of sugars appended to complex natural-product scaffolds.2 Building upon this concept, we recently reported that simple synthetic aromatic glycosides could serve as efficient donors in such reactions by dramatically altering the overall equilibrium in favor of sugar nucleotide formation (i.e., the ‘reverse’ of a conventional GT-catalyzed reaction).3 This pilot study also revealed that GT-driven sugar nucleotide synthesis could be directly coupled to another GT-catalyzed reaction to ultimately afford a transglycosylation reaction from the synthetic glycoside donor to a complex target scaffold (Figure 1C). To extend the prior pilot study, herein we describe the synthesis of 2-chloro-4-nitrophenyl 6′-deoxy-6′-methoxyamino-β-D-glucopyranoside and its application as a novel donor for: i) direct NDP-6′-deoxy-6′-methoxyamino-D-glucose synthesis, and ii) in situ two GT-catalyzed attachment of 6′deoxy-6′-methoxyamino-D-glucose to the vancomycin aglycon for subsequent chemoselective neoglycosylation through a selective reaction between free reducing sugar and the alkoxyamine handle.4 Cumulatively, this model study illustrates a new general platform to generate disaccharide analogs of complex natural products with anticipated broad applicability.
Figure 1.
(A) Classical GT reaction in which glycoside formation is thermodynamically favored. (B) GT reaction where an appropriately ‘activated’ glycoside donor (Donor*) shifts the reaction thermodynamics to favor sugar nucleotide formation. (C) One-pot coupled dual-GT-catalyzed reaction which combines GT-driven (OleD) sugar nucleotide synthesis with a subsequent GT-catalyzed (GtfE) glycosylation reaction to ultimately accomplish transglycosylation from a synthetic aromatic glycoside to a targeted aglycon (in this case, the vancomycin aglycon 3).
The activated alkoxyaminosugar donor 1 for this study was synthesized in 8 steps with an overall yield of 46% (see supporting information and supplementary figure S1). Donor 1 was subsequently assessed as a substrate for the GT-catalyzed production of 2 in the presence of UDP and the enhanced GT (OleD TDP16) (see supplementary figure S2).5 This reaction was conducted with a 1:1 ratio of glycoside donor to UDP and, after HPLC purification, provided 68 mg (88% isolated yield) of the desired product (2) (see supporting information for full characterization). This illustrated efficiency of the OleD TDP16-catalyzed reverse reaction with the non-native donor 1 sugar sets the stage for a potential one-pot procedure for alkoxyaminosugar attachment to the model vancomycin aglycon as described below and also provides a facile route to a potentially new reagent for glycobiology.
The complex natural product model selected for this study (the glycopeptide vancomycin) is a treatment of last resort for certain antibiotic-resistant Gram-positive pathogens.6 The selection for this model was based upon the known impact of sugar modification upon improving the activity of vancomycin analogs against vancomycin-resistance bacteria,4f,7 and the permissive nature of the vancomycin GT GtfE.8 Vancomycin aglycon (3) was readily obtained by acid hydrolysis of vancomycin.4f,9 and with the simple activated 2-chloro-4-nitrophenyl glycoside (1) in hand we attempted to form the monoglycosylated vancomycin derivative (4) using a dual-GT-catalyzed coupled reaction (Figure 1C). The optimized single-pot reaction (50 mM Tris-HCl buffer, pH 8.5; 1.2 mM donor 1; 1 mM UDP; 1 mM vancomycin aglycon 3; 4 μM OleD TDP16; 10 μM GtfE; 30 °C) was followed by analytical HPLC (see supplementary figure S5). Notably, conversion to the desired product (4) in this one pot reaction (47%) was comparable to the GtfE-catalyzed production of 4 directly from UDP-Glc and 3 (53%).3 To generate sufficient material for downstream neoglycosylation, the dual-GT-catalyzed reaction was scaled (200 mL reaction volume) which, after deproteination and simple purification, afforded 150 mg of the desired product 4 (35% isolated yield, see supplementary material for full characterization) for subsequent neoglycosylation using representative sugars found within the bacterial cell wall or appended to glycopeptides.
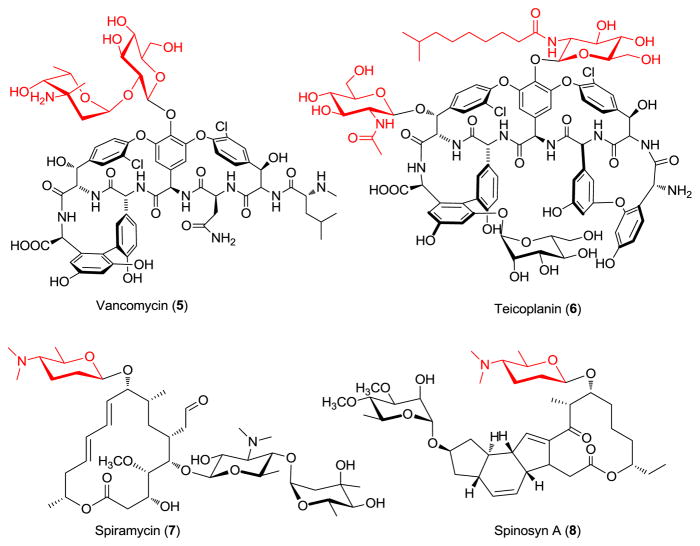
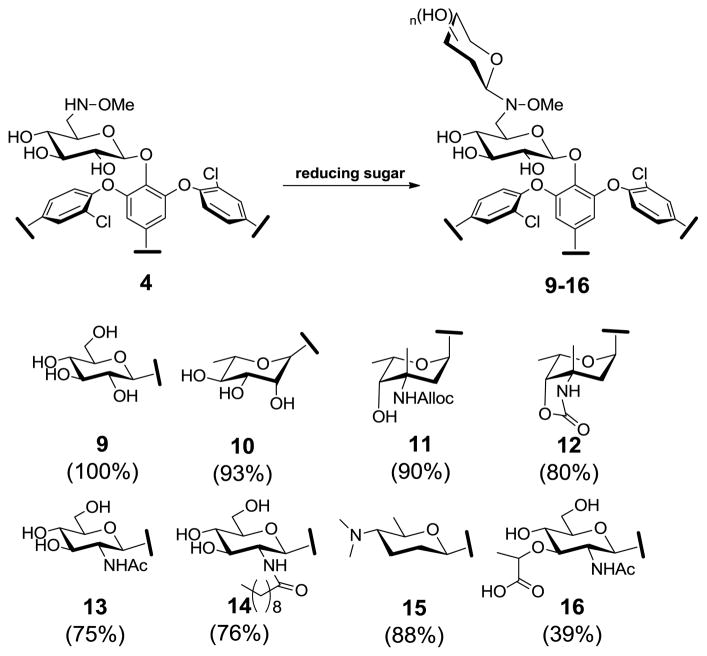
A wide variety of uniquely functionalized carbohydrates decorate the peptide backbone of naturally-occurring glycopeptides and other antibiotics. Most of these sugars fall within the hexo- and 6-deoxyhexopyranosides and include D-glucose, L-rhamnose, L-vancosamine, D-glucosamine and the 2′-N-acyl-D-glucolipid found in teicoplanin (6).10 In addition to these four sugars represented among glycopeptides, we also selected the highly deoxygenated D-forosamine from the macrolide spiramycin (7) and insecticidal spinosyns (8) (Figure 2)11 and N-acetyl-muramic acid, a main component of bacterial peptidoglycan. Among this series, D-glucose, L-rhamnose, D-glucosamine and N-acetyl-muramic acid were commercially available, a representative 2′-N-acyl-D-glucolipid was synthesized as previously described,4f while D-forosamine and L-vancosamine were generated via acid hydrolysis of spiramycin and alloc-protected vancomycin, respectively.9 For the pilot reaction, compound 4 was dissolved in DMSO/AcOH, followed by the addition of a 10-fold excess of D-glucose. The reaction was allowed to proceed at 40 °C and monitored by HPLC. Within 16 h, the starting material was fully consumed, and the newly generated material (9) was subsequently isolated by preparative HPLC. Neoglycosides 10 to 16 were generated in identical fashion (isolated yields from 31 to 95%, see supplementary materials for full characterization) and, with the exception of compound 10 which was formed as an α/β (1:2) mixture, exclusive formation of a single anomer was observed. HMBC and ROESY experiments revealed a strong correlation between the anomeric proton of the neoglycoside with C-6 and H-6 of the alkoxyaminosugar respectively, supporting the regioselectivity of attachment (see supplementary Figure S8).
Figure 2.
Structures of the natural glycopeptide antibiotics vancomycin (5) and teicoplanin (6) and known natural products containing D-forosamine.
Neoglycosides 9–16 and aglycon 3 were found to inhibit growth of two methicillin-resistant (33491 and MW2) bacteria at a concentration between 0.25 to 16 μg/mL (MIC of vancomycin 5 = 0.5 and 0.25 μg/mL respectively), whereas no significant growth inhibition of vancomycin-resistant strains (Van A 256, VA-21) by 9–16 was observe (MIC = 64 μg/mL) (see supplemental Table S3). Thus, while the current range of analogs were unable to circumvent resistance mechanisms, this analysis suggests that 1,6-neoglycosyl modification does not infringe upon the standard vancomycin mechanism of action. More importantly, this model study illustrates an enabling platform for the disaccharide modification of complex biomolecules for which permissive glycosyltransferases are available.12
Supplementary Material
Figure 3.
The neoglycosylation of the 6′-alkoxyaminosugar-substituted vancomycin neoaglycon (4) (upper). The products of the neoglycosylation reaction with corresponding conversions based upon HPLC are illustrated (lower). Full characterization of 9–16 is presented in supporting information.
Acknowledgments
We thank the School of Pharmacy Analytical Instrumentation Center (University of Wisconsin-Madison) for analytical support and Dr. M. Zhou (University of Wisconsin-Madison) for providing forosamine. This work was supported by funding from the NIH (AI52218).
Footnotes
Supporting Information Available: Materials and methods, synthetic procedures and products characterizations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Gantt RW, Peltier-Pain P, Thorson JS. Nat Prod Reports. 2011;28:1811. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]; (b) Chang A, Singh S, Phillips GN, Jr, Thorson JS. Curr Opin Biotechnol. 2011;22:800. doi: 10.1016/j.copbio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Palcic M. Curr Opin Chem Biol. 2011;15:226. doi: 10.1016/j.cbpa.2010.11.022. [DOI] [PubMed] [Google Scholar]; (d) Lairson L, Henrissat B, Davies GJ, Withers SG. Annu Rev Biochem. 2008;77:521. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]; (e) Erb A, Weiss H, Harle J, Bechthold A. Phytochemistry. 2009;70:1812. doi: 10.1016/j.phytochem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 2.(a) Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee I-K, Li L, Thorson JS. Science. 2006;313:1291. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]; (b) Modolo LV, Escamilla-Treviño LL, Dixon RA, Wang X. FEBS Lett. 2009;583:2131. doi: 10.1016/j.febslet.2009.05.046. [DOI] [PubMed] [Google Scholar]; (c) Zhang C, Moretti R, Jiang J, Thorson JS. ChemBioChem. 2008;9:2506. doi: 10.1002/cbic.200800349. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Minami A, Kakinuma K, Eguchi T. Tetrahedron Lett. 2005;46:6187. [Google Scholar]; (e) Zhang C, Albermann C, Fu X, Thorson JSJ. Am Chem Soc. 2006;128:16420. doi: 10.1021/ja065950k. [DOI] [PubMed] [Google Scholar]; (f) Lairson LL, Wakarchuk WW, Withers SG. Chem Commun. 2007:365. doi: 10.1039/b614636h. [DOI] [PubMed] [Google Scholar]; (g) Lougheed B, Ly HD, Wakarchuk WW, Withers SG. J Biol Chem. 1999;274:37717. doi: 10.1074/jbc.274.53.37717. [DOI] [PubMed] [Google Scholar]
- 3.Gantt RW, Peltier-Pain P, Cournoyer WJ, Thorson JS. Nat Chem Biol. 2011;7:685. doi: 10.1038/nchembio.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Peri F, Dumy P, Mutter M. Tetrahedron. 1998;54:12269. [Google Scholar]; (b) Peri F, Deutman A, La Ferla B, Nicotra F. Chem Commun. 2002;1504 doi: 10.1039/b203605c. [DOI] [PubMed] [Google Scholar]; (c) Peri F, Jimenez-Barbero J, Garcia-Aparicio V, Tvaroska I, Nicotra F. Chem-Eur J. 2004;10:1433. doi: 10.1002/chem.200305587. [DOI] [PubMed] [Google Scholar]; (d) Griffith BR, Langenhan JM, Thorson JS. Curr Opin Biotechnol. 2005;16:622. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]; (e) Langenhan JM, Peters NR, Guzei IA, Hoffman FM, Thorson JS. Proc Natl Acad Sci U S A. 2005;102:12305. doi: 10.1073/pnas.0503270102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Griffith BR, Krepel C, Fu X, Blanchard S, Ahmed A, Edmiston CE, Thorson JS. J Am Chem Soc. 2007;129:8150. doi: 10.1021/ja068602r. [DOI] [PubMed] [Google Scholar]; (g) Ahmed A, Peters NR, Fitzgerald MK, Watson JA, Jr, Hoffman FM, Thorson JS. J Am Chem Soc. 2006;128:14224. doi: 10.1021/ja064686s. [DOI] [PubMed] [Google Scholar]; (h) Goff RD, Thorson JS. J Med Chem. 2010;53:8129. doi: 10.1021/jm101024j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Goff RD, Thorson JS. Chem Med Chem. 2011;6:7774. [Google Scholar]; (j) Langenhan JM, Engle JM, Slevin LK, Fay LR, Lucker RW, Smith KR, Endo MM. Bioorg Med Chem Lett. 2008;18:670. doi: 10.1016/j.bmcl.2007.11.058. [DOI] [PubMed] [Google Scholar]; (k) Peltier-Pain P, Timmons SC, Grandemange A, Benoit E, Thorson JS. Chem Med Chem. 2011;6:1347. doi: 10.1002/cmdc.201100178. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Goff RD, Thorson JS. Org Lett. 2009;11:461. doi: 10.1021/ol8025704. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Goff RD, Thorson JS. Org Lett. 2012;14:2454. doi: 10.1021/ol300703z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams GJ, Yang J, Zhang C, Thorson JS. ACS Chem Biol. 2011;6:95. doi: 10.1021/cb100267k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Khane D, Leimkuhler C, Lu W, Walsh CT. Chem Rev. 2005;105:425. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]; (b) Butler MS, Cooper MA. J Antibiotics. 2011;64:413. doi: 10.1038/ja.2011.44. [DOI] [PubMed] [Google Scholar]; (c) Butler MS. Nat Prod Report. 2008;25:475. doi: 10.1039/b514294f. [DOI] [PubMed] [Google Scholar]; (d) Mishra BB, Tiwari VK. Eur J Med Chem. 2008;46:4769. doi: 10.1016/j.ejmech.2011.07.057. [DOI] [PubMed] [Google Scholar]
- 7.(a) Ge M, Chen Z, Onishi HR, Kohler J, Silver LL, Kerns R, Fukuzawa S, Thompson C, Khane D. Science. 1999;284:507. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]; (b) Jabès D, Candiani C, Romano G, Brunati C, Riva M, Cavaleri M. Antimicrob Agents Chemother. 2004;48:1118. doi: 10.1128/AAC.48.4.1118-1123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Patti GJ, Kim SJ, Yu T-Y, Dietrich E, Tanaka KSE, Parr TR, Jr, Rafai Far A, Schaefer J. J Biol Chem. 2009;392:1178. doi: 10.1016/j.jmb.2009.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Ritter TK, Mong KKT, Liu H, Nakatani T, Wong CH. Angew Chem Int Ed. 2003;42:4657. doi: 10.1002/anie.200351534. [DOI] [PubMed] [Google Scholar]; (e) Liu YC, Li YS, Lyu SY, Hsu LJ, Chen YH, Huang YT, Chan HC, Huang CJ, Chen GH, Chou C-C, Tsai MD, Li TL. Nat Chem Biol. 2011;7:304. doi: 10.1038/nchembio.556. [DOI] [PubMed] [Google Scholar]; (f) Oh TJ, Kim DH, Kang SY, Yamaguchi T, Sohng JK. J Antiobiot. 2011;64:103. doi: 10.1038/ja.2010.131. [DOI] [PubMed] [Google Scholar]
- 8.(a) Fu X, Albermann C, Zhang C, Thorson JS. Org Lett. 2005;7:1513. doi: 10.1021/ol0501626. [DOI] [PubMed] [Google Scholar]; (b) Losey HC, Jiang J, Biggins JB, Oberthür M, Ye XY, Dong SD, Kahne D, Thorson JS, Walsh C. Chem Biol. 2002;9:1305. doi: 10.1016/s1074-5521(02)00270-3. [DOI] [PubMed] [Google Scholar]; (c) Fu X, Albermann C, Jiang J, Lia J, Zhang C, Thorson JS. Nat Biotechnol. 2003;21:1467. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]; (d) Solenberg PJ, Matsushima P, Stack DR, Wilkie SC, Thompson RC, Baltz RH. Chem Biol. 1997;4:195. doi: 10.1016/s1074-5521(97)90288-x. [DOI] [PubMed] [Google Scholar]
- 9.Thompson C, Ge M, Kahne D. J Am Chem Soc. 1999;121:1237. [Google Scholar]
- 10.Nicolaou KC, Boddy CNC, Bräse S, Winssinger N. Angew Chem Int Ed. 1999;38:2096. doi: 10.1002/(sici)1521-3773(19990802)38:15<2096::aid-anie2096>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 11.(a) Nguyen HC, Karray F, Lautru S, Gagnat J, Lebrihi A, Ho Huynh TD, Pernodet J-L. Antimicrob Agents Chemother. 2010;54:2830. doi: 10.1128/AAC.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Waldron C, Matsushima P, Rosteck PR, Jr, Broughton MC, Turner J, Madduri K, Crawford KP, Merlo DJ, Baltz RH. Chem Biol. 2001;8:487. doi: 10.1016/s1074-5521(01)00029-1. [DOI] [PubMed] [Google Scholar]
- 12.Williams GJ, Thorson JS. Adv Enzymol Relat Areas Mol Biol. 2009;76:55. doi: 10.1002/9780470392881.ch2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.