Abstract
Within the nucleus of each cell lies DNA—an unfathomably long, twisted, and intricately coiled molecule—segments of which make up the genes that provide the instructions that a cell needs to operate. As we near the 60th anniversary of the discovery of the DNA double helix, crucial questions remain about how the physical arrangement of the DNA in cells affects how genes work. For example, how a cell stores the genetic information inside the nucleus is complicated by the necessity of maintaining accessibility to DNA for genetic processing. In order to gain insight into the roles played by various proteins in reading and compacting the genome, we have developed new methodologies to simulate the dynamic, three-dimensional structures of long, fluctuating, protein-decorated strands of DNA. Our a priori approach to the problem allows us to determine the effects of individual proteins and their chemical modifications on overall DNA structure and function. Here, we present our recent treatment of the communication between regulatory proteins attached to precisely constructed stretches of chromatin. Our simulations account for the enhancement in communication detected experimentally on chromatin compared to protein-free DNA of the same chain length, as well as the critical roles played by the cationic ‘tails’ of the histone proteins in this signaling. The states of chromatin captured in the simulations offer new insights into the ways that the DNA, histones, and regulatory proteins contribute to long-range communication along the genome.
Keywords: Chromatin, Histone tails, DNA looping, Monte-Carlo simulations, Protein communication
Introduction
Although the genetic messages in DNA are stored in a linear sequence of base pairs, the genomes of living species do not function in a linear fashion. Gene expression is regulated by DNA elements that often lie far apart along the genomic sequence but come close together during genetic processing. The intervening residues form loops, mediated by proteins, which bind at distant sites along the DNA. The DNA in eukaryotes is further organized into nucleosomes, assemblies of four duplicated histone proteins and ∼150 base pairs of DNA. The known high-resolution structures of nucleosomes are conserved across species ranging from yeast to humans, with the DNA following similar left-handed superhelical pathways around the histone protein core in the ∼70 presently solved high-resolution structures (Xu and Olson 2010). The nucleosomes, in turn, assemble into chromatin fibers, and the strings of nucleosomes in chromatin often form extremely long loops, e.g., up to hundreds of thousands of base pairs in HeLa cells (Jackson et al. 1990). How proteins communicate across these distances has long been a mystery. In order to clarify the role of histones and DNA in chromatin looping, we have developed a structurally based model of chromatin at the resolution of a single base pair and performed Monte Carlo simulations to explore the relative ease of long-distance communication between regulatory proteins at the ends of nucleosome-bound and protein-free DNA chains (Kulaeva et al. 2012). This review describes our computational approach and the new insights into chromatin architecture and folding gained from the correspondence between our simulations of protein-mediated loop formation and new quantitative biochemical measurements of long-range protein communication. The combination of simulation and experiment gives us the ability to relate local changes in nucleosome composition to global chromatin architecture and to tie the looping propensities of stretches of chromatin to biological activity.
Computational challenges and solutions
Long-range communication on chromatin
The level of gene products formed upon the association of transcriptional proteins, bound at the ends of saturated, nucleosome-decorated DNA arrays, provides a novel measure of the looping propensities of chromatin (Polikanov et al. 2007; Polikanov and Studitsky 2009). Molecular configurations that bring a bound activator protein into direct contact with the RNA polymerase assembly lead to the production of RNA transcripts. The amounts of RNA detected upon association of these proteins reveal surprising new information about the intervening DNA. In particular, the presence of nucleosomes along the DNA strongly facilitates distant communication between the regulatory proteins and suggests that the observed enhancement of gene expression over that found on nucleosome-free DNA is more than simple compaction of the double helix by the nucleosomes (Rubtsov et al. 2006). Furthermore, the communication between the regulatory proteins is more pronounced on chromatin constructs made up of intact rather than tailless nucleosomes (Kulaeva et al. 2012). In other words, the positively charged N-terminal tails of the histone proteins somehow influence gene transcription.
Interpretation of these findings requires a model of chromatin that incorporates the chemical and spatial features of the nucleosome as well as the natural properties of the protein-free segments of DNA, or linkers, between successive nucleosomes. The size of the genetic system precludes atomic-level simulations of the observed long-range communication. The DNA constructs include 4–25 nucleosomes, separated by 30-bp linkers, and the binding sites of two comparably sized molecular assemblies—a 5′-terminal glnAp2 promoter site that recognizes the RNA-polymerase-σ54 complex and a 3′-terminal enhancer site specific to the bacterial nitrogen regulatory protein C (NtrC). The currently best-resolved structure of the nucleosome core particle, a roughly cylindrical assembly with two copies of each of four Xenopus laevis histones surrounded by a 147-bp fragment of human DNA (Davey et al. 2002) (Fig. 1), contains over 13,000 heavy (non-hydrogen) atoms, and those of the RNA polymerase (Murakami et al. 2002) and NtrC (De Carlo et al. 2006) assemblies an additional ∼10,000 and ∼20,000 atoms, respectively. The numbers of atoms in the chromatin constructs greatly exceed those incorporated to date in simulations of large biomolecular systems with customized, state-of-the-art computers (Jensen et al. 2012). Routine analysis of long-range protein signaling along stretches of chromatin clearly calls for a much simpler molecular model.
Fig. 1.
Molecular images highlighting the superhelical wrapping of DNA, the locations of charged amino-acid atoms, and the pseudo-symmetric halves of the nucleosome in the currently best-resolved crystal structure (Davey et al. 2002). The DNA is depicted in gold with the phosphorus atoms on complementary strands joined by ribbons and the bases drawn as stick figures. The charge centers of cationic amino acids (arginine Cζ, lysine Nζ) are depicted in blue and those of anionic amino acids (aspartic acid Cγ, glutamic acid Cδ) in red. The H2A·H2B·H3·H4 tetramers are color-coded to denote the directionality of the complex with the tetramer bound at the 5′ end of the sequence-bearing DNA strand in blue and that at the 3′ end in green. The charges in the histone interior are distinguished by smaller, more darkly shaded spheres. Views looking a down the superhelical axis with the structural dyad shown at the top center, b perpendicular to both the dyad and the superhelical axes with the dyad shown on the top, and c, d perpendicular to the helical axis with the dyad located, respectively, in the middle foreground and on the opposite side of the assembly
Mesoscale treatment of chromatin
The simplifications introduced in modeling long-range protein communication along chromatin must capture the molecular features controlled experimentally (Kulaeva et al. 2012). These include the capability to neutralize or delete specific histone residues in the constructs, typically cationic amino acids on the histone tails, and to position nucleosomes and regulatory proteins at precise locations along DNA. The spacing along the double helix controls both the distance of separation and the relative orientation of the proteins. The bound proteins, in turn, place constraints on the pathway of DNA.
Until our recent work (Czapla et al. 2006, 2008, 2011; Swigon et al. 2006; Swigon and Olson 2008), most treatments of protein-mediated DNA looping ignored the complexity of the double-helical structure, such as the large distortions of DNA found in high-resolution protein–DNA complexes. The effects of bound proteins on loop formation were typically subsumed in the parameters of a simplified model, such the bending and twisting moduli and the double-helical repeat of the classic ideal elastic rod representation of DNA (Shimada and Yamakawa 1984). Any connections between observed macromolecular behavior and protein/DNA structure were lost.
Our new methodologies treat long, protein-decorated DNA chains at a ‘realistic’ level—a mesoscale representation that connects detailed features of local chemical architecture to the classic coarse-grained description of DNA as an ideal elastic rod. The approach allows us to establish the role of nucleosome structure and DNA deformation on the organization and expression of genes (Kulaeva et al. 2012). We model DNA at the level of base-pair steps (Fig. 2), using six rigid-body parameters to position one base pair with respect to its neighbor (Lu and Olson 2003; Lu and Olson 2008) and elastic potentials to take account of the variability in both the intrinsic structure and the elastic moduli of individual base pair steps (Olson et al. 1998). The model allows for the incorporation of intrinsic sequence-dependent effects, such as the greater deformability of pyridmiine–purine versus purine–pyrimidine base-pair steps, but can be reduced, if desired, to the conventional description of double-helical DNA as an ideal, naturally straight, inextensible elastic rod. In contrast to earlier models of chromatin that reduce the sequence of DNA base pairs to a small number of beads (Arya and Schlick 2006; Langowski 2006), we keep account of every base pair, including those bound to protein. This information is needed to keep precise account of the positions of regulatory proteins on the DNA chain ends. The DNA bound to protein is kept rigid and assigned the set of rigid-body parameters observed in the currently best-determined structure (Davey et al. 2002). The latter parameters can be varied, if desired, to simulate nucleosomal distortions, such as the peeling of DNA off the histone core.
Fig. 2.
Top Schematic illustration of the six rigid-body ‘step’ parameters (Dickerson et al. 1989) used to generate three-dimensional models of long fragments of protein-decorated DNA at base-pair-level resolution. The block images were obtained with 3DNA (Lu and Olson 2003; Lu and Olson 2008) and illustrated with RasMol (Sayle and Milnerwhite 1995). Bottom Variation of the Roll angle in nucleosome-bound versus linker DNA illustrating the different treatments of the two types of DNA. Whereas nucleosome-bound DNA is assigned the step parameters found in the currently best-resolved core-particle structure (Protein Data Bank entry 1kx5) (Davey et al. 2002), linker DNA is treated as an ideal, inextensible elastic rod subject to fluctuations guided by an assumed potential. The values of Roll in the rigid nucleosomal fragment are plotted as a function of superhelical position, i.e., the number of double-helical turns that each base-pair step is displaced from the structural dyad (here denoted by 0). The 60 base-pair steps bound to the (H3·H4)2 tetramer are highlighted by the heavy lines at the center of the plotted data. The distribution of Roll in the linker DNA, plotted on the right side of the figure, is scaled with respect to a value of unity at the equilibrium rest state (0°)
Computational system
DNA linkers
The variables used to generate configurations of chromatin reside in the DNA linkers. Simulations of constructs with 25 evenly spaced nucleosomes separated by 30-bp linkers entail approximately 25 × 30 = 750 deformable base-pair steps. The binding of regulatory proteins to the ends of the array requires roughly 150 extra base pairs. If the DNA is approximated as an ideal inextensible elastic rod, the system comprises nearly 3,000 independent structural variables—specifically, the Tilt, Roll, and Twist angles at each protein-free step (Fig. 2). Each angle, in turn, undergoes room-temperature fluctuations, guided by a harmonic potential consistent with the solution properties of DNA (Czapla et al. 2006). Trial moves of the protein-free segments are generated with Gaussian sampling, i.e., a standard Gaussian random number generator is used to produce structural variables for a randomly selected base-pair step on an arbitrary DNA linker.
Nucleosomes
The presence of histones complicates the simulations in that internucleosomal interactions may trap the system in small, deep energy wells. Although computation of the overlaps between cylinders enclosing each nucleosome core particle helps to avoid chain self-intersection, i.e., excluded-volume effects, the interactions of charged atoms may stabilize tight arrangements of the protein-decorated DNA. The histones contain cationic and anionic amino-acid residues which are thought to be important to the organization of the nucleosome core particle (Ramakrishnan 1997) and to the interactions of nucleosomes in chromatin (Dorigo et al. 2003; Chodaparambil et al. 2007; Zhou et al. 2007). As a first approximation, the amino acids that associate as ion pairs inside the folded histone core (closely spaced red and blue spheres in Fig. 1) are ignored and the uncompensated amino-acid charges at and beyond the surface of the core are clustered into groups of closely spaced charges, 18 at the histone-DNA interface and 16 on the histone tails (Fig. 3), each containing about six charged amino acids. The positions of the charged clusters are varied at random, with the allowed movements based on the observed spatial dispersion of the charge-bearing atoms that comprise each cluster. The rest states of the clusters are the mean coordinates of the atoms in each grouping and the magnitudes of the point charges in the clusters correspond to the net charges of the grouped atoms. The anionic charges along nucleosomal and linker DNA are clustered in groups comparable in size to those of the histone atoms and spaced at regular increments along the chain, i.e., point charges on every third base pair. The electrostatic interactions are estimated with a Debye-Hückel potential using a screening length of 8.3 Å (consistent with the ionic composition of the transcription buffer), a dielectric constant of 80, and charges on DNA reduced by ∼75 % in accordance with predictions of counterion condensation theory (Manning 1978). This simplified treatment was designed to underestimate electrostatic interactions so that any computed effects of histone and DNA charges on overall chromatin folding would likely be meaningful. The excluded volume is evaluated for the proposed moves of the DNA linkers and the acceptance or rejection of the moves is based on the difference in the electrostatic energies between the proposed, sterically feasible state and the current configuration using the standard Metropolis criterion (Metropolis et al. 1953).
Fig. 3.
Snapshot of four-nucleosomal chromatin fragments in simulated chains with intact (top) and tailless (bottom) histones illustrating the different local interactions and orientations of nearby complexes. The small blue spheres depict the charge-cluster sites on histone tails described in the text. See Fig. 1 for color codes. Nucleosomes spaced at 177-bp increments (147-bp histone-bound DNA and 30-bp linkers)
Regulatory proteins
In contrast to earlier simulations of enhancer-promoter communication along DNA (Schulz et al. 2000), which ignore the interactions of NtrC and RNA polymerase-σ54, we bind to the nucleosome-decorated DNA simplified protein models—a cylinder (124 Å diameter × 40 Å high) and a prolate ellipsoid (150 Å × 110 Å × 115 Å) consistent with the dimensions of the respective molecular complexes (Murakami et al. 2002; De Carlo et al. 2006), i.e., displace these objects by an appropriate distance from the central base pairs of the enhancer and promoter sequences (see Fig. 4). The data reported below do not take precise account of the DNA associated with these proteins. The pathways imposed on the DNA by the regulatory proteins can be estimated, if desired, from known high-resolution structures.
Fig. 4.
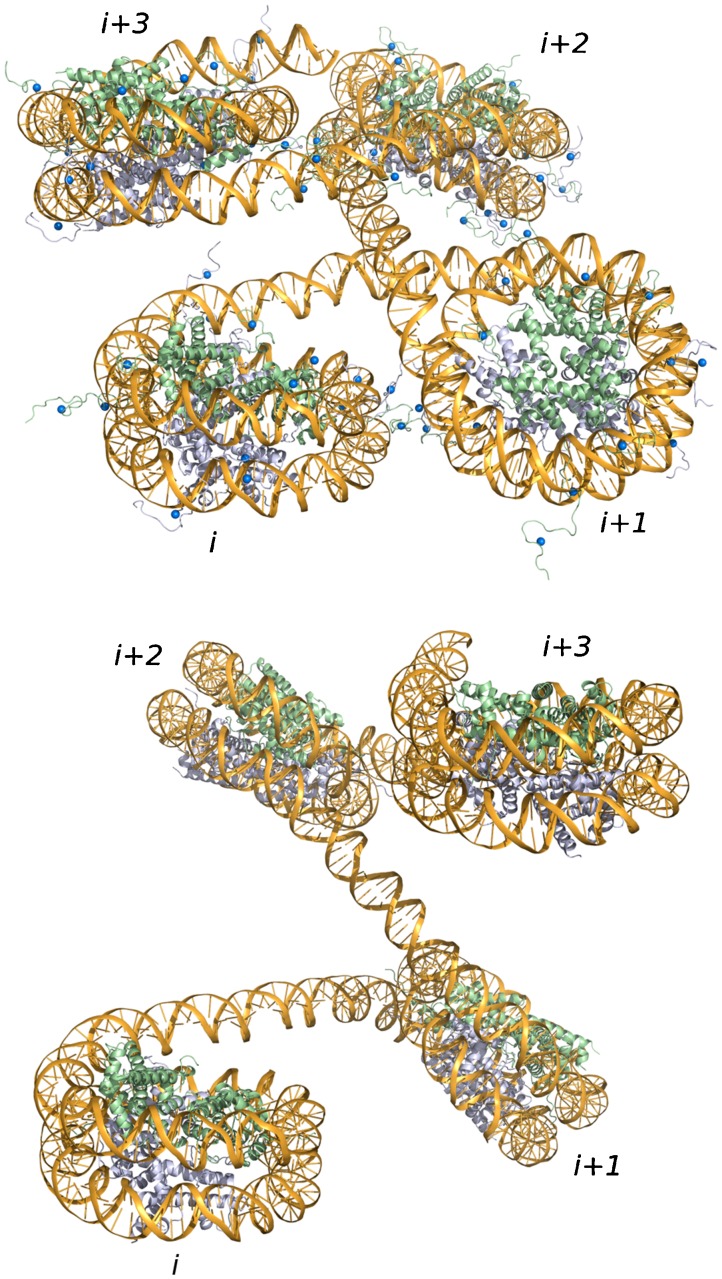
Snapshots of communication between transcriptional proteins attached to the ends of chromatin fragments bearing 13 evenly spaced nucleosomes with intact (top) or tailless (bottom) histones. The NtrC assembly, bound to the enhancer site at the 3′ end of the chain, is denoted by the yellow cylinder and the RNA polymerase-σ54 complex, positioned on the promoter at the 5′ end, by the russet ellipsoid. The smaller cylinders enclose the folded histone cores of the nucleosomes and the small blue spheres occupy the sites of clustered charges on the histone tails. The twisted gold ‘ribbon’ traces the pathway taken by the DNA base pairs. The color-coding of three successive nucleosomes—in mauve, light blue, and green, respectively—illustrates the higher-order wrapping of nucleosomes in the simulated structures. Images created with Mathematica™ and rendered with Blender. Note the more effective communication of regulatory proteins on the construct with nucleosomes bearing intact versus tailless histones. The terminal proteins adopt closed configutations like these, with the long axis of the RNA polymerase-σ54 ellipsoid approximately parallel to the cylindrical axis of the activator protein, i.e., r ≤ 300 Å and φ ≤ 30°, in 0.24 and 0.08 % of the simulated configurations of the respective nucleosome-decorated molecules
Loop formation
We model the enhancement in long-range communication detected in gene expression studies by computing the probabilities that nucleosome-decorated and nucleosome-free DNA constructs of the same chain length adopt looped configurations that bring the regulatory proteins into close spatial contact. We deduce the looping propensity from the fraction of states in a simulated configurational ensemble that meets selected chain-closure criteria (Czapla et al. 2008), and express the communication enhancement in terms of the ratio of the predicted looping probability for the nucleosome-bound DNA compared to that of the nucleosome-free chain. The number of sampled states depends upon the number of configurational variables, with ensembles of ∼109 non-self-intersecting configurations required to capture the looping propensities of the longest (∼4,700-bp) nucleosome-free DNA chains.
In view of the lack of information about the ways in which NtrC physically associates with the RNA polymerase-σ54 assembly, we consider how the predicted enhancement levels depend upon the spatial disposition of the two proteins. We describe the orientation of the two proteins in terms of the spherical polar angles, φ and θ, that respectively define the azimuthal angle (latitude) and the angle of precession (longitude) of the long axis of the RNA polymerase-σ54 ellipsoid with respect to the cylindrical axis of NtrC (see the schematic illustration in Fig. 6, below). We identify looped states as those in which the distances between the geometric centers of the RNA polymerase-σ54 and the NtrC are 300 Å or less. The computed likelihood of loop formation is thus analogous to classic estimates of polymer cyclization as a product of probabilities (Jacobson and Stockmayer 1950; Flory et al. 1976). That is, the predicted occurrence of looping is proportional to the likelihood that the ends of the DNA lie sufficiently close to one another multiplied by conditional probabilities that the interacting proteins are correctly oriented when the chain ends come together.
Fig. 6.
Effects of regulatory protein orientation on the predicted looping propensities of DNA decorated with 13 evenly spaced nucleosomes compared to those computed for protein-free chains of the same length. Values determined for closely spaced regulatory proteins (center-to-center distances ≤300 Å) on chromatin constructs bearing nucleosomes with and without histone tails (red and blue lines) as a function of the azimuthal angle φ and the polar angle θ between the long axis of the RNA polymerase-σ54 ellipsoid and the cylindrical axis of the NtrC assembly. Measured transcript levels are denoted in the plot on the left by the color-coded ribbons of thickness corresponding to twice the observed standard deviation. The looping enhancement predicted on the basis of distance alone is denoted by the dashed lines of the same colors in the plot on the right. The schematic illustration of the protein orientation angles was created with Adobe Illustrator
Prediction versus experiment
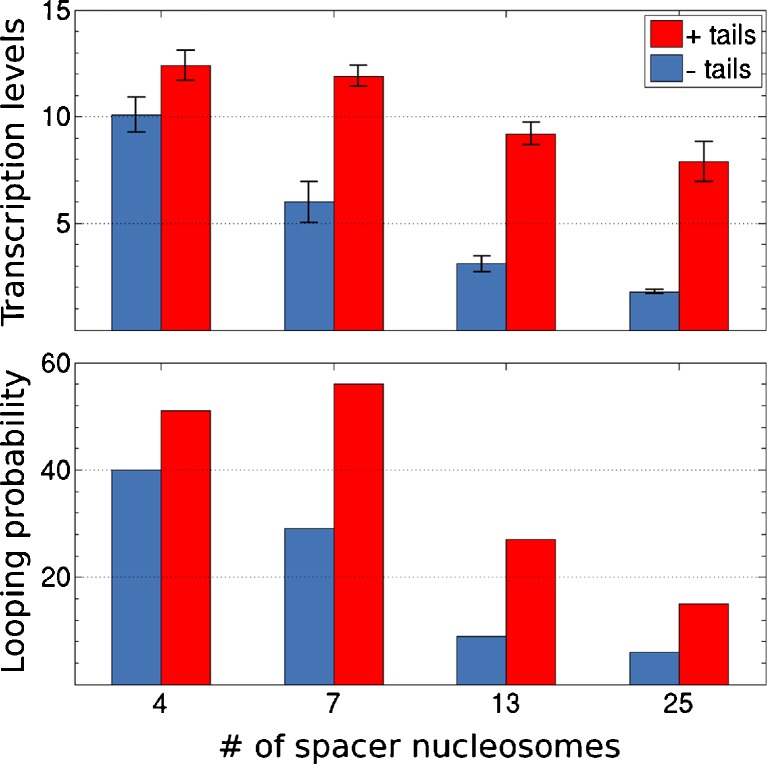
Simulations of looping performed with the above protocols capture the enhancement in long-range communication between regulatory proteins detected experimentally on chromatin compared to bare DNA of the same chain length (observed vs. predicted data plotted, respectively, in the upper and lower histograms in Fig. 5). An ideal DNA chain decorated by arrays of nucleosomes is substantially more prone to looping mediated by other proteins. The simulated likelihood of close contact between a cylindrical NtrC activator protein assembly located on the enhancer at the 3′ end of the chain and an ellipsoidal RNA polymerase-σ54 complex bound at the promoter on the 5′ end is more than an order of magnitude greater for duplexes with 4–25 intervening nucleosomes than for ideal, protein-free DNA of the same chain length.
Fig. 5.
RNA transcripts formed upon the interaction of regulatory proteins (NtrC and RNA polymerase-σ54) bound at the 3′ and 5′ ends of short, precisely defined chromatin fragments (top) compared with the predicted enhancement in loop formation between proteins located at the same positions on simulated nucleosome-decorated chains (bottom). Numbers are expressed as ratios relative to observed and computed values for nucleosome-free DNA chains of the same length. Red and blue bars refer respectively to constructs with native and tailless histones. The values along the abscissa denote the number of positioned nucleosomes on the different constructs. The error bars in the upper plot correspond to the standard deviation of the mean of three independent experiments
The computations also mimic the observed sensitivity of the long-range interactions to chain length and to changes in the local chemical environment. The relative looping propensities of the DNA molecules decorated with arrays of nucleosomes that contain intact histone tails versus those made up of tailless histones mirror the relative levels of RNA transcripts (Fig. 5). Moreover, as found in the gene expression studies, the positive impact of the histone tails on the efficiency of looping in chromatin becomes greater as the size of the loop increases. That is, the likelihood of chromatin looping falls off less rapidly with chain length in the presence of histone tails.
The calculated estimates of enhancer-promoter communication on the basis of the distances between proteins attached to the ends of chromatin versus free DNA exceed the measured levels by roughly fourfold (note the different scales for observed vs. simulated data in the upper and lower plots in Fig. 5). The predicted enhancement of loop formation in nucleosome-bound versus free DNA, however, changes if constraints are placed on the orientation of closely spaced regulatory proteins. The precise level of enhancement depends upon the assumed spatial disposition of the NtrC and polymerase-σ54 complexes. For example, arrangements that orient the long axis of the polymerase-σ54 ellipsoid roughly parallel to the cylindrical axis of the NtrC complex, i.e., values of the azimuthal angle φ near zero, lower both the predicted looping propensities of chains bearing 13 nucleosomes and the predicted looping enhancement over free DNA (Fig. 6). Other azimuthal orientations raise the predicted enhancement to values higher than those determined on the basis of the end-to-end distance alone. Although restrictions on the polar rotation θ of the polymerase ellipsoid with respect to the cylindrical axis of the NtrC cylinder reduce the simulated looping propensities, they have little effect on the predicted looping enhancement. In other words, whereas the looping propensities are sensitive to the azimuthal orientation of the proteins, they are nearly independent of the polar rotation (Fig. 6).
Molecular insights from computations
The correspondence between the predicted and observed looping propensities of nucleosome-decorated DNA lends credence to the model of chromatin used in our computations and to the molecular insights gleaned from the simulated structures. The superhelical wrapping of DNA on the surface of the nucleosome enhances the looping of chromatin over that of bare DNA by simply bringing the ends of the chain into closer contact. The degree of superhelical folding on the nucleosome, in combination with the length of the protein-free linker DNA, gives rise to a higher-order wrapping of the nucleosomes along the chromatin constructs. Every second or third nucleosome lies on roughly the same face of the simulated structures (Fig. 4).
The deformability of the linker DNA contributes to irregularities in the modeled structures and allows for large-scale fluctuations that bring the ends of longer constructs into close contact. The simulated communication between regulatory proteins also reflects the motions of the free DNA segments that lie outside the nucleosome arrays. The deformability of these chain ends appears to offset the greater contour length of constructs with seven versus four nucleosomes. That is, even though the average distance between terminal nucleosomes is greater for the DNA with seven nucleosomes, the frequency of contact between regulatory proteins bound to the flexible ends is comparable to that on the shorter chain.
The enhanced looping propensities of constructs with intact versus tailless nucleosomes seemingly stem from subtle differences in the simulated higher-order chromatin structure. Nucleosomes made up of intact histones make fewer and different types of contacts with nearby parts of the chain than do those with tailless histones (Fig. 3). The latter nucleosomes are not only more tightly packed locally but also more likely to come into long-range contact (Figs. 5, 6). The configurations of DNA chains decorated with intact nucleosomes also show greater curvature than do those with tailless nucleosomes. Preliminary calculations suggest that the effects of histone tails on long-range communication may be lower, if not counterproductive, in chromatin fragments with different numbers of nucleosomes between regulatory proteins. Deformations of nucleosomal DNA and changes in nucleosome spacing that affect higher-order chromatin structure may also change the relative looping propensities of chains with intact versus tailless nucleosomes.
Finally, the computations suggest that the structures of the regulatory proteins themselves may contribute to long-range communication through a preferred mode of association. The current work does not consider the highly charged nature of these proteins, the precise pathways taken by the protein-bound DNA, and the locations of protein-protein recognition sites, all of which may contribute to the observed long-range communication levels.
Acknowledgements
We thank Drs. Vasily Studitsky and Anirvan Sengupta for stimulating discussions and for sharing their findings, and Mr. Michael Smith for input on the presentation. The U.S. Public Health Service under research grant GM34809 and instrumentation grant RR022375 has generously supported this work.
Conflict of Interest
None
Footnotes
Special issue: Computational Biophysics.
References
- Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc Natl Acad Sci USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat Struct Mol Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czapla L, Swigon D, Olson WK. Sequence-dependent effects in the cyclization of short DNA. J Chem Theor Comp. 2006;2:685–695. doi: 10.1021/ct060025+. [DOI] [PubMed] [Google Scholar]
- Czapla L, Swigon D, Olson WK. Effects of the nucleoid protein HU on the structure, flexibility, and ring-closure properties of DNA deduced from Monte-Carlo simulations. J Mol Biol. 2008;382:353–370. doi: 10.1016/j.jmb.2008.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czapla L, Peters JP, Rueter EM, Olson WK, Maher LJ., III Understanding apparent DNA flexibility enhancement by HU and HMGB proteins: experiment and simulation. J Mol Biol. 2011;409:278–289. doi: 10.1016/j.jmb.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon T. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson RE, Bansal M, Calladine CR, Diekmann S, Hunter WN, Kennard O, von Kitzing E, Lavery R, Nelson HCM, Olson WK, Saenger W, Shakked Z, Sklenar H, Soumpasis DM, Tung C-S, Wang AH-J, Zhurkin VB. Definitions and nomenclature of nucleic acid structure parameters. J Mol Biol. 1989;208:787–791. [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/S0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- Flory PJ, Suter UW, Mutter M. Macrocyclization equilibria. 1. Theory. J Am Chem Soc. 1976;98:5733–5739. doi: 10.1021/ja00435a001. [DOI] [Google Scholar]
- Jackson DA, Dickinson P, Cook PR. The size of chromatin loops in HeLa cells. EMBO J. 1990;9:567–57. doi: 10.1002/j.1460-2075.1990.tb08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson H, Stockmayer WH. Intramolecular reaction in polycondensations. I. The theory of linear systems. J Chem Phys. 1950;18:1600–1606. [Google Scholar]
- Jensen MØ, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- Kulaeva OI, Zheng G, Polikanov YS, Colasanti AV, Clauvelin N, Mukhopadhyay S, Sengupta AM, Studitsky VM, Olson WK. Internucleosomal interactions mediated by histone tails allow distant communication in chromatin. J Biol Chem. 2012;287:20248–20257. doi: 10.1074/jbc.M111.333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski J. Polymer chain models of DNA and chromatin. Eur Phys J E Soft Matter. 2006;19:241–249. doi: 10.1140/epje/i2005-10067-9. [DOI] [PubMed] [Google Scholar]
- Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding, and visualization of three-dimensional nucleic-acid structures. Nature Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/S0033583500002031. [DOI] [PubMed] [Google Scholar]
- Metropolis NA, Rosenbluth AW, Rosenbluth MN, Teller H, Teller E. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Olson WK, Gorin AA, Lu X-J, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Studitsky VM. Analysis of distant communication on defined chromatin templates in vitro. Methods Mol Biol. 2009;543:563–576. doi: 10.1007/978-1-60327-015-1_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Rubtsov MA, Studitsky VM. Biochemical analysis of enhancer-promoter communication in chromatin. Methods. 2007;41:250–258. doi: 10.1016/j.ymeth.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu Rev Biophys Biomol Struct. 1997;26:83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- Rubtsov MA, Polikanov YS, Bondarenko VA, Wang YH, Studitsky VM. Chromatin structure can strongly facilitate enhancer action over a distance. Proc Natl Acad Sci USA. 2006;103:17690–17695. doi: 10.1073/pnas.0603819103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle RA, Milnerwhite EJ. RasMol: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/S0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Schulz A, Langowski J, Rippe K. The effect of the DNA conformation on the rate of NtrC-activated transcription of Escherichia coli RNA polymerase.σ54 holoenzyme. J Mol Biol. 2000;300:709–725. doi: 10.1006/jmbi.2000.3921. [DOI] [PubMed] [Google Scholar]
- Shimada J, Yamakawa H. Ring-closure probabilities for twisted wormlike chains. Application to DNA. Macromolecules. 1984;17:689–698. doi: 10.1021/ma00134a028. [DOI] [Google Scholar]
- Swigon D, Olson WK. Mesoscale modeling of multi-protein-DNA assemblies: the role of the catabolic activator protein in Lac-repressor-mediated looping. Intl J Non-linear Mechanics. 2008;43:1082–1093. doi: 10.1016/j.ijnonlinmec.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigon D, Coleman BD, Olson WK. Modeling the Lac repressor-operator assembly: the influence of DNA looping on Lac repressor conformation. Proc Natl Acad Sci USA. 2006;103:9879–9884. doi: 10.1073/pnas.0603557103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Olson WK. DNA architecture, deformability, and nucleosome positioning. J Biomol Struct Dyn. 2010;27:725–739. doi: 10.1080/073911010010524943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]