Abstract
Plastoquinone and tocopherols are the two major quinone compounds in higher plant chloroplasts and are synthesized by a common pathway. In previous studies we characterized two loci in Arabidopsis defining key steps of this biosynthetic pathway. Mutation of the PDS1 locus disrupts the activity of p-hydroxyphenylpyruvate dioxygenase (HPPDase), the first committed step in the synthesis of both plastoquinone and tocopherols in plants. Although plants homozygous for the pds1 mutation could be rescued by growth in the presence of homogentisic acid, the product of HPPDase, we were unable to determine if the mutation directly or indirectly disrupted HPPDase activity. This paper reports the isolation of a cDNA, pHPPD, encoding Arabidopsis HPPDase and its functional characterization by expression in both plants and Escherichia coli. pHPPD encodes a 50-kD polypeptide with homology to previously identified HPPDases, including 37 highly conserved amino acid residues clustered in the carboxyl region of the protein. Expression of pHPPD in E. coli catalyzes the accumulation of homogentisic acid, indicating that it encodes a functional HPPDase enzyme. Mapping of pHPPD and co-segregation analysis of the pds1 mutation and the HPPD gene indicate tight linkage. Constitutive expression of pHPPD in a pds1 mutant background complements this mutation. Finally, comparison of the HPPD genomic sequences from wild type and pds1 identified a 17-bp deletion in the pds1 allele that results in deletion of the carboxyterminal 26 amino acids of the HPPDase protein. Together, these data conclusively demonstrate that pds1 is a mutation in the HPPDase structural gene.
Plastoquinone and tocopherols are the two major classes of chloroplastic, lipid-soluble quinone compounds in higher plants. Plastoquinone is best known for its role as an electron carrier between PSII and the Cyt b6/f complex, and to a lesser extent as an electron carrier for NAD(P)H-plastoquinone oxidoreductases (Berger et al., 1993). In mammals, which cannot synthesize plastoquinone or tocopherols, α-tocopherol (vitamin E) is an essential dietary component (Mason, 1980) and has a well-documented role as a membrane-associated free radical scavenger (for review, see Liebler, 1993). In plants, tocopherols are also presumed to function as membrane-associated antioxidants and as structural components of membranes, although evidence supporting these roles is limited (for review, see Hess, 1993).
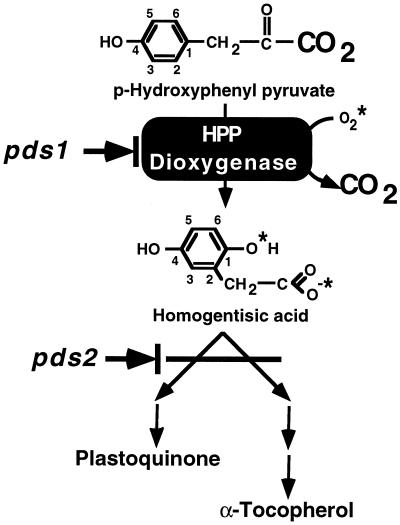
Figure 1 shows the pathway for plastoquinone and tocopherol biosynthesis in plants. The first step of this pathway, common to the synthesis of both plastoquinone and tocopherol, is the formation of HGA from HPP by the enzyme HPPDase (EC 1.13.11.27). HPPDase catalyzes a complex, irreversible reaction involving the introduction of two molecules of oxygen, and decarboxylation and rearrangement of the side chain (Fig. 1). HPPDase is generally present at low levels in plant tissues and has only recently been purified to homogeneity from a plant source (Garcia et al., 1997).
Figure 1.
The plastoquinone and α-tocopherol biosynthetic pathway in higher plants. For clarity, not all biosynthetic steps are shown and only the HPPDase reaction is shown in detail. The CO2 lost and molecular oxygen introduced by HPPDase are indicated with a larger font and asterisks, respectively. The conjugated rings of HPP and HGA are numbered to indicate rearrangement of the side chain. The locations of the pds1 and pds2 mutations in the pathway are indicated.
Although mammals and nonphotosynthetic bacteria cannot synthesize plastoquinone or tocopherols, they do nonetheless contain HPPDase enzymatic activity. This activity is often present at very high levels and is involved in Phe and Tyr degradation. HPPDase has been purified from several mammalian and bacterial sources (Wada et al., 1975; Lindstedt et al., 1977; Roche et al., 1982; Endo et al., 1992), and in all cases the active enzyme was found to be a homodimer or, less commonly, a homotetramer, with subunits of approximately 40 to 48 kD. As a result of the central role HPPDase serves in aromatic amino acid metabolism in mammals and plastidic quinone synthesis in plants, a class of competitive inhibitors of HPPDases collectively known as triketones has been developed and used for a variety of clinical and agricultural purposes (Lindstedt et al., 1992; Schultz et al., 1993; Secor, 1994). In humans, the triketone 2-(2-nitro-4-trifluromethylbenzoyl)-1,3-cyclohexanedione and related compounds are used as an alternative to liver transplantation in patients with the otherwise fatal hereditary disorder tyrosinemia type I. This disorder results from a deficiency in the last enzyme of Tyr catabolism (Lindstedt et al., 1992; Gibbs et al., 1993) and 2-(2-nitro-4-trifluromethylbenzoyl)-1,3-cyclohexanedione treatment inhibits liver HPPDase activity, blocking formation of HGA and its subsequent breakdown to the toxic intermediates succinylacetoacetate and succinylacetone. In plants, triketones such as sulcotrione (2-[4-chloro-2-nitrobenzoyl]-5,5-dimethylcyclohexane-1,3-dione) are effective bleaching herbicides. Their mode of action arises from a direct inhibition of plastoquinone and tocopherol synthesis and an indirect inhibition of carotenoid desaturation (Mayonado et al., 1989; Schultz et al., 1993; Secor, 1994). The latter results in accumulation of the carotenoid biosynthetic intermediate phytoene and photooxidation of the plastid.
A genetic basis for the effects of triketones on plant carotenoid synthesis was suggested by the identification of two Arabidopsis mutations that disrupt phytoene desaturation (pds mutations) but do not map to the phytoene desaturase enzyme locus (Norris et al., 1995). Previous work demonstrated that mutations in either the PDS1 or PDS2 loci resulted in plants deficient in tocopherol and plastoquinone biosynthesis and, as a secondary effect of this deficiency, disruption of carotenoid desaturation (Norris et al., 1995). The pds mutations thus provide genetic evidence that plastoquinone is an essential component for carotenoid biosynthesis in plants and provide insight into plastidic quinone synthesis and function. The biochemical basis of the pds1 mutation was hypothesized to be a disruption in the HPPDase structural gene because the mutant phenotype could be biochemically complemented by growth on medium supplemented with the product (HGA) but not the substrate (HPP) of the HPPDase enzyme. However, despite this compelling evidence, it could not be determined whether the pds1 mutation directly or indirectly affected the HPPDase enzyme (Norris et al., 1995).
To functionally test the hypothesis that the Arabidopsis pds1 mutation is the result of a lesion in the structural HPPDase gene, it is necessary to isolate and functionally characterize Arabidopsis HPPDase cDNAs and the corresponding wild-type and mutant HPPDase alleles. In this paper we report the isolation and characterization of a cDNA encoding HPPDase from Arabidopsis and demonstrate the activity of the protein when expressed in Escherichia coli. Linkage of the HPPD gene and the pds1 mutant was demonstrated by both mapping and co-segregation analysis. Sequence analysis of the wild-type and mutant HPPDase genomic sequences identified a small deletion that produces a truncated protein in the mutant. Finally, we show functional complementation of the pds1 mutant phenotype when the HPPDase cDNA is constitutively expressed. Combined, these data conclusively demonstrate that pds1 is a mutation of the HPPDase gene.
MATERIALS AND METHODS
cDNA and Genomic DNA Isolation and DNA Sequence Analysis
A BLAST search (Altschul et al., 1990) of plant DNA sequence databases with various bacterial and mammalian HPPDase sequences identified a truncated Arabidopsis cDNA (accession no. T20952) with homology to the carboxy terminus of human HPPDase (accession no. X72389). The 460-bp insert from this expressed sequence tag was used as a probe to screen 4 × 105 plaques of the Arabidopsis PRL2 library (Newman et al., 1994). Eighty-one individual plaques were collected for further evaluation and detailed characterization was performed on 32 isolates, of which four full-length clones were sequenced. Isolate 18 was chosen for further studies and renamed pHPPD. A pHPPD probe was made by labeling a SalI/NotI fragment of pHPPD using the Random Prime kit (Boehringer Mannheim).
Genomic DNAs for use as substrates for PCR were isolated from wild-type and pds1 genotypes (both are ecotype Wassilewskija [Ws]) by the modified minipreparation method (DellaPorta et al., 1983). Two sets of primers were used to amplify genomic copies of the HPPDase gene from wild-type Ws tissue: SN418T7+10 (5′-CGTCCGAGTTTTAGCAGAGTTGG-3′) and SN418MF+11 (5′-AGAGCCAGATGTTGTAGCCC-3′) for the first 1000 bp of the gene, and SN418T7+4 (5′-CCAATTCGCAGAGTTC-3′) and SN418MF+12 (5′-CGTTTTAAATGAGATGTTGTATAAC-3′) for the last 700 bp of the gene. Similarly, for the pds1 mutant, two sets of primers were used: SN418T7+10 and SN418MF+1b (5′-CAGATGTTGTAGCCCT-3′) for the first 1000 bp of the gene, and SN418T7+4 and SN418MF+12 for the last 700 bp of the gene. In both cases, the two amplified genomic fragments overlap by about 200 bp.
Three independent sets of PCR reactions were performed for each fragment amplification. PCR products were analyzed by gel electrophoresis, and equal concentrations of each were pooled, purified, and used directly for sequencing. DNA sequencing was performed using a dye deoxy terminator cycle sequencing kit (Applied Biosystems) and an automated DNA sequencer (model 310, Applied Biosystems). DNA-sequence analysis was done using both DNAStar and MacVector (International Biotechnologies, Inc., New Haven, CT).
Protein Overexpression
For cloning purposes a NcoI site was introduced 5′ of the ATG start codon by changing the A at position −1 to a C using PCR-based mutagenesis with the two oligonucleotides 5′-TGTAAAACGACGGCCAGT-3′ and 5′-GTTGGTGAAATCCATGGGCCACCAAAACGC-3′. The amplified product was ligated into the pCRII vector (Invitrogen, San Diego, CA), generating clone SN507. A 1.49-kb NcoI/BamHI fragment from SN507 was ligated into the pET15b vector (Novagen, Madison, WI), generating pET-HPPD. pET15b and pET-HPPD were transformed into Escherichia coli cell line BL21(DE3) (Novagen) via electroporation. HPLC analysis of bacterial cultures for the presence of HGA was performed according to published procedures (Denoya et al., 1994). HGA was identified in extracts based on comparison of retention time and spectra to a HGA (Sigma) standard with a Hewlett-Packard series 1100 chromatograph and photodiode array detector.
Linkage Analysis
Co-segregation of the pds1 and HPPDase loci was determined by restriction fragment-length polymorphism linkage analysis using pHPPD as probe. F2 progeny heterozygous for the pds1 mutation were selected from a cross between PDS1/pds1 (ecotype Ws) and PDS1/PDS1 (ecotype Columbia [Col]). Digestion of Ws and Col genomic DNA with NcoI gave a restriction fragment-length polymorphism for the pHPPD probe. Genomic DNA for co-segregation analysis was isolated from F2 progeny by the modified minipreparation method (DellaPorta et al., 1983). The digested DNA was separated on a 0.6% agarose gel and transferred to a nylon membrane (Micron Separations, Westborough, MA). The blots were hybridized with the pHPPD probe and washed two times at room temperature for 15 min with 2× SSC, 0.1% SDS and two times at 55°C for 25 min in 1× SSC, 0.1% SDS.
Plant Transformation
Clone SN500 was generated by subcloning a 1.5-kb KpnI/HindIII fragment containing the complete coding region of pHPPD into the plant-transformation shuttle vector pART7 (Gleave, 1992). After partial digestion of SN500 with NotI, a 4.4-kb fragment containing the cauliflower mosaic virus promoter, pHPPD coding sequences, and an OCS terminator was isolated and ligated into the binary plant-transformation shuttle vector pART27 (Gleave, 1992), generating clone SN506. SN506 was electroporated into Agrobacterium tumefaciens strain C58 and used to transform wild-type Arabidopsis (ecotype Ws) via vacuum infiltration (Bent et al., 1994). Seed was collected from individual T1 plants, surface sterilized, and plated on MS2 medium (Norris et al., 1995) with 100 mg/L carbenicillin, 60 mg/L kanamycin, and 10 mg/L benomyl. Kanamycin-resistant T2 seedlings were transferred to soil and grown to maturity, and T3 seed was harvested. For complementation analysis, kanamycin-resistant T2 plants were crossed with PDS1/pds1 heterozygotes. The resulting F1 seeds were surface sterilized and plated on MS2 medium with 60 mg/L kanamycin. Kanamycin-resistant F1 seedlings were transferred to soil and grown as described above. Developing F2 seeds in siliques of mature F1 plants were scored for the homozygous albino mutant pds1 phenotype as described previously (Norris et al., 1995). The F2 seeds were also collected at maturity, surface sterilized, and plated on MS2 medium with and without 60 mg/L kanamycin and then scored for both kanamycin resistance and the pds1 mutant phenotype.
RESULTS
Isolation and Characterization of an Arabidopsis HPPDase cDNA
Genes and cDNAs encoding HPPDase have been identified from several mammalian, fungal, bacterial, and plant sources (Gershwin et al., 1987; Endo et al., 1992, 1995; Hummel et al., 1992; Ruetschi et al., 1993; Coon et al., 1994; Denoya et al., 1994; Wilson et al., 1994; Wintermeyer et al., 1994; Kaneko et al., 1995; Wyckoff et al., 1995; Garcia et al., 1997) and show between 25% and 95% identity at the amino acid level. A computer search of the plant DNA databases, including 20,000 random Arabidopsis cDNAs (Newman et al., 1994), was conducted using human and bacterial HPPDase sequences as the query. This search identified a 460-bp truncated Arabidopsis cDNA (single-underlined DNA sequence in Fig. 2) with significant homology to the carboxy terminus of previously identified HPPDases. This partial cDNA was used as a probe to isolate a full-length cDNA that was named pHPPD. The first ATG of pHPPD begins an open-reading frame encoding a 50-kD protein of 445 amino acids (Fig. 2). The putative Arabidopsis HPPDase protein has from 17% to 27% amino acid identity with bacterial, fungal, and animal HPPDases and between 58% and 70% amino acid identity with two other plant HPPDases. The estimated 50-kD size of the Arabidopsis HPPDase protein closely approximates that reported for other HPPDases, which range from 40 to 48 kD.
Figure 2.
Nucleotide and deduced amino acid sequence of the Arabidopsis HPPDase cDNA pHPPD. The protein sequence is shown in boldface underneath the nucleotide sequence (accession no. AF000228). The nucleotide sequence of the originally identified, truncated expressed sequence tag (accession no. T20952) is indicated by a single underline. Alignments were performed to13 other HPPDase proteins (accession nos. AJ000693, D64004, L38493, U11864, U87257, S69666, M59289, M59429, Z50016, X72389, D29987, M18405, and D13390). Arabidopsis HPPDase amino acid residues showing identity in 9 of the other 13 HPPDase proteins are indicated with shaded boxes. Amino acid residues identical in all 14 HPPDase sequences are denoted with black boxes. The five conserved Tyr and His residues postulated to form the HPPDase ferric iron center are indicated by filled dots. The location of the single 107-bp intron in the HPPDase genomic sequences of Ws and pds1 is denoted by an inverted, filled triangle. The 17-bp deletion in the HPPDase gene in pds1 is denoted by a boldface, italic DNA sequence and two overhead lines. The Ws and pds1 HPPDase gene and protein sequences are identical up to the deletion. The consequence of this mutation at the protein level is indicated in the box below the deletion.
Functional Analysis of the Arabidopsis HPPDase Protein
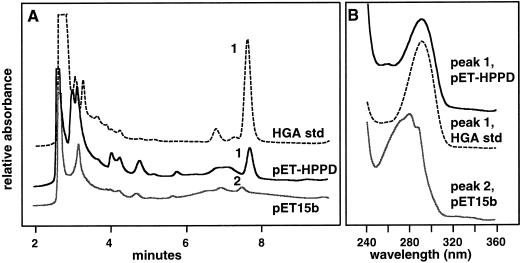
The protein-sequence homology of the putative Arabidopsis HPPDase to other HPPDases suggested that it encodes an HPPDase enzyme. To test this hypothesis, pHPPD was overexpressed in E. coli and functionally analyzed. E. coli harboring the pET-HPPD construct developed a dark-brown color, whereas cultures containing the empty pET15b vector did not (data not shown). A similar dark-brown coloration was reported when the gene encoding HPPDase from Streptomyces avermitilis was expressed in E. coli (Denoya et al., 1994). This brown coloration is caused by the accumulation of ochronotic pigment, which forms upon the oxidative polymerization of HGA. To verify that the brown coloration in E. coli expressing pET-HPPD was the result of plasmid-mediated HGA production, cell-free supernatants from E. coli cultures containing the empty pET15b vector and pET-HPPD were analyzed by HPLC for the presence of HGA (Fig. 3). A HGA standard eluted at 7.9 min and had the spectra and absorbance maximum (291 nm) shown in Figure 3B. The pET-HPPD culture filtrate had a prominent peak that co-migrated with the HGA standard (Fig. 3A) and had a spectrum and absorbance maximum that were identical to those of the HGA standard (Fig. 3B). The pET15b control culture lacked a peak at 7.9 min and had a minor peak at 7.7 min, with a spectrum and absorbance maxima (271, 280, and 287 nm) that indicated that it was not HGA (Fig. 3B). These results indicate that Arabidopsis pHPPD encodes a functional HPPDase enzyme.
Figure 3.
Expression of Arabidopsis HPPDase cDNA in E. coli. A, HPLC analysis of a HGA standard in Luria-Bertani broth is shown in the top plot. The middle and bottom plots are cell-free extracts from cultures of E. coli harboring the pET-HPPD construct and the pET15b construct, respectively. B, Absorption spectra of peaks 1 and 2 from A. Peak 1, HGA standard and co-migrating peak in medium of pET-HPPD; peak 2, unidentified compound in pET15b.
Mapping, Molecular Complementation, and Genomic Sequence Analysis
In previous work we demonstrated that the biochemical basis of the Arabidopsis pds1 mutation is an inability to convert HPP to HGA (Fig. 1; Norris et al., 1995). The PDS1 gene product could therefore be the HPPDase enzyme, a regulator of HPPDase expression or activity, or a cofactor required for HPPDase activity. Three complementary approaches were undertaken to determine whether the gene identified by the pds1 mutation encodes HPPDase: co-segregation of the pds1 mutation and HPPD gene, functional complementation of the pds1 mutant with the wild-type pHPPD cDNA, and DNA-sequence analysis of the wild-type and mutant HPPD alleles.
The pds1 mutation was previously mapped to chromosome 1 between distorted1 and chlorina1 (Norris et al., 1995). Recombinant inbred lines (Lister and Dean, 1993) were used to determine the chromosomal location of the HPPDase gene, which was localized in the region of PDS1 on chromosome 1 (data not shown). For finer mapping, segregation analysis of the pds1 mutation and a restriction fragment-length polymorphism for the HPPDase gene showed no recombinations in 38 PDS1/pds1 lines, indicating that the two were linked within 4 centimorgans (data not shown). Together, these data indicate that the PDS1 locus and HPPDase gene are linked in the Arabidopsis genome.
Molecular complementation of the pds1 mutation with the Arabidopsis pHPPD cDNA was undertaken to determine if the pds1 mutant could be rescued by constitutive overexpression of the wild-type HPPDase protein. A transcriptional fusion of the cauliflower mosaic virus 35S promoter and the full-length pHPPD cDNA in the sense orientation was used to transform wild-type Arabidopsis (Ws) plants. Three independent transgenic lines constitutively overexpressing HPPDase were selected and crossed with PDS1/pds1 heterozygotes. Fifty percent of the resulting kanamycin-resistant F1 progeny from these crosses were also heterozygous for the pds1 mutation. These kanamycin-resistant, pds1 heterozygous F1 plants were then selfed, and segregation of their F2 progeny for both kanamycin resistance and the pds1 phenotype was determined (Table I). χ2 analysis shows that the ratio of green to white embryos in each line is statistically significant for a 15:1 ratio (Table I), indicating that the pds1 mutant phenotype was complemented by the presence of the overexpressed pHPPD cDNA in all plant lines analyzed. Loss of the transgene should restore a 3:1 green:white ratio to such plants. This hypothesis was verified by analyzing the F1 plants that were 100% kanamycin sensitive; one-half of which contained F2 progeny segregating 100% green (PDS1/PDS1) and half of which segregated 3:1 green:white (PDS1/pds1) (data not shown). These data demonstrate that overexpression of a wild-type HPPDase protein in the pds1 mutant background complements the mutation and suggest that the molecular basis of the pds1 mutation is a disruption in the HPPDase gene.
Table I.
Segregation analysis of progeny from plants heterozygous for both an HPPDase transgene and the pds1 mutation
| Transgenic Plant Line | Green:White | χ2a | Kanamycin Resistant: Kanamycin Sensitive | χ2b |
|---|---|---|---|---|
| 105 -1-6 | 125 :8 | 0.9c | 135 :35 | 1.76d |
| 107 -2-2 | 95 :10 | 1.38e | 79 :30 | 0.37c |
| 113 -1-8 | 187 :10 | 0.80c | 133 :51 | 0.72c |
χ2 values were calculated for a 15:1 ratio.
χ2 values were calculated for a 3:1 ratio.
P > 0.3.
P > 0.1.
P > 0.2.
To determine the molecular basis of the pds1 mutation, the HPPDase genomic DNA sequences from wild-type Ws Arabidopsis (accession no. AF060481) and pds1 mutant tissues were determined by direct sequencing of PCR-amplified products. Both HPPDase genomic sequences contain a single 107-bp intron of identical sequence between positions 1162 and 1163 of the HPPD cDNA sequence in Figure 2. The coding frames of the wild-type and pds1 HPPDase alleles were completely identical with the exception of a 17-bp deletion (5′-TTTTGGCAAAGGCAATT-3′) in the pds1 HPPD gene from nucleotides 1254 to 1270 of the wild-type cDNA sequence in Figure 2. This deletion causes a frame shift and substitution of a Leu for the conserved Phe at position 419, followed immediately by a stop codon (Fig. 2). This stop codon results in the deletion of the remaining 26 amino acids from the carboxyterminal end of the protein. This result defines the molecular basis of the pds1 mutation as a mutation in the structural HPPD gene.
DISCUSSION
The plastids of higher plants accumulate large amounts of two biosynthetically related quinone compounds: plastoquinones and tocopherols. Plastoquinones are fundamentally important components of the photosynthetic electron-transport chain, whereas tocopherols are thought to be important for free radical scavenging and protection from oxidative stress. Plastoquinone and tocopherols share a common biosynthetic pathway that has been elucidated for some time (Fig. 1). Recently, genetic insight into the pathway has been obtained, primarily because of the isolation and characterization of mutations in Arabidopsis that disrupt two key steps of plastidic quinone biosynthesis (Norris et al., 1995). One of these mutations, pds1, was shown to affect the activity of HPPDase, the committed step of plastidic quinone biosynthesis (Fig. 1). To further understand the nature of the pds1 mutation, we have isolated and functionally analyzed cDNAs and genomic clones encoding HPPDase from Arabidopsis.
Computer database searches with mammalian and bacterial HPPDase sequences identified a single truncated Arabidopsis expressed sequence tag with significant homology to the carboxyl domains of other HPPDases. This expressed sequence tag was used to isolate a full-length Arabidopsis cDNA clone, pHPPD. Comparison of the putative Arabidopsis HPPDase protein sequence with HPPDase protein sequences from 13 other diverse species identified 37 conserved residues clustered primarily in the carboxy region of the protein (Fig. 2). Presumably, these highly conserved residues are important for substrate binding or the catalytic mechanism of HPPDases. Five Tyr and His residues, postulated to form a ferric iron center in HPPDases (Denoya et al., 1994), are also conserved in the putative Arabidopsis HPPDase (Fig. 2).
To determine whether the putative Arabidopsis HPPDase cDNA encoded a functional HPPDase enzyme, the open-reading frame of this cDNA was expressed in E. coli. As shown in Figure 3, E. coli cultures expressing pHPPD accumulate a compound that co-migrates with, and has a spectrum identical to, the HGA standard (Fig. 3). E. coli containing a control plasmid without the HPPDase open-reading frame lacks this peak (Fig. 3A). In addition to HPPDase-dependent HGA accumulation, pHPPD expression in E. coli resulted in accumulation of ochronotic pigment, an oxidative polymerization product of HGA. Similar results were reported when a S. avermitilis HPPDase was expressed in E. coli (Denoya et al., 1994). These data demonstrate that the Arabidopsis cDNA pHPPD encodes a functional HPPDase enzyme.
As discussed previously, Arabidopsis plants homozygous for the pds1 mutation are unable to synthesize both plastoquinone and tocopherols because of an inability to convert HPP to HGA (Fig. 1). Although it was clear from previous work that the pds1 mutation affected HPPDase activity, we could not determine whether the pds1 mutation directly or indirectly affected the HPPDase enzyme (Norris et al., 1995). Isolation of Arabidopsis pHPPD provided the means for directly testing the hypothesis that pds1 is a disruption in the HPPDase gene by mapping, molecular complementation, and DNA sequence analysis.
Linkage analysis indicated that the gene corresponding to Arabidopsis pHPPD maps near (±4 centimorgans) the pds1 mutation (data not shown). Transgenic plants overexpressing the pHPPD cDNA were generated in a wild-type background and crossed with plants heterozygous for the pds1 mutation. Kanamycin-resistant F1 plants were selected and selfed, and the resulting F2 plants were scored. Failure of the transgene to functionally complement the pds1 mutation would result in F2 progeny that segregate 3:1 green:white (wild type to mutant), whereas functional complementation by the transgene would result in F2 progeny that segregate 15:1 green:white, assuming that the transgene and the pds1 mutation were not linked. Table I shows that the F2 green-to-white segregation ratios from crosses of pds1 heterozygotes to three independent, parental transgenic lines are statistically significant for a 15:1 ratio. These data provide genetic evidence that constitutive expression of the pHPPD transgene complements the pds1 mutation.
Sequence analysis of the HPPDase gene from both wild-type and homozygous pds1 mutant plants was performed to define the molecular basis of the pds1 mutation. As shown in Figure 2, the wild-type and pds1 HPPD alleles are identical in sequence with the exception of a 17-bp deletion in the pds1 HPPD allele. This deletion results in a substitution of Leu for the highly conserved Phe at position 419, followed immediately by a stop codon. The consequence of this deletion at the protein level is the loss of 26 carboxy-terminal amino acids from the HPPDase protein, including a tight cluster of several amino acids that are conserved in all HPPDase proteins in the database. The null phenotype of the pds1 mutant suggests that these 26 carboxy-terminal residues are essential for HPPDase enzymatic activity. Most significantly, these data define the molecular basis of the pds1 mutation as a lesion in the structural HPPD gene.
In conclusion, we have identified and characterized Arabidopsis cDNA and genomic clones encoding HPPDase. The functional expression of the Arabidopsis HPPDase cDNA in E. coli demonstrates that it encodes a functional HPPDase enzyme. Constitutive expression of the protein encoded by the Arabidopsis HPPDase cDNA is sufficient to restore wild-type pigmentation to plants homozygous for the pds1 mutation. Finally, and most significantly, we have shown that the pds1 HPPD gene contains a small deletion that results in the elimination of a portion of the carboxy terminus of the protein. These results demonstrate conclusively that the nature of the pds1 mutation in Arabidopsis is a mutation in the gene encoding the HPPDase enzyme. Future studies will determine the consequences of overexpressing the wild-type HPPDase enzyme in plants. Continued analysis of other plastoquinone and tocopherol biosynthetic mutants, such as pds2 (Norris et al., 1995), will also provide valuable information concerning the subcellular location and regulation of tocopherol and plastoquinone synthesis in plants.
Abbreviations:
- HGA
homogentisic acid
- HPP
p-hydroxyphenylpyruvate
- HPPDase
p-hydroxyphenylpyruvate dioxygenase
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers E, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bent A, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Berger S, Ellersiek U, Westoff P, Steinmuller K. Studies on the expression of NDH-H, a subunit of the NAD(P)H-plastoquinone-oxidoreductase of higher plant chloroplasts. Planta. 1993;190:25–31. [Google Scholar]
- Coon SL, Kotob S, Jarvis BB, Wang S, Fuqua WC, Weiner RM. Homogentisic acid is the product of MelA, which mediates melanogenesis in the marine bacterium Shewanella colwelliana D. Appl Environ Microbiol. 1994;60:3006–3010. doi: 10.1128/aem.60.8.3006-3010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPorta S, Wood J, Hicks J. A plant DNA minipreparation: version 2. Plant Mol Biol Rep. 1983;1:19–22. [Google Scholar]
- Denoya CD, Skinner DD, Morgenstern MR. A Streptomyces avermitilis gene encoding a 4-hydroxyphenylpyruvic acid dioxygenase-like protein that directs the production of homogentisic acid and an ochronotic pigment in Escherichia coli. J Bacteriol. 1994;176:5312–5319. doi: 10.1128/jb.176.17.5312-5319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo F, Awata H, Katoh H, Matsuda I. A nonsense mutation in the 4-hydroxyphenylpyruvic acid dioxygenase gene (Hpd) causes skipping of the constitutive exon and hypertyrosinemia in mouse strain III. Genomics. 1995;25:164–169. doi: 10.1016/0888-7543(95)80122-3. [DOI] [PubMed] [Google Scholar]
- Endo F, Awata H, Tanoue A, Ishiguro M, Eda Y, Titani K, Matsuda I. Primary structure deduced from complementary DNA sequence and expression in cultured cells of mammalian 4-hydroxyphenylpyruvic acid dioxygenase. J Biol Chem. 1992;267:24235–24240. [PubMed] [Google Scholar]
- Garcia I, Rodgers M, Lenne C, Rolland A, Sailland A, Matringe M. Subcellular localization and purification of a p-hydroxyphenylpyruvate dioxygenase from cultured carrot cells and characterization of the corresponding cDNA. Biochem J. 1997;325:761–769. doi: 10.1042/bj3250761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershwin ME, Coppel RL, Bearer E, Peterson MG, Sturgess A, MacKay IR. Molecular cloning of the liver-specific rat F antigen. J Immunol. 1987;139:3828–3833. [PubMed] [Google Scholar]
- Gibbs TC, Payan J, Brett EM, Lindstedt S, Holme E, Clayton PT. Peripheral neuropathy as the presenting feature of tyrosinaemia type I and effectively treated with an inhibitor of 4-hydroxyphenylpyruvate dioxygenase. J Neurol Neurosurg Psychiatry. 1993;56:1129–1132. doi: 10.1136/jnnp.56.10.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Hess JL. Vitamin E, α-tocopherol. In: Alscher R, Hess J, editors. Antioxidants in Higher Plants. Boca Raton, FL: CRC Press; 1993. pp. 111–134. [Google Scholar]
- Hummel R, Norgaard P, Andreasen PH, Neve S, Skjodt K, Tornehave D, Kristiansen K. Tetrahymena gene encodes a protein that is homologous with the liver-specific F-antigen and associated with the membranes and Golgi apparatus and transport vesicles. J Mol Biol. 1992;228:850–861. doi: 10.1016/0022-2836(92)90869-l. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 1995;2:153–166. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- Liebler DC. The role of metabolism in the antioxidant function of vitamin E. Crit Rev Toxicol. 1993;23:147–169. doi: 10.3109/10408449309117115. [DOI] [PubMed] [Google Scholar]
- Lindstedt S, Holome E, Lock EA, Hjalmarson O, Strandvik B. Treatment of hereditary tyrosinemia type I by inhibition of 4 hydroxyphenylpyruvate dioxygenase. Lancet. 1992;340:813–817. doi: 10.1016/0140-6736(92)92685-9. [DOI] [PubMed] [Google Scholar]
- Lindstedt S, Odelhog B, Rundgren M. Purification and some properties of 4-hydroxyphenylpyruvate dioxygenase from Pseudomonas sp. P.J. 874. Biochemistry. 1977;16:3369–3377. doi: 10.1021/bi00634a013. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Mason K (1980) The first two decades of vitamin E history. In LJ Machlin, ed, Vitamin E: A Comprehensive Treatise. Marcel Dekker, New York, pp 1–6
- Mayonado DJ, Hatzios KK, Orcutt DM, Wilson HP. Evaluation of the mechanism of action of the bleaching herbicide SC-0051 by HPLC analysis. Pestic Biochem Physiol. 1989;35:138–145. [Google Scholar]
- Newman T, De Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M and others. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;7:2139–2148. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche PA, Moorehead TJ, Hamilton GA. Purification and properties of hog liver 4-hydroxyphenylpyruvate dioxygenase. Arch Biochem Biophys. 1982;216:62–73. doi: 10.1016/0003-9861(82)90188-6. [DOI] [PubMed] [Google Scholar]
- Ruetschi U, Dellsen A, Sahlin P, Stenman G, Rymo L, Lindstedt S. Human 4 hydroxyphenylpyruvate dioxygenase: primary structure and chromosomal localization of the gene. Eur J Biochem. 1993;213:1081–1089. doi: 10.1111/j.1432-1033.1993.tb17857.x. [DOI] [PubMed] [Google Scholar]
- Schultz A, Ort O, Beyer P, Kleinig H. SC-0051, a 2-benzoylcyclohexane-1,3-dione bleaching herbicide, is a potent inhibitor of the enzyme p-hydroxyphenylpyruvate dioxygenase. FEBS Lett. 1993;318:162–166. doi: 10.1016/0014-5793(93)80013-k. [DOI] [PubMed] [Google Scholar]
- Secor J. Inhibition of barnyardgrass 4-hydroxyphenylpyruvate dioxygenase by sulcotrione. Plant Physiol. 1994;106:1429–1433. doi: 10.1104/pp.106.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada GH, Fellman JH, Fujita TS, Roth ES. Purification and properties of avian liver p-hydroxyphenylpyruvate hydroxylase. J Biol Chem. 1975;250:6720–6726. [PubMed] [Google Scholar]
- Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J and others. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Wintermeyer E, Fluegel M, Ott M, Steinert M, Rdest U, Mann KH, Hacker J. Sequence determination and mutational analysis of the lly locus of Legionella pneumophila. Infect Immun. 1994;62:1109–1117. doi: 10.1128/iai.62.3.1109-1117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff EE, Pishko EJ, Kirkland TN, Cole GT. Cloning and expression of a gene encoding a T-cell reactive protein from Coccidioides immitis: homology to a 4-hydroxyphenylpyruvate dioxygenase and the mammalian F antigen. Gene. 1995;161:107–111. doi: 10.1016/0378-1119(95)00250-a. [DOI] [PubMed] [Google Scholar]