Abstract
Purpose
Early diagnosis of primary immunodeficiency disorders (PIDD) is critical for maximizing patient survival and clinical outcomes. Consequently, there is significant interest in developing broad-based, high-throughput, screening approaches capable of utilizing small blood volumes to identify patients with PIDD.
Experimental Design
We developed a novel proteomic screening approach using tandem mass spectrometry to simultaneously identify specific signature peptides derived from the transmembrane protein CD3ε and the intracellular proteins WASP and BTK as markers of three life-threatening PIDDs; Severe Combined Immunodeficiency (SCID), Wiskott-Aldrich syndrome (WAS) and X-Linked Agammaglobulinemia (XLA). Signature peptides were analyzed by LC/MS-MS in proteolytically digested lysates from cell lines and white blood cells. The amount of each peptide was determined by the ratio of the signature peptide peak area to that of a known amount of labeled standard peptide. Peptide concentrations were normalized to Actin.
Results
We show that signature peptides from CD3ε, WASP, and BTK were readily detected in proteolytically digested cell lysate and their absence could correctly identify PIDD patients.
Conclusions and clinical relevance
This proof of concept study demonstrates the applicability of this approach to screen for PIDD and raises the possibility that it could be further multiplexed to identify additional PIDDs and potentially other disorders.
Keywords: Primary Immunodeficiency, Severe Combined Immunodeficiency, X-linked Agammaglobulinemia, Wiskott-Aldrich Syndrome, Peptide Analysis, Tandem Mass Spectrometry
1. INTRODUCTION
Primary immunodeficiency disorders (PIDD) are a diverse group of clinical syndromes. Many of these are associated with the absence of a particular protein or a particular immune cell subset that can provide clues to a diagnosis. Because of the susceptibility to severe infections caused by the immunodeficiency, early diagnosis is critical to initiate patients on appropriate therapy and optimize clinical outcomes. The identification of more than 180 single gene defects that cause various forms of PIDD has dramatically expanded our knowledge of immune mechanisms in humans, but poses new challenges for diagnosing these disorders. High-throughput sequencing techniques are now being utilized to simultaneously screen for genetic defects in a large number of genes or even throughout the genome. These may identify genetic alterations in a number of genes in a single individual. The evaluation of protein expression or function will then be required to determine which of these are relevant and pathogenic. Development of a high-throughput, proteomic approach specifically focused on diagnosis and screening for PIDD would, therefore, be of significant utility. We have developed a novel proteomic technique using tandem mass spectrometry (MS/MS) to identify specific proteolytic “signature peptides” derived from key marker proteins involved in the function of immune cells. We have performed a pilot study to evaluate this approach for the identification of three life threatening immunodeficiency disorders: Severe Combined Immunodeficiency (SCID), Wiskott-Aldrich Syndrome (WAS) and X-Linked Agammaglobulinemia (XLA).
Severe Combined Immunodeficiency (SCID) is a congenital immunodeficiency disorder in which affected children have minimal immune function and are very susceptible to bacterial, viral, and fungal pathogens. Patients with SCID typically become ill within the first months of life. The definitive treatment for SCID is hematopoietic stem cell transplantation. If transplanted before the acquisition of severe viral or fungal infections (typically within the first 3 months of life), the outcome is excellent and survival is now >90% for most types of donor sources [1, 2]. Mutations in 21 different genes have been associated with development of a SCID phenotype [3–7]. The one unifying feature of patients with SCID is a lack of T lymphocytes. Only T-cells express the cell surface CD3 complex, so this can be used as a protein biomarker for SCID. CD3 is composed of multiple subunits (γ, δ, ε, and ζ) that are associated with the T cell receptor [8]. The epsilon subunit (CD3ε) is arguably the best characterized subunit of the CD3 complex and is the target of the majority of monoclonal antibodies generated to detect CD3 on the surface of cells.
Wiskott-Aldrich syndrome (WAS; OMIM 300392) is caused by mutations in the WAS gene that encodes the WASP protein [9, 10]. Patients have a combined immunodeficiency and demonstrate susceptibility to recurrent infections [11]. They also have small, dysfunctional platelets that lead to bleeding problems and thrombocytopenia. X-Linked Agammaglobulinemia (XLA; OMIM 300755) is caused by mutations in the gene encoding Bruton’s Tyrosine Kinase (BTK) [12–14]. Patients with pathogenic mutations in this kinase exhibit a block in B cell development, and suffer recurrent bacterial upper and lower respiratory tract infections, gastrointestinal infections, and sepsis. The disorders presented above are potentially treatable if diagnosed early.
BTK, WASP, and CD3ε are low abundance transmembrane or cytoplasmic proteins expressed only in hematopoietic cells. These proteins are not detectable in plasma. We hypothesize that these proteins can be observed in proteolytically digested extract from white blood cells (WBC), and that analyzing the resultant peptides by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) would allow detection of patients with little or no expression of BTK, WASP, and CD3ε. This is a novel approach, exploiting established genetic information and applying it to a proteomics based assay.
2. METHOD
2.1 Materials
ProteaseMAX™ Surfactant was purchased from Promega Corporation (Madison, WI). Proteomics grade trypsin, bovine serum albumin protein standard (200 mg/mL), and dextran from Leuconostoc mesenteroides were from Sigma Life Science (St. Louis, MO). Dithiothreitol (DTT) was purchased from Fermantas Inc. (Glen Burnie, Maryland). HPLC grade water and acetonitrile (ACN) Optima were from Fisher Scientific. Formic acid (FA), glacial acetic acid, ammonium bicarbonate, and sodium chloride were purchased from Fluka. Coomassie Plus-The Better Bradford Assay™ Reagent was from Thermo Scientific. Custom peptides listed in Table 1 were obtained from AnaSpec (Fremont, CA). Peptide sequences were synthesized as unmodified peptides with free N-terminal and C-terminal amino acids. The stable isotope label (13C, 15N) was incorporated at the lysine or arginine position, resulting in a mass shift of +8 or +10 Da, respectively. The purity of all synthetic peptides was >95% except for BTK 526-536 (90.7%) as measured by HPLC.
Table 1.
List of Candidate Peptides
| Peptide | Sequence | Molecular Weight | Parent Ion | Daughter Ions |
|---|---|---|---|---|
| Actin 31-41 | AVFPSIVGRPR | 1197.7 | 599.9 | 584.4 (y5), 697.4 (y6), 784.5 (y7), 881.5 (y8) |
| Actin 53-63* | DSYVGDEAQSK | 1197.5 | 599.8 | 366.1 (b3), 465.2 (b4), 734.3 (y7), 833.4 (y8) |
| BTK 407-417* | ELGTGQFGVVK | 1133.6 | 567.8 | 549.3 (y5), 734.4 (y7), 835.5 (y8), 892.5 (y9) |
| BTK 526-536* | NCLVNDQGVVK | 1187.6 | 594.8 | 645.4 (y6), 759.4 (y7), 858.47 (y8), 971.6 (y9) |
| BTK 545-558* | YVLDDEYTSSVGSK | 1561.7 | 781.9 | 828.4 (y8), 957.5 (y9), 1072.5 (y10), 1187.5 (y11) |
| CD3ε 74-85 | NIGSDEDHLSLK | 1326.7 | 664.3 443.2 |
1043.5 (y9), 1100.5 (y10) 347.2 (y3), 460.3 (y4) |
| CD3ε 86-101* | EFSELEQSGYYVCYPR | 1968.8 | 985.4 | 1107.5 (y9), 1235.6 (y10), 1364.6 (y11), 1477.7 (y12) |
| CD3ε 197-205* | DLYSGLNQR | 1064.5 | 533.3 | 417.2 (y3), 587.3 (y5), 674.4 (y6), 837.4 (y7) |
| WASP 14-34* | GAPAVQQNIPSTLLQDHENQR | 2315.2 | 772.7 | 683.3 (y5), 719.4 (y12+2), 926.4 (y7), 1039.5 (y8) |
| WASP 274-288* | AGISEAQLTDAETSK | 1519.7 | 760.9 | 751.3 (y7), 864.4 (y8), 992.5 (y9), 1063.5 (y10) |
| WASP 289-304* | LIYDFIEDQGGLEAVR | 1836.9 | 613.3 | 474.3 (y4), 644.4 (y6), 701.4 (y7), 1073.5 (y10) |
Peptides marked with an asterisk are confirmed as signature peptides. The ion type for daughter ions are in parenthesis.
2.2 Selection of Peptides
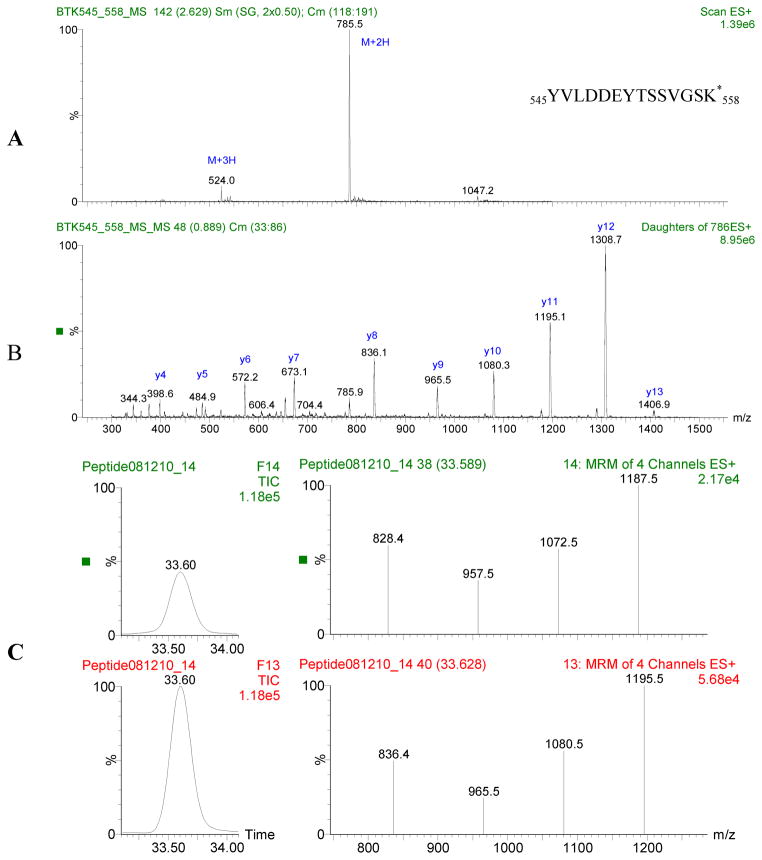
In silico trypsin digestion modeling was performed using the web-based program Protein Prospector (Baker, P.R. and Clauser, K.R. http://prospector.ucsf.edu). Signature peptides were selected for use in MRM experiments based on the following criteria: Peptides containing methionine were excluded because methionine is frequently oxidized. Peptides with single nucleotide polymorphisms (SNPs) were also excluded. The remaining peptides were evaluated for suitability based on sequence. Specifically, the presence of polar amino acids that promote fragmentation and the absence of multiple prolines which inhibit fragmentation were preferable characteristics. Peptide length was also taken into consideration, with 13 to 16 residues considered optimal because such peptides are doubly or triply changed when using electrospray ionization (ESI), which is ideal for fragmentation. The hydrophobicity of the peptide was also taken into account, since very hydrophobic peptides are difficult to synthesize and require long HPLC gradients [15]. Candidate peptides were submitted to a BLAST search to insure that the sequences are unique within the human genome. Candidate peptides were synthesized, and both full MS and MS/MS spectrums were performed on each peptide to ascertain the best MRM transitions and conditions (Figure 1). The “signature” peptide candidates and their MRM transitions are listed in Table 1. The final “signature” peptides were selected by evaluating the MRM chromatogram of an isotopically labeled version of the peptide and comparing this to the mixed peptide pool from proteolytically digested WBC.
Figure 1.
A: Mass spectrum of labeled BTK 545:558. B: Tandem mass spectrum of 2+2H2+. Abundant fragment ions are selected and optimized for MRM analysis. C: Chromatogram and MRM spectrum for BTK 545:558 peptide. Top panel is signature peptide found in lymphocyte cell line extract, bottom panel is the isotopically labeled internal standard. Chromatographic peaks overlap and MRM patterns are comparable.
2.3 Sample Preparation
WBCs from normal individuals and PIDD patients were isolated from 2.5 – 5 mL of anticoagulated whole blood (EDTA or ACD) using a dextran gradient. The study was approved by the institutional review board of Seattle Children’s Hospital. WBCs were then dissolved in 500 μL 0.1% ProteaseMAX surfactant in 50 mM ammonium bicarbonate by vortexing for 30 min. At this point aliquots were reserved for a Bradford assay. Disulfide bond reduction and trypsin digestion were performed in a single step with 2 M DTT and ACN added to final concentrations of 5 mM and 15% respectively. Trypsin was used in a 1:20 enzyme to protein ratio (w:w). The mixture was incubated in a 37 °C water bath overnight. After digestion, samples were dried under a stream of nitrogen to approximately 500 μL. At this point, 1 mL acetonitrile was added and the sample was vortexed for 1 minute then placed in an ice bath for 30 min. After a 5 minute centrifugation, the supernatant was transferred to a new tube, and dried completely under a nitrogen stream. The dried sample was reconstituted in 5% ACN in water with 0.1% FA containing a 25 nM mixture of labeled internal standard (IS) peptides. The samples were centrifuged at 13000 rpm for 5 minutes to remove impurities left from the digestion.
2.4 Liquid Chromatography-Mass Spectrometry
LC-MS/MS analyses were performed on a Waters Acquity UPLC system coupled to a Waters Quattro Premier XE triple quadrupole mass spectrometer equipped with an electrospray ion source. The peptides were separated on a Waters Acquity UPLC BEH C18 column (2.1 × 150 mm, 1.7 μm particles). Elution was performed at a flow rate of 100 μL/min with water containing 0.1% FA (v/v) as solvent A and acetonitrile containing 0.1% FA (v/v) as solvent B. A linear gradient of 5–25% solvent B was applied over 60 minutes followed by column washing and reconditioning. MS/MS analysis was performed in positive ion mode with the capillary voltage set to 3.00 kV. The desolvation gas was 700 L/hr and the cone gas was 100 L/hr. The collision energy and cone voltage were optimized for each peptide by directly infusing the isotopically labeled peptide in 20% solvent B at 100 μL/min. The resolutions of Q1 and Q3 were, respectively, 14.0/14.0 (HM/LM) and 12/12 (HM/LM). The dwell times were 0.05 s for BTK 407 and CD3ε 197, 0.07 s for BTK 526 and WASP 274, 0.1 s for Actin 53, 0.13 s for BTK 545 and WASP 14, 0.15 s for CD3 ε 86, and 0.2 s for WASP 289 transitions.
2.5 Protein Quantification
The chromatographic peaks for the signature peptide and isotopically labeled peptide were integrated using QuanLynx (Waters) software. The most abundant transition for each pair was used for quantification unless interference from the matrix observed. The concentrations of BTK, WASP, CD3ε, and actin, as represented by their respective signature peptides are reported, normalized to actin unless otherwise noted. The amount of each peptide in a particular WBC sample was determined by calculating the ratio of the peak areas for the signature peptide to that of its labeled IS present at a known concentration.
2.6 Precision and Linearity
WBCs from individual normal donors were isolated from 2.5 ml aliquots of fresh blood. These were used to validate the precision and linearity of the LC-MS/MS assay. Inter-assay precision was evaluated by analyzing 3 WBC pellets from single whole blood donors. Protein extraction, digestion, and MRM analysis were performed independently for each sample. Intra-assay precision was evaluated using 10 WBC pellets from a single donor. Protein extraction, digestion, and MRM analysis were performed in a single experiment.
Linearity was established by loading between 1 μg and 100 μg of digested protein onto the UPLC column for analysis. A stock protein solution was prepared by extracting protein from a WBC pellet isolated from 8 mL of whole blood. The amount of total protein, determined by a Bradford assay, was used to determine the amount of trypsin necessary for complete digestion and for calculating the amount of digested protein (μg) loaded onto the UPLC column. A seven point curve spanning two orders of magnitude was acquired. The concentrations of labeled IS were held constant at 25 nM throughout the experiment in order to assess ion suppression.
2.7 Control and Patient Protein Concentration Ranges
The normal concentration ranges for each protein were established using 45 control samples. Samples consisted of de-identified EDTA blood left over from routine blood testing in the hospital laboratory. When 2.5 mL of whole blood was available, the sample was labeled with gender and age. When less than 2.5 mL was available, samples were pooled into 3 groups based on age (< 1 year old, 1–5 years old, and > 5 years old).
In blinded trial, various cell lines (Jurkat, Daudi, WAS (−) BLCL, HUT78, and THP-1) lacking one or more of the target proteins (WASP, CD3ε, BTK) were used to establish the absence of signature peptides for WAS, SCID, and XLA. Each pellet contained approximately 1 million cells and was digested using the procedure described for WBC. Likewise, 16 de-identified PBMC samples from 5 WAS, 5 SCID, and 6 XLA patients were also analyzed in a blinded fashion. Lastly, as an additional approach to mimic the situation in SCID, CD3+ T cells were depleted from whole blood using the HLA whole blood positive selection kit (Stemcell Technologies, Vancouver, Canada). CD3 depleted and non-depleted samples were prepared from the same normal individuals. Depletion of CD3+ T cells was confirmed by flow cytometry (data not shown). The procedure for protein extraction, digestion and analysis was identical to the control samples.
3. RESULTS
3.1 Selection of Signature Peptides
Signature peptide candidates for each target protein (WASP, CD3ε, BTK) were identified using in silico analysis with the criteria outlined above. To determine which of the candidates would be useful as signature peptides, an isotopically labeled version of each candidate peptide was synthesized. WBC pellets from normal controls were solubilized and digested with trypsin to yield a complex peptide mixture. This mixture was analyzed for the presence of each candidate peptide by simultaneously scanning for the candidate and labeled peptide transitions. A candidate was confirmed as a signature peptide only when the chromatographic peak and MRM pattern for the candidate peptide and labeled peptide were comparable. This is demonstrated in Figure 1; in this chromatogram, unlabeled BTK 545:558 derived from the WBC, and isotopically labeled BTK 545:558, which was added to the WBC extract, co-elute and had comparable MRM patterns. This confirms BTK 545-558 as a signature peptide.
Three isotopically labeled standard peptides each were purchased for BTK, WASP, and CD3ε, and two for actin. Each peptide was evaluated as described and at least one peptide from each protein was chosen as its signature peptide. During the UPLC chromatographic separation, 9 signature peptides and their isotopically labeled internal standard (IS) were analyzed by MRM. The peptides selected as signature peptides are indicated in Table 1 with an asterisk and their chromatograms and mass spectrum were shown in Figure S1.
3.2 Quantitation of Signature Peptides
To determine if the chosen signature peptides could be used to accurately identify cells lacking one or more of the three target proteins, five different cell lines were analyzed in a blinded fashion for the presence of WASP, CD3ε, and BTK. The results, summarized in Table 2, indicate which proteins were detected in each cell line. As shown, this assay accurately identified the predicted phenotype of each cell line studied (Table 2). Based on these results, the signature peptides that consistently performed best in terms of sensitivity and specificity (WAS 274, CD3ε 197, and BTK 526) were chosen for further analysis. We next determined whether the assay could be used to accurately identify patients with immunodeficiency by analyzing 16 blinded PBMC pellets from 5 SCID, 5 WAS and 6 XLA patients. In addition to the proteins of interest, one housekeeping protein, actin, was included in the analysis as an internal control for both normalizing protein concentration and sample integrity. We found that when actin level from control samples was very low, none of target peptides was observed, indicating potential sample degradation. One sample from a WAS patient was eliminated from further analysis due to very low level of actin. Of the 15 blinded PBMC samples, 12 samples were correctly identified (Table 3). The 3 patients that were not correctly identified highlight some of the potential weaknesses of this method. Patient SCID5 had X-linked SCID caused by mutations in the IL2RG gene. Under normal circumstances, patients with X-SCID lack both T cells and NK cells but have normal to elevated numbers of B cells (TnegB+NKneg SCID). This patient was however engrafted with maternal T cells so that his T cell counts were actually elevated. The CD3ε peptide was therefore readily detected as was the WAS peptide however the BTK peptide was not detected. We believe that this was due to the majority of lymphocytes in the sample being T cells which diluted-out the BTK peptide below the detection limit of this assay. The other 2 patients that could not be characterized were WAS3 and WAS4, brothers who were both moderately lymphopenic. We believe that this may be the reason that the peptides were all below the detection limit of the assay. For each of these samples, the Actin peptide was detected and found to be in an acceptable range suggesting that there may have been contaminating non-lymphoid cells in the sample, possibly myeloid lineage cells. In PBMC with CD3+ T cell artificially depleted (samples #1 and #5), no CD3ε or very low amount of CD3ε were detected compared to the normal PBMC sample (#3) as expected (Table 4).
Table 2.
Signature Peptide Expression in Cell Lines
| Name | Cell Type | Actin 53 | BTK 526 | BTK 407 | BTK 545 | WASP 274 | WASP 14 | WASP 289 | CD3ε 197 | CD3ε 86 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Normalized to Cell Counta (x10^8) | Jurkat | T-cell leukemiac | 1.8 × 103 | NDb | ND | ND | 1.2 | 2.2 | 3.4 | 17.3 | 41.1 |
| HUT78 | T-cell lymphomac | 2.3 × 103 | ND | ND | ND | 11.0 | 12.8 | 12.8 | 5.0 | 14.6 | |
| Daudi | B-cell lymphomad | 3.0 × 103 | 9.0 | 6.3 | 5.3 | 10.9 | 15.1 | 17.7 | ND | ND | |
| WAS(−) BLCL | B lymphoblastse | 8.7 × 103 | 6.4 | 6.3 | 4.5 | ND | 0.5 | ND | ND | ND | |
| THP-1 | Myelomonocyticd | 1.8 × 104 | 62.7 | 7.7 | 3.3 | 3.7 | 15.3 | 71.4 | ND | ND | |
|
| |||||||||||
| Normalized to Actin (x10^3)a | Jurkat | T-cell leukemia | ND | ND | ND | 0.7 | 1.2 | 1.9 | 9.6 | 22.7 | |
| HUT78 | T-cell lymphoma | ND | ND | ND | 4.8 | 5.6 | 5.6 | 2.2 | 6.4 | ||
| Daudi | B-cell lymphoma | 3.0 | 2.1 | 1.8 | 3.6 | 5.0 | 5.9 | ND | ND | ||
| WAS(−) BLCL | B lymphoblasts | 0.7 | 0.7 | 0.5 | ND | 0.1 | ND | ND | ND | ||
| THP-1 | Myelomonocytic | 3.5 | 0.4 | 0.2 | 0.2 | 0.9 | 4.0 | ND | ND | ||
Relative ratio of peak area for signature peptide to peak area for isotopically labeled internal standard, normalized to cell count or actin concentration.
ND: Not detected.
Predicted phenotypes: T-cell lines: BTKnegWASP+CD3ε+, B-cell lines: BTK+WASP+CD3εneg, WAS(−) BLCL: BTK+WASPnegCD3εneg.
Table 3.
Signature Peptide Concentrations in 15 Blinded PBMC Samples from PIDD Patients
| Sampleb | Normalized to Actina | |||
|---|---|---|---|---|
| BTK 526 | WASP 274 | CD3ε 197 | Gene (Mutation) | |
| SCID1 | 1.4 | 0.5 | ND | IL2RG p.L293Q |
| SCID2 | 2.0 | 0.3 | ND | IL2RG p.R285Q |
| SCID3 | 1.6 | 0.3 | ND | RAG1 p.delM661 |
| SCID4 | 4.4 | 1.0 | ND | IL2RG p.E110X |
| SCID5 | ND | 0.3 | 4.4 | IL2RG p.R224W |
| WAS1 | 2.6 | ND | 2.1 | WASP c.del-719_273 |
| WAS2 | 1.2 | ND | 0.8 | WASP p.D485N |
| WAS3 | 1.8 | ND | ND | WASP c.471_476insA |
| WAS4 | ND | ND | ND | WASP c.471_476insA |
| XLA1 | ND | 5.0 | 3.3 | BTK p.R520X |
| XLA2 | ND | 3.1 | 2.4 | BTK p.R520X |
| XLA3 | ND | 3.4 | 4.8 | BTK p.E589G |
| XLA4 | ND | 1.1 | 1.9 | BTK p.R525X |
| XLA5 | ND | 0.8 | 3.5 | BTK c.974+4A>G |
| XLA6 | ND | 0.6 | 3.4 | BTK p.W251X |
| Control Samplesc | ||||
| < 1 year old (24 samples) | 0.47 ± 0.3 | 0.29 ± 0.1 | 0.12 ± 0.1 | |
| 1–5 years old (5 samples) | 0.53 ± 0.4 | 0.57 ± 0.7 | 0.35 ± 0.2 | |
| > 5 years old (16 samples) | 0.37 ± 0.3 | 0.31 ± 0.2 | 0.18 ± 0.2 | |
|
Precision (% C.V.)
| ||||
| Inter-assay (3) | 21.0% | 20.3% | 14.1% | |
| Intra-assay (10) | 19.6% | 18.1% | 24.3% | |
Relative ratio to isotopically labeled internal standards normalized to actin concentration,
PBMC samples,
Control ranges established using proteolytically digested WBC
Table 4.
CD3ε signature peptide concentrations in T-cell depleted PBMC
| Normalized to Actin | ||
|---|---|---|
|
| ||
| Sample | CD3ε86 | CD3ε197 |
| CD3ε depletion #1 | ND | ND |
| CD3ε depletion #3 | 12.4 | 1.1 |
| CD3ε depletion #5 | 0.8 | 0.1 |
3.3 Validation Results
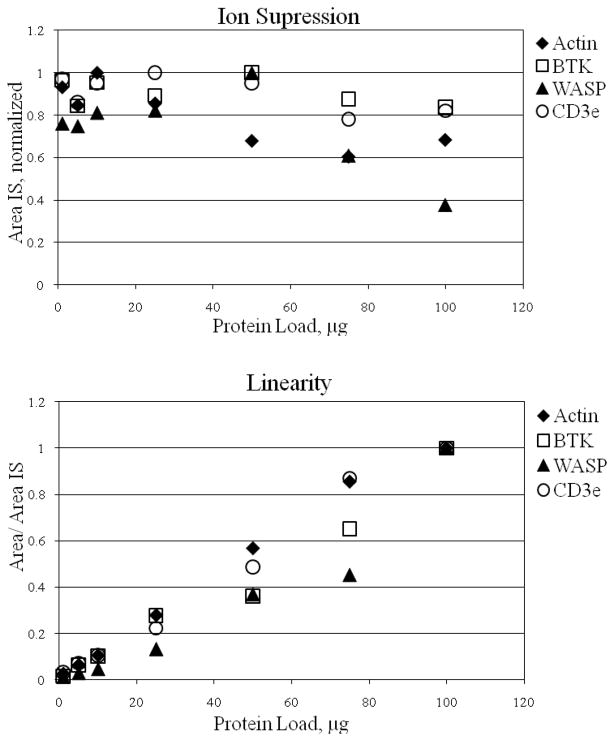
The intra-assay and inter-assay coefficients of variation (%CV) are listed in Table 3, ranging from 13.8% to 24.3% CV. Each labeled peptide demonstrated modest ion suppression with increasing sample load, but the ratio of signature peptide to labeled peptide was linear over two orders of magnitude for each peptide except WASP, which was linear over only one order of magnitude (Figure 2). The WASP peptide proved to be very sensitive to total protein load. As expected, the quality of the chromatography decreased with higher sample loads; this led to fat peaks and drifting retention times. The limit of detection, measured using the isotopically labeled standards, was 280 fmole/mg protein for each peptide except BTK 526-536, for which it was 1.4 pmole/mg protein.
Figure 2.
Top: Intensity of 25 nM labeled peptide expressed as a function of total protein load. Bottom: Ratio of signature peptide to labeled standard is linear over a range from 1 to 50 μg protein load for all peptides and up to 100 μg protein load for except WASP.
4. DISCUSSION
We have developed a proteomic method of screening for primary immunodeficiency disorders using proteolytic “signature” peptides and tandem mass spectrometry. We have demonstrated that our method can readily identify genetic disorders that are characterized by a complete absence or marked decrease of a specific protein. In our blinded cell line study, 5 different cell lines were analyzed, probing peptides from 3 proteins that are not expressed in the disease state. The results from the cell line study were very clear; we observed the peptides from the proteins expected to be present, and peptides from the proteins not expected to be in the sample were undetectable. We did detect a small amount of WASP 14-34 in the WAS (−) BLCL sample. The concentration was significantly lower than what was observed in the other PBMC samples, specifically, it was only 2% of the concentration observed in the other B-cell line. The other WASP peptides were not detectable and it is possible that the observed peak is an artifact.
In efforts to further demonstrate that our method can be used as a diagnostic for genetic disorders, we first analyzed fresh PBMC’s from three BTK deficient patients. The results clearly indicate that CD3ε and WASP are present and BTK is not detectable. This is encouraging because 60% of BTK mutations result in the absence of the BTK protein in B cells and platelets. It appears that this method could quickly allow the detection of BTK proteins. We also had successfully identified CD3ε and WASP deficient cell lines and also documented that CD3ε was missing in T-cell depleted cell line. As most disease causing mutations are functionally null in these conditions, it is feasible that we could easily detect the patient lacking CD3ε or WASP with our method. To further investigate and validate the targeted peptides in affected patients, we were able to retrieve 13 additional cryopreserved samples for analysis. One of them was removed from the data as the sample had non-detectable actin peak likely degraded. In one patients (WAS4), we did not see any detectable targeted peaks though substantial amount of actin peak was present. In WAS3 sample, correct diagnosis couldn’t be also reached due to two absent peptide peaks. This possibly indicates both B and T cell populations were too low to be quantitated indicating the amount of peptide was below the detection limit. In SCID5 sample, the result was inconsistent with a known diagnosis. We saw significant amount of CD3e in this sample, while the BTK peak was absent which also mislead to the diagnosis of XLA. In addition, this method can miss some patients who would present with functional deficiency but intact protein concentration. We believe this method requires further study on important groups such as atypical SCID (Omenn’s syndrome or those with maternal-fetal engraftment) or those lymphopenic patients secondary to drugs or untreated HIV.
XLA and WAS are X-linked conditions, however, in quantitating these proteins, we did not observe any difference between female and male control samples. The control sample numbers in this study are quite limited, so we would need to analyze more samples, including proven carriers, to determine more accurate reference and disease ranges. When we prepared WBC pellet from 2.5 mL of whole blood, we observed considerable variation in the size of the pellets and the amount of total protein extracted. This is not surprising as there is significant variability in WBC counts between healthy individuals. We observed that the concentrations of our marker proteins, normalized to total protein, varied significantly from sample to sample. We believe that this may be the result of contamination from varying amounts of serum proteins. To address this issue, we normalized our marker proteins to actin, a ubiquitous intracellular structural protein. The %CV when normalizing to actin is below 20% for BTK and WASP, and 24.3% for CD3ε.
Although this CV may still be too high for large scale testing, we believe that this can be improved with modest optimization for sample preparation. This variability is due to multiple factors, including trypsin digestion efficiency, peptide recovery in the process, and sample handling. One way to control for variable digestion and peptide recovery efficiency would be to use stable isotope labeled protein standards since the stable isotope labeled protein standards undergoes the same processing as endogenous target proteins, resulting in improvement in the precision. Moreover, the use of automated sample preparation would increase the throughput of the system, decrease sample handling errors, and increases reproducibility.
Linearity studies performed for each of the signature peptides provide insight into the complex nature of our samples. When choosing how much sample to load it is important to consider both limit of detection and ion suppression. Our data indicate that loading 25 μg of digested protein strikes a balance between LOD and ion suppression. Below 5 μg total digest protein, all signature peptides fell below an acceptable signal to noise ratio. At protein loads greater than 50 μg, the response of all of the labeled peptides decrease.
Analysis of low abundance proteins using this method presents some unique challenges: First, sufficient sample needs to be loaded to allow detection of low abundance proteins, but increasing sample load causes distortions to the chromatography and ion suppression. A popular approach for low abundance protein analysis is depletion of high abundance proteins from whole blood protein mixtures. Several high abundance protein depletion kits are available, but these kits are aimed at depleting plasma proteins [21–23]. Given that the plasma proteome is different from the WBC proteome, we do not believe these would significantly improve our current results. We are, however, refining this method in order to use whole blood or dried blood spots as the starting material. Commercially available depletion kits which deplete hemoglobin, albumin, and other abundant proteins in blood may be of limited benefit. Alternatively, we believe that using a peptide affinity-based approach could allow us to enrich signature peptides while simultaneously removing unwanted high abundance peptides [24, 25].
Recently, the Secretary’s Advisory Committee for Heritable Disorders in Newborns and Children unanimously agreed to recommend the addition of SCID to the uniform newborn screening panel [26]. The purpose of newborn screening is to identify patients with treatable genetic conditions early enough to prevent them from developing permanent complications. With the development of tandem mass spectrometry in the early 1990s, the number of detectable diseases expanded considerably, and included fatty acid oxidation defects and organic acid disorders [27]. The absence of T-cell receptor gene excision circles appears a sensitive marker of profound T lymphocytopenia and currently is the most developed screening method for SCID. Our ultimate goal is to transition from white blood cell pellets to dried blood spots so this technique may be used both for diagnosis of suspected PIDD patients and for newborn screening. While this will be challenging, we believe that with further optimizing and improvements to the peptide enrichment process, this is a possible goal.
In summary, we developed and validated a method to quantify proteolytic signature peptides for BTK, WASP and CD3ε allowing us to rapidly and simultaneously screen for the absence of these proteins in various cell lines and patient WBC samples. Of note, these proteins are either cytoplasmic or membrane bound low-abundance proteins. There are numerous immunodeficiency disorders that are associated with decreased or absent expression of a particular protein or absence of a specific cell subset that can be detected by absence of a particular marker protein. Since this approach has the potential to be highly multiplexed, we believe that it may allow for simultaneous screening for numerous conditions. With further optimization and enrichment, this method can be ultimately applied to newborn screening for primary immunodeficiency.
Supplementary Material
Clinical Implications.
Signature peptides from three primary immunodeficiency disorders were observed in proteolytically digested white blood cells. BTK, WASP, and CD3ε are low abundance transmembrane or cytoplasmic proteins, not detectable in plasma. This is the first report demonstrating the large proteins within the WBC can be targeted for proteomic clinical applications. Quantification of these peptides provides the basis for a multiplexed screening and diagnostic tool. This approach can provide a testing methodology that is viable both for newborn screening and for rapid diagnostics for severe immunodeficiency and other congenital disorders. This will provide opportunities to identify patients early in life and provide life-saving therapies.
Acknowledgments
This study was supported by the grant from NIH (5R21AI85488-2).
Abbreviations
- ACD
Acid Citrate Dextrose
- BTK
Bruton’s Tyrosine Kinase
- CD3
Cluster of Differentiation 3
- DTT
Dithiothreitol
- FA
Formic Acid
- HLA
Human Leukocyte Antigen
- LC-MS/MS
Liquid Chromatography Tandem Mass Spectrometry
- PBMC
Peripheral Blood Mononuclear Cells
- PIDD
Primary Immunodeficiency Disorders
- SCID
Severe Combined Immunodeficiency
- SNP
Single Nucleotide Polymorphism
- UPLC
Ultra Performance Liquid Chromatography
- WAS
Wiskott-Aldrich Syndrome
- WASP
Wiskott-Aldrich Syndrome Protein
- WBC
White Blood Cell
- XLA
X-Linked Agammaglobulinemia
References
- 1.Grunebaum E, Mazzolari E, Porta F, Dallera D, et al. Bone marrow transplantation for severe combined immune deficiency. Jama. 2006;295(5):508–18. doi: 10.1001/jama.295.5.508. [DOI] [PubMed] [Google Scholar]
- 2.Roifman CM, Grunebaum E, Dalal I, Notarangelo L. Matched unrelated bone marrow transplant for severe combined immunodeficiency. Immunol Res. 2007;38(1–3):191–200. doi: 10.1007/s12026-007-0042-y. [DOI] [PubMed] [Google Scholar]
- 3.Corneo B, Moshous D, Gungor T, Wulffraat N, et al. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or Omenn syndrome. Blood. 2001;97(9):2772–6. doi: 10.1182/blood.v97.9.2772. [DOI] [PubMed] [Google Scholar]
- 4.Kalman L, Lindegren ML, Kobrynski L, Vogt R, et al. Mutations in genes required for T-cell development: IL7R, CD45, IL2RG, JAK3, RAG1, RAG2, ARTEMIS, and ADA and severe combined immunodeficiency: HuGE review. Genet Med. 2004;6(1):16–26. doi: 10.1097/01.GIM.0000105752.80592.A3. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz K, Gauss GH, Ludwig L, Pannicke U, et al. RAG mutations in human B cell-negative SCID. Science. 1996;274(5284):97–9. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 6.Tabori U, Mark Z, Amariglio N, Etzioni A, et al. Detection of RAG mutations and prenatal diagnosis in families presenting with either T-B- severe combined immunodeficiency or Omenn’s syndrome. Clin Genet. 2004;65(4):322–6. doi: 10.1111/j.1399-0004.2004.00227.x. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Z, Yannone SM, Dunn E, Cowan MJ. A novel missense RAG-1 mutation results in T-B-NK+ SCID in Athabascan-speaking Dine Indians from the Canadian Northwest Territories. Eur J Hum Genet. 2009;17(2):205–12. doi: 10.1038/ejhg.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumberg RS, Ley S, Sancho J, Lonberg N, et al. Structure of the T-cell antigen receptor: evidence for two CD3 epsilon subunits in the T-cell receptor-CD3 complex. Proc Natl Acad Sci U S A. 1990;87(18):7220–4. doi: 10.1073/pnas.87.18.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78(4):635–44. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- 10.Lemahieu V, Gastier JM, Francke U. Novel mutations in the Wiskott-Aldrich syndrome protein gene and their effects on transcriptional, translational, and clinical phenotypes. Hum Mutat. 1999;14(1):54–66. doi: 10.1002/(SICI)1098-1004(1999)14:1<54::AID-HUMU7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan KE, Mullen CA, Blaese RM, Winkelstein JA. A multiinstitutional survey of the Wiskott-Aldrich syndrome. J Pediatr. 1994;125(6 Pt 1):876–85. doi: 10.1016/s0022-3476(05)82002-5. [DOI] [PubMed] [Google Scholar]
- 12.Bruton OC. Agammaglobulinemia. Pediatrics. 1952;9(6):722–8. [PubMed] [Google Scholar]
- 13.Rawlings DJ, Witte ON. Bruton’s tyrosine kinase is a key regulator in B-cell development. Immunol Rev. 1994;138:105–19. doi: 10.1111/j.1600-065x.1994.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 14.Vetrie D, Vorechovsky I, Sideras P, Holland J, et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature. 1993;361(6409):226–33. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 15.Cagney G, Amiri S, Premawaradena T, Lindo M, Emili A. In silico proteome analysis to facilitate proteomics experiments using mass spectrometry. Proteome Sci. 2003;1(1):5. doi: 10.1186/1477-5956-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derry JM, Kerns JA, Weinberg KI, Ochs HD, et al. WASP gene mutations in Wiskott-Aldrich syndrome and X-linked thrombocytopenia. Hum Mol Genet. 1995;4(7):1127–35. doi: 10.1093/hmg/4.7.1127. [DOI] [PubMed] [Google Scholar]
- 17.Jin Y, Mazza C, Christie JR, Giliani S, et al. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood. 2004;104(13):4010–9. doi: 10.1182/blood-2003-05-1592. [DOI] [PubMed] [Google Scholar]
- 18.Qasim W, Gilmour KC, Heath S, Ashton E, et al. Protein assays for diagnosis of Wiskott-Aldrich syndrome and X-linked thrombocytopenia. Br J Haematol. 2001;113(4):861–5. doi: 10.1046/j.1365-2141.2001.02832.x. [DOI] [PubMed] [Google Scholar]
- 19.Wengler GS, Notarangelo LD, Berardelli S, Pollonni G, et al. High prevalence of nonsense, frame shift, and splice-site mutations in 16 patients with full-blown Wiskott-Aldrich syndrome. Blood. 1995;86(10):3648–54. [PubMed] [Google Scholar]
- 20.Greer WL, Shehabeldin A, Schulman J, Junker A, Siminovitch KA. Identification of WASP mutations, mutation hotspots and genotype-phenotype disparities in 24 patients with the Wiskott-Aldrich syndrome. Hum Genet. 1996;98(6):685–90. doi: 10.1007/s004390050285. [DOI] [PubMed] [Google Scholar]
- 21.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Cellar NA, Karnoup AS, Albers DR, Langhorst ML, Young SA. Immunodepletion of high abundance proteins coupled on-line with reversed-phase liquid chromatography: a two-dimensional LC sample enrichment and fractionation technique for mammalian proteomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(1–2):79–85. doi: 10.1016/j.jchromb.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Qian WJ, Kaleta DT, Petritis BO, Jiang H, et al. Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Mol Cell Proteomics. 2008;7(10):1963–73. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whiteaker JR, et al. Evaluation of large scale quantitative proteomic assay development using peptide affinity-based mass spectrometry. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiteaker JR, Zhao L, Abbatiello SE, Burgess M, et al. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362(1):44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipstein E, Knapp AA, Perrin JM. EVIDENCE REVIEW: Severe Combined Immunodeficiency (SCID) ADVISORY COMMITTEE ON HERITABLE DISORDERS IN NEWBORNS AND CHILDREN 2010 [Google Scholar]
- 27.Millington DS, Kodo N, Norwood DL, Roe CR. Tandem mass spectrometry: a new method for acylcarnitine profiling with potential for neonatal screening for inborn errors of metabolism. J Inherit Metab Dis. 1990;13(3):321–4. doi: 10.1007/BF01799385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.