Abstract
ERD2s (ER luminal protein receptors)-mediated retrograde transport is one of the most substantial processes to maintain the endoplasmic reticulum (ER) homeostasis. It is completed by the recognition of the escaped ER luminal proteins, the gathering into COP I vesicle, and the fusion and releasing into the ER. ERD2s can recognize HDEL/KDEL motifs at the C-terminal of the escaped ER luminal proteins at the Golgi to initiate the retrograde transport. However, these mechanisms remain largely unknown in plants. We recently found that two Nicotiana benthamiana homologs, ERD2a and ERD2b, functioned as ER luminal protein receptors, were required for both HDEL/KDEL motifs-mediated ER retrieval and participated in cell death triggered by ER stress and nonhost pathogens. Here, we provide a set of new data that ERD2a/2b can form homo- or hetero-oligomerization and interact with both the ADP-ribosylation factor 1 (ARF1) and its potential GTPase-activating proteins (GAP) indicated by the firefly luciferase complementation imaging assay (LCI). These evidences further support the ER luminal protein receptor function of ERD2a/2b in plants and suggest their evolutionarily conserved mechanism during the retrograde trafficking. We also analyze the characteristics of ERD2s within a species and among different species.
Keywords: ADP-ribosylation factor 1 (ARF1), ER luminal protein receptor, GTPase-activating proteins (GAP), Nicotiana benthamiana, oligomerization
The existence of intracelluar compartments in the eukaryotic cells is one of the most important characteristics compared with that in the prokaryotic cells. These compartments temporarily contact with each other or are interconnected by the vesicles to complete the cargo transport or the components exchange.1 Secretory proteins enter into the ER for synthesis and modification, followed by the COP II vesicle-mediated delivery to the Golgi for sorting.2-4 In plants, the soluble proteins appear to be transported to the Golgi dependent on COP II vesicle in the manner of bulk flow.5,6 Along this transport, the ER chaperones are also packaged into COP II vesicle and escaped to the Golgi. To maintain the ER function, these escaped ER chaperones should be retrieved, which is mediated by COP I vesicle. The ER luminal protein receptors (ERD2s) are the key component to initiate this process.
ERD2 was first identified through the screening of ER-retention defective (erd) yeast mutants.7 Followed is the description of two human KDEL receptors (as KDEL is the typical motif in human).8,9 In 1993, Arabidopsis ERD2 gene was also isolated and had the ability to complement the lethal phenotype of yeast erd2 deletion mutant.10 Further experiments to screen the suppressors that allow yeast to growth without ERD2 gene result in the identification of six suppressors of erd2-deletion (SED) genes.11 Together with the evidences of these SED genes, two important roles were suggested for ERD2: one is for the retention of HDEL/KDEL-tagged proteins in the ER; the other is to maintain the intracellular membrane homeostasis. Direct findings to support the role of ERD2s in the COP I vesicle-mediated retrograde transport from the Golgi to the ER is the consequential identification of the physical interaction partners, such as GAP for ARF1, one of the key components to initiate the trafficking.12,13 Besides the function of ER retrieval, ERD2s could also act as the signaling components. First, KDEL receptors can form oligomerization.13 Second, KDEL receptor was found to modulate the ER stress through mitogen-activated protein kinase (MAPK) signaling cascades.14 Recent research found that chaperone-bound KDEL receptor triggered the activation of Src family kinases on the Golgi complex which was essential in the regulation of Golgi-to-plasma membrane transport.15 All these evidences lead to further explore the signaling roles of ERD2 in the future.
Arabidopsis genome has seven ERD2-like proteins which can be grouped into two classes, two belonging to class I and the other five belonging to class II.16 However, little is known about plant ERD2s until recently. We found that N. benthamiana ERD2a (GU388433) and ERD2b (GU388432) have the activity of ER luminal protein receptor.17 By a high-throughput virus-induced gene silencing (VIGS) screen, we found that knocking down ERD2b delayed the nonhost pathogen Xanthomonas oryzae pv oryzae (Xoo)-induced cell death. To investigate the class I ERD2s, we cloned the other member ERD2a. Both ERD2a and ERD2b can function as the ER luminal protein receptor in yeast. They localized to both the ER and the Golgi, consistent with their predicted role to shuttle between these two compartments. Further, we used GFP carrying with C-terminal HDEL and KDEL motifs to moniter the activity of ER retrieval process. We demonstrated that ERD2a and ERD2b were required for HDEL/KDEL-mediated retrieval of ER luminal proteins as ER luminal protein receptors in plants with functional addative or redundancy. Silencing ERD2b eliminated most of the GFP fluorescence for GK plants (GFP-KDEL transgenic plants) but not for 16c plants (GFP-HDEL transgenic plants), suggesting that ERD2b could have preferential recognition for KDEL-tagged proteins in plants.
Here, we provide another data to support plant ERD2a and ERD2b can function as ER luminal protein receptor. Firefly luciferase is split into N domain (Nluc) and C domain (Cluc) which are fused to the tested pairwise proteins. Upon interaction, the complement luciferase is reconstituted and can be detected easily, and this method is called firefly luciferase complementation imaging assay (LCI).18 Using this method, we found the pairwise interaction among ERD2, ARF1 (DQ531849) and the potential ARF1-GAP (TC136734) (Fig. 1A). Oligomerization has been reported to be a conserved and commom interaction form for both soluble proteins and membrane proteins to transduce signals and regulate different cellular events. We also showed that ERD2a/2b can oligomerize homologously or heterologously (Fig. 1B), suggesting that ERD2s may also act as signaling components in plants though downstream targets remain unknown. All the results suggest a conserved role of ERD2 may exist evolutionarily.
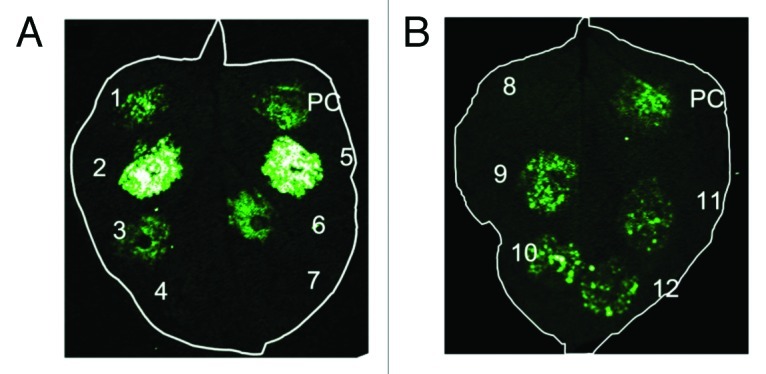
Figure 1. LCI assay: Tested pairwise genes were fused to N-terminal domain of firefly luciferase (Nluc) or C-terminal domain of firefly luciferase (Cluc) and were cloned into T-DNA vector. Agrobacterium cultures harboring the tested pairwise plasmids were mixed at a 1:1 ratio and were co-infiltrated into N. benthamiana. 48 h later, leaves were detached and sprayed with 1 mM luciferin. Fluorescence chemical signal was captured with a low-light cooled CCD camera (Andor iXon CCD camera, Andor Technology Ltd.). Pictures were taken after 10 min explosure. (A). ERD2s interact with ARF1 and ARF1-GAP. “1” represents tested pairwised proteins ARF1-Cluc/GAP-Nluc; “2”: ERD2a-Cluc/ARF1-Nluc; “3”: ERD2a-Cluc/GAP-Nluc; “4”: ERD2a-Cluc/Nluc; “5”: ERD2b-Cluc/ARF1-Nluc; “6”: ERD2b-Cluc/GAP-Nluc; “7”: ERD2b-Cluc/Nluc; “4” and “7” are negative control; (B). ERD2a/2b oligomerize homologously or heterologously. “8” is negative control expressing Nluc and Cluc; “9”: ERD2a-Cluc/ERD2a-Nluc; “10”: ERD2a-Cluc/ERD2b-Nluc; “11”: ERD2b-Cluc/ERD2b-Nluc; “12”: ERD2a-Nluc/ERD2b-Cluc; “PC” is positive control expressing Cluc-AtRar1/AtSgt1a-Nluc (At: Arabidopsis).
Though the typical roles of ERD2s may be conserved, there are still some differences among family members of some speciesor different organisms. First, in yeast, HDEL is the typical retrieval motif carried by ER luminal proteins while KDEL is adpoted in mammals. These two motifs are commonly found in plants though with other diverse motifs, like KAEL for (UDP)-glucose:glycoprotein glucosyltransferase (UGGT). It is worth noting that the distribution of HDEL/KDEL among the plant ER luminal proteins appear not random: HDEL is mostly carried by the luminal binding proteins (BiPs)/calreticulins (CRTs) family while KDEL is mostly contained by endoplasmin and most protein disulfide isomerases (PDIs).19 Among different species, the ligands of ERD2s could be encoded by different numbers of genes and have different retrieval motifs. For example, three BiP genes are found in Arabidopsis and more than seven in tobacco with HDEL motif, while only one gene encode BiP in yeast and human with HDEL and KDEL respectively. Since ERD2s are responsible for their recognition and retrieval, the variation of the retrieval motifs among different organisms indirectly points out the functional difference between ERD2s and their cargo. Second, the family members of ERD2 orthologs are different among different species. three KDEL receptors (human ERD2s) are identified in human genome vs. only one ERD2 in yeast. Arabidopsis genome contains seven ERD2 like proteins. Our blast results against Nicotiana tabacum EST database also indicated seven ERD2 like proteins. It remains largely unknown whether the amount of ERD2s homologs is correlated with the recognition specificity to the different retrieval motifs. Third, the activity of ER luminal protein receptor may be organism-specific. Human KDEL receptor 1 cannot complement the lethal phenotype of yeast erd2 deletion mutant.8 In contrasts, Arabidopsis ERD2a, N. benthamiana ERD2a and ERD2b have been found to function in yeast.10,17 Further, ERD2a and ERD2b have been proved to function as ER luminal protein receptors in plants.17 Whether the other five class II ERD2-like proteins in plants can function as ER luminal protein receptor needs to be investigated further. Fourth, mutation of Arabidopsis ERD2b specifically affected CRT3, but not CRT1 or CRT2 or BiP, although they all carry a C-terminal HDEL signal.16 However, we found that silencing N. benthamiana ERD2a and/or ERD2b affected BiP accumulation in the ER, suggesting BiP may be a client of N. benthamiana ERD2a and ERD2b. The difference may be species-specific and raise the question to determine the biological significance of these differences. Fifth, silencing N. benthamiana ERD2b caused more loss of GFP with KDEL motif than GFP with HDEL motif from the ER, suggesting a preferred characteristic for different retrieval motifs.
We found that in most cases, ERD2a and ERD2b functioned addatively to regulate the biological events including ER retrieval, ER stress induced cell death, R genes triggered cell death. Silencing N. benthamiana ERD2a and/or ERD2b enhance R genes triggered cell death, suggesting ER stress is also one part of hypersensitive response. We found only two different effects: one is the retrieval motif selection as mentioned above; the other one is their contrast response to two tested nonhost pathogens Xanthomonas oryzae pv oryzae (Xoo) and Pseudomonas syringae pv tomato DC3000 (Pst DC3000) induced cell death. Human KDEL receptor was reported to be involved in the activation of Src family kinases and may act as signaling component besides ER retrieval.20,21 Thus, we postulated that the different effects on nonhost cell death of ERD2a and ERD2b may be attribute to pathogen-induced ER stress, ER quality control of the cell death related proteins or the downstream signaling pathways to be identified.
Taken together, ERD2 mediated ER retrieval are highly conserved among different species with few functional differences. Multiple genes for ERD2s in plants provide the possibility to verify the specificity between the ERD2s and their clients, and further to illustrate the evolutionary divergence.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21217
References
- 1.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/S0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.Hanton SL, Matheson LA, Brandizzi F. Seeking a way out: export of proteins from the plant endoplasmic reticulum. Trends Plant Sci. 2006;11:335–43. doi: 10.1016/j.tplants.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Wachter RM, King BA, Heim R, Kallio K, Tsien RY, Boxer SG, et al. Crystal structure and photodynamic behavior of the blue emission variant Y66H/Y145F of green fluorescent protein. Biochemistry. 1997;36:9759–65. doi: 10.1021/bi970563w. [DOI] [PubMed] [Google Scholar]
- 4.Marti L, Fornaciari S, Renna L, Stefano G, Brandizzi F. COPII-mediated traffic in plants. Trends Plant Sci. 2010;15:522–8. doi: 10.1016/j.tplants.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Matheson LA, Hanton SL, Brandizzi F. Traffic between the plant endoplasmic reticulum and Golgi apparatus: to the Golgi and beyond. Curr Opin Plant Biol. 2006;9:601–9. doi: 10.1016/j.pbi.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Phillipson BA, Pimpl P, daSilva LL, Crofts AJ, Taylor JP, Movafeghi A, et al. Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell. 2001;13:2005–20. doi: 10.1105/TPC.010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza JC, Hardwick KG, Dean N, Pelham HR. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990;61:1349–57. doi: 10.1016/0092-8674(90)90698-E. [DOI] [PubMed] [Google Scholar]
- 8.Lewis MJ, Pelham HR. A human homologue of the yeast HDEL receptor. Nature. 1990;348:162–3. doi: 10.1038/348162a0. [DOI] [PubMed] [Google Scholar]
- 9.Lewis MJ, Pelham HR. Sequence of a second human KDEL receptor. J Mol Biol. 1992;226:913–6. doi: 10.1016/0022-2836(92)91039-R. [DOI] [PubMed] [Google Scholar]
- 10.Lee HI, Gal S, Newman TC, Raikhel NV. The Arabidopsis endoplasmic reticulum retention receptor functions in yeast. Proc Natl Acad Sci U S A. 1993;90:11433–7. doi: 10.1073/pnas.90.23.11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick KG, Boothroyd JC, Rudner AD, Pelham HR. Genes that allow yeast cells to grow in the absence of the HDEL receptor. EMBO J. 1992;11:4187–95. doi: 10.1002/j.1460-2075.1992.tb05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majoul I, Straub M, Hell SW, Duden R, Söling HD. KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic: measurements in living cells using FRET. Dev Cell. 2001;1:139–53. doi: 10.1016/S1534-5807(01)00004-1. [DOI] [PubMed] [Google Scholar]
- 13.Aoe T, Cukierman E, Lee A, Cassel D, Peters PJ, Hsu VW. The KDEL receptor, ERD2, regulates intracellular traffic by recruiting a GTPase-activating protein for ARF1. EMBO J. 1997;16:7305–16. doi: 10.1093/emboj/16.24.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto K, Hamada H, Shinkai H, Kohno Y, Koseki H, Aoe T. The KDEL receptor modulates the endoplasmic reticulum stress response through mitogen-activated protein kinase signaling cascades. J Biol Chem. 2003;278:34525–32. doi: 10.1074/jbc.M304188200. [DOI] [PubMed] [Google Scholar]
- 15.Capitani M, Sallese M. The KDEL receptor: new functions for an old protein. FEBS Lett. 2009;583:3863–71. doi: 10.1016/j.febslet.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci U S A. 2009;106:15973–8. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G, Li S, Xie K, Zhang Q, Wang Y, Tang Y, et al. Plant ERD2-like Proteins Function as ER Luminal Protein Receptors and Participate in Programmed Cell Death during Innate Immunity. Plant J. 2012 doi: 10.1111/j.1365-313X.2012.05053.x. In press. [DOI] [PubMed] [Google Scholar]
- 18.Chen HM, Zou Y, Shang YL, Lin HQ, Wang YJ, Cai R, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–76. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadlington JL, Denecke J. Sorting of soluble proteins in the secretory pathway of plants. Curr Opin Plant Biol. 2000;3:461–8. doi: 10.1016/S1369-5266(00)00114-X. [DOI] [PubMed] [Google Scholar]
- 20.Sallese M, Giannotta M, Luini A. Coordination of the secretory compartments via inter-organelle signalling. Semin Cell Dev Biol. 2009;20:801–9. doi: 10.1016/j.semcdb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Pulvirenti T, Giannotta M, Capestrano M, Capitani M, Pisanu A, Polishchuk RS, et al. A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat Cell Biol. 2008;10:912–22. doi: 10.1038/ncb1751. [DOI] [PubMed] [Google Scholar]


