Abstract
Receptor-like kinases (RLKs) play important roles in multiple aspects of plant growth and development. As a member of the TNFR-like RLK subfamily, rice Crinkly4 (OsCR4) functions mainly in epidermal cell differentiation in many organs. Here we show that in addition to its essential role in epidermal cell differentiation in the palea and lemma, OsCR4 positively regulates rice culm elongation, similar to maize CR4. Although OsCR4 is an active kinase, like CR4 in maize and ACR4 in Arabidopsis, the conserved amino acid K532 in OsCR4 is not essential for its kinase activity in vitro. Whether other conserved amino acids are required for its kinase activity and the relationship between its activity and function in plant development remain to be investigated.
Keywords: OsCR4, TNFR, culm, kinase activity, rice
The Receptor-Like Kinase (RLK) Rice Crinkly4 (OsCR4) Positively Regulates Culm Elongation
The tumor necrosis factor receptor (TNFR)-like subfamily is a small one; there are eight members in Arabidopsis and seven in rice.1 Only three orthologs, CRINKLY4 (CR4) in Zea mays, ACR4 in Arabidopsis, and OsCR4 in Oryza sativa, contain the classical structure of this subfamily in their extracellular region, including seven crinkly repeats and one cysteine-rich TNFR-like domain.2
The function of the CR4 subfamily was first reported in maize, where it is mainly involved in epidermal cell differentiation in the leaves and aleurone.3 cr4 mutant plants also displayed defects in floral organs, including the glumes, anthers, and silks,3,4 and they exhibited very short stature.3 ACR4 also participates in epidermal cell differentiation in many tissues or organs, including leaf epidermis, as well as sepal margin, ovule integument, seed coat development and morphogenesis of embryo.2,5-7 The stature of the acr4 mutant was almost the same as that of wild type,7 perhaps due to the redundancy of ACR4 homologs. Another elegant work demonstrated that ACR4 promotes formative cell divisions and constrains the number of dividing cells at the onset of organogenesis in the lateral root meristem.8 This work suggested that the members of the TNFR-like subfamily not only regulate epidermal cell differentiation, but also keep the balance between stem cell maintenance and differentiation.
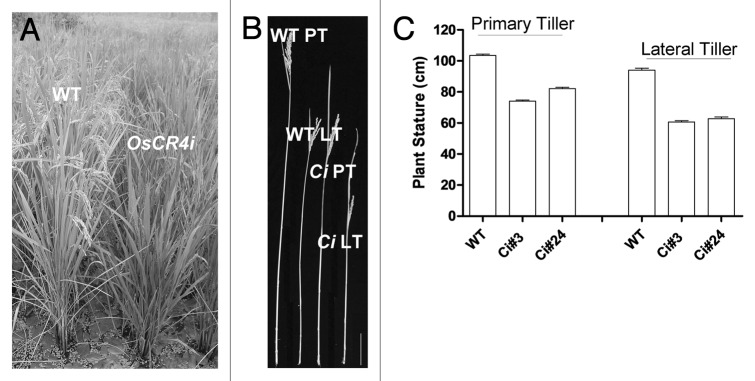
Similar to ZmCR4 and ACR4, OsCR4 functions to promote epidermal cell differentiation in the palea and lemma to maintain the interlocking structure of the spikelet; OsCR4i plants exhibit defects in the leaf epidermis and aleurone, as well as defects in fertility and seed development.9 Moreover, we found that the stature of OsCR4i plants was obviously shorter than that of wild-type plants at the harvest stage (Fig. 1A). Statistical analysis of two independent OsCR4i transgenic lines showed that the heights of their primary and lateral tillers were approximately two-thirds those in wild type (Fig. 1B and C), which is consistent with the obvious activity of the OsCR4 promoter in the stems of OsCR4::GUS transgenic plants.9 This phenotype suggested that OsCR4 positively regulates culm elongation, similar to ZmCR4. Regardless, mutant oscr4 plants will be a good material for fully recovering the function of OsCR4 in plant growth.
Figure 1. The stature of OsCR4i plants is shorter than that of wild-type plants. (A) Wild-type and OsCR4i plants in a natural field at the harvest stage. (B) Photograph of a single tiller from an OsCR4i plant and a wild-type plant at the harvest stage. (C) Statistical analysis of the stature of primary and lateral tillers from the two independent OsCR4i plants shown in (B). Bar = 10 cm in (B). WT, wild type; Ci,OsCR4i; PT, primary tiller; LT, lateral tiller.
The Conserved Amino Acid Lysine (K)532 is Not Required for the Kinase Activity of OsCR4 in Vitro
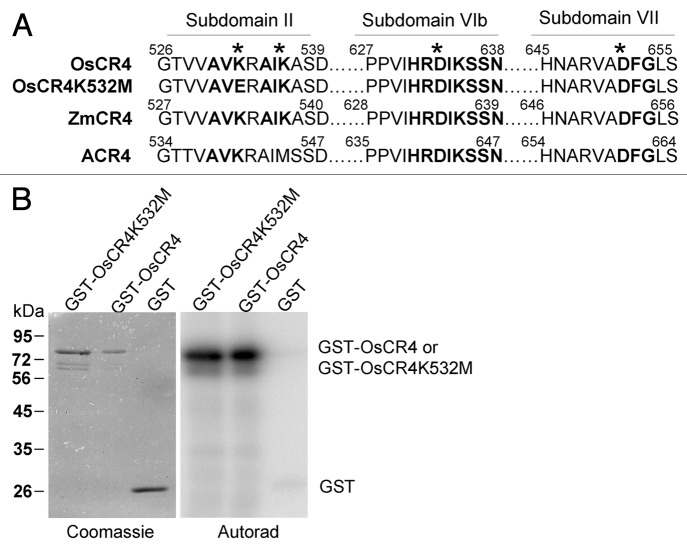
The kinase activity of three classical members of the TNFR-like subfamily was demonstrated in vitro.2,4,6,7,9 Whether the same conserved amino acid serves the same essential function is unknown. The primary structure of eukaryotic protein kinases is divided into 12 subdomains, and each contains conserved motifs and amino acids. In Subdomain II, the K residue in the AXK motif is considered to be essential for the activity of the kinase; in Subdomain VIb, the asparagine (D) in the HRDLKXXN motif is a candidate catalytic base; and in Subdomain VII, the D in the DFG motif is involved in chelating positive ions (e.g., Mg 2+) to orient the γ-phosphate of ATP for transfer.10 Classical plant TNFR-like RLKs also harbor these important conserved motifs and amino acids (Fig. 2A), and some have been confirmed in vitro. For example, substitution of the D652 (bold) in CR4 Subdomain VII by alanine (A) destroyed its kinase activity.4 In addition, the mutation of residue K540 (bold) in ACR4 Subdomain VIb to methionine (M),6 tryptophan (W),7,11 or A2 also destroyed the kinase activity of the enzyme. In summary, residues D652 in CR4 and K540 in ACR4 are essential for the activity of these kinases, respectively, in vitro. Unexpectedly, OsCR4K532M containing a point mutation in the AXK motif (asterisk) of Subdomain II produced the same autophosphorylation signal as wild-type OsCR4 in vitro (Fig. 2B); therefore, it appears that K532 is not essential for the kinase activity of the enzyme in vitro. Thus, we conclude that the K residues in the AVK (asterisk) and AIK (asterisk) motifs are important for the kinase activity of OsCR4. However, this should be validated by additional kinase assays.
Figure 2. Amino acid K532 in the kinase domain is not essential for OsCR4 kinase activity. (A) Amino acid alignment of the conserved kinase motif from OsCR4,OsCR4K532M, ZmCR4 and ACR4. (B) In vitro kinase assay of the recombination proteins glutathione S-transferase (GST)-OsCR4 and point-mutated GST-OsCR4K532M, and the negative control GST. In contrast to GST, both purified GST-OsCR4 and GST-OsCR4K532M exhibited autophosphorylation activity. Part of this figure was previously published.9
In assessing whether the kinase activity of TNFR-like family members is required for their functions, Gifford et al.12 proposed that the entire kinase domain and not the kinase activity of ACR4 was required for its signaling function. However, in our opinion, the K540M point mutant of ACR4 made by Gifford et al.12 using to complement the acr4 mutant may possess kinase activity in vivo, similar to OsCR4. Thus, we propose that there is a connection between the kinase activity of CR4s and their functions in plant development; however, this should be investigated further.
Materials and Methods
The point-mutated vector pGST-OsCR4K532M was generated from pGST-OSCR49 using a Muta-directTM kit (SBS Genetech Co., Ltd., Beijing, China) and the primers OsCR4K532M-F (5′-CAGTTGTTGCCGTGATGCGTGCAATTAAGGC-3′) and OsCR4K532M-R (5′-GCCTTAATTGCACGCATCACGGCAACAACTG-3′) according to the manufacturer’s protocol.
Acknowledgments
This research was supported by the National Key Program on the Development of Basic Research in China (2007CB108702) and the National Science Foundation of China (30570153).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21106
References
- 1.Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–34. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao X, Li K, Suh SG, Guo T, Becraft PW. Molecular analysis of the CRINKLY4 gene family in Arabidopsis thaliana. Planta. 2005;220:645–57. doi: 10.1007/s00425-004-1378-3. [DOI] [PubMed] [Google Scholar]
- 3.Becraft PW, Stinard PS, McCarty DR. CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science. 1996;273:1406–9. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- 4.Jin P, Guo T, Becraft PW. The maize CR4 receptor-like kinase mediates a growth factor-like differentiation response. Genesis. 2000;27:104–16. doi: 10.1002/1526-968X(200007)27:3<104::AID-GENE30>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka H, Watanabe M, Watanabe D, Tanaka T, Machida C, Machida Y. ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant Cell Physiol. 2002;43:419–28. doi: 10.1093/pcp/pcf052. [DOI] [PubMed] [Google Scholar]
- 6.Gifford ML, Dean S, Ingram GC. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development. 2003;130:4249–58. doi: 10.1242/dev.00634. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe M, Tanaka H, Watanabe D, Machida C, Machida Y. The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 2004;39:298–308. doi: 10.1111/j.1365-313X.2004.02132.x. [DOI] [PubMed] [Google Scholar]
- 8.De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322:594–7. doi: 10.1126/science.1160158. [DOI] [PubMed] [Google Scholar]
- 9.Pu C-X, Ma Y, Wang J, Zhang Y-C, Jiao X-W, Hu Y-H, et al. Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J. 2012;70:940–53. doi: 10.1111/j.1365-313X.2012.04925.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–96. [PubMed] [Google Scholar]
- 11.Tanaka H, Watanabe M, Sasabe M, Hiroe T, Tanaka T, Tsukaya H, et al. Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development. 2007;134:1643–52. doi: 10.1242/dev.003533. [DOI] [PubMed] [Google Scholar]
- 12.Gifford ML, Robertson FC, Soares DC, Ingram GC. ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell. 2005;17:1154–66. doi: 10.1105/tpc.104.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]