Abstract
Salinity negatively affects plant growth and disturbs chloroplast integrity. Here, we aimed at identifying salt-responsive translation-related genes in Arabidopsis thaliana with an emphasis on those encoding plastid-located proteins. We used quantitative real-time PCR to test the expression of 170 genes after short-term salt stress (up to 24 h) and identified several genes affected by the stress including: PRPL11, encoding plastid ribosomal protein L11, ATAB2, encoding a chloroplast-located RNA-binding protein presumably functioning as an activator of translation, and PDF1B, encoding a peptide deformylase involved in N-formyl group removal from nascent proteins synthesized in chloroplasts. These genes were previously shown to have important functions in chloroplast biology and may therefore represent new targets for biotechnological optimization of salinity tolerance.
Keywords: Arabidopsis thaliana, gene expression, salt stress, ribosomal protein, translation, variation of information, clustering, dynamic time warping
Introduction
Plants as sessile organisms have developed sophisticated regulatory systems to respond to changing environmental conditions and overcome different stresses.1 Plants respond to abiotic stresses by various cellular processes that involve stress sensing, different signaling pathways and changes in gene expression controlled and modulated by transcription factors.1,2 Altered gene expression contributes to establishing a new cellular state that may increase the tolerance to the stress, allowing plants to survive under unfavorable conditions.2-4
Salt stress is a major abiotic stress that affects plant growth and development and decreases yield and production. Moreover, it affects metabolism, protein synthesis, and the energy household.4,5 Like other abiotic stresses, salinity leads to changes at the transcription level.6,7 In addition, the protein synthesis machinery is sensitive to NaCl,8 and increased protein synthesis ability has been reported to contribute to salt tolerance.9-12 Salt stress affects de novo protein synthesis,13 and the synthesis of ribosomes itself is required for protein synthesis and closely correlates with growth. Therefore, changes in growth rate, affected by different factors, including abiotic stresses, is tightly coupled with ribosome biosynthesis rate.14 Ribosomes are potential targets of control mechanisms that generate signals and activate adaptive regulons or developmental programs.15 Transcription of ribosome precursor genes represents a starting point for the synthesis of ribosomal subunits and is known to be highly regulated.14,16,17 Moreover, it has been reported that ribosomal subunit transcription is affected by biological factors and drugs.18 Although the ribosomal proteins are essential for growth and development in all organisms, our knowledge about expression regulation of genes encoding ribosomal proteins in plants under stress conditions remains fragmented.19 Many ribosomal proteins have been reported to be up- or downregulated under various stress conditions,19-22 and it has been proposed that the synthesis of ribosomal proteins plays a role for restructuring the protein synthesis apparatus under abiotic stress.22
Comparative transcriptomics performed on Arabidopsis thaliana, a glycophyte, and salt cress (Thellungiella halophila), a halophyte, suggested that salt-sensitive and salt-tolerant species have a common set of salt responses whose regulation differs.23 In salt cress, a strong increase of several proteins involved in protein synthesis and mainly ribosomal proteins and proteins involved in translation apparatus (eIF3A) was found.11 Recently, a model has been proposed whereby halophytes have a more efficient system for the regulation of transcription, protein synthesis and processing compared with glycophytes, which in turn affects the levels of different proteins to achieve homeostasis under salt stress condition.11 As the protein synthesis apparatus has an important role for plant growth and development under both stress and normal conditions, we studied the transcriptomic response of translation-related genes to salt stress in Arabidopsis.
Results and Discussion
Identifying salt-responsive genes
The aim of this study was to a gain deeper understanding of the short- (up to 8 h) and medium-term (24 h) response of leaf translation-related genes to salt stress in Arabidopsis seedlings. To this end, 16-d-old seedlings were salt-stressed (150 mM NaCl) for different times (2, 4, 6, 8 and 24 h) and RNA was extracted for gene expression analysis by quantitative real-time polymerase chain reaction (qRT-PCR). Control seedlings without salt stress were harvested at identical time points. For this experiment, 170 genes were selected from a set of genes related to protein synthesis in Arabidopsis leaves (see Methods). The experiment was performed in three biological replicates; additionally, each sample was measured in two technical replications.
The workflow for the analysis used in this study is illustrated in Figure 1. We used the results from preprocessing the obtained data, including normalization, qualification and filtering (see Methods), in two types of analyses - inferential and descriptive. With the inferential analysis, we set out to identify differentially expressed genes by performing the following comparisons: between time points in one condition, between consecutive time points across conditions, and over all time points, resulting in determination of global regulators. The first and the second steps in the inferential analysis were performed by using linear models. This is a first choice of method when analyzing differential expression, especially in the case when few replicates are available. The results are shown in Supplementary file 1, Table S1, and Fig. S1. Different statistical tools, available in existing R packages (see Methods), were then used to perform the third analysis to identify any global regulators (Supplementary file 1, Table S2 and S3). In addition, we performed a descriptive analysis based on clustering to identify genes which contribute to the changes in the cluster structure between control and treatment conditions, which we term global effectors (Supplementary file 1, Table S4).
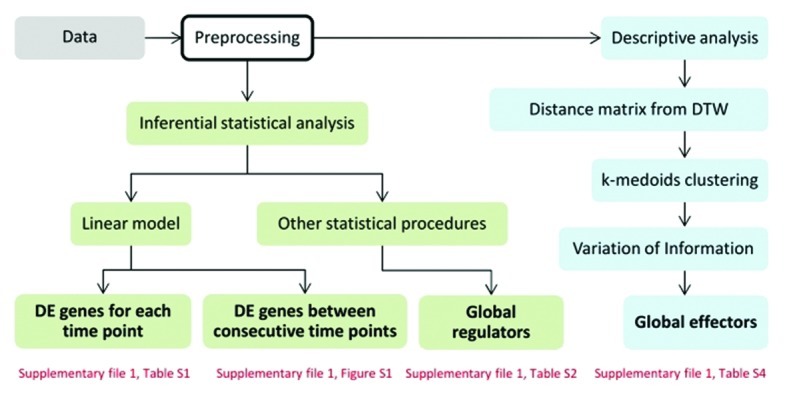
Figure 1. Workflow of the analysis. Two types of analyses were conducted on the preprocessed control and treatment data - inferential and descriptive - depicted by green and blue color, respectively. Text in bold shows the result of the respective process and the text in red indicate tables and figures containing the results.
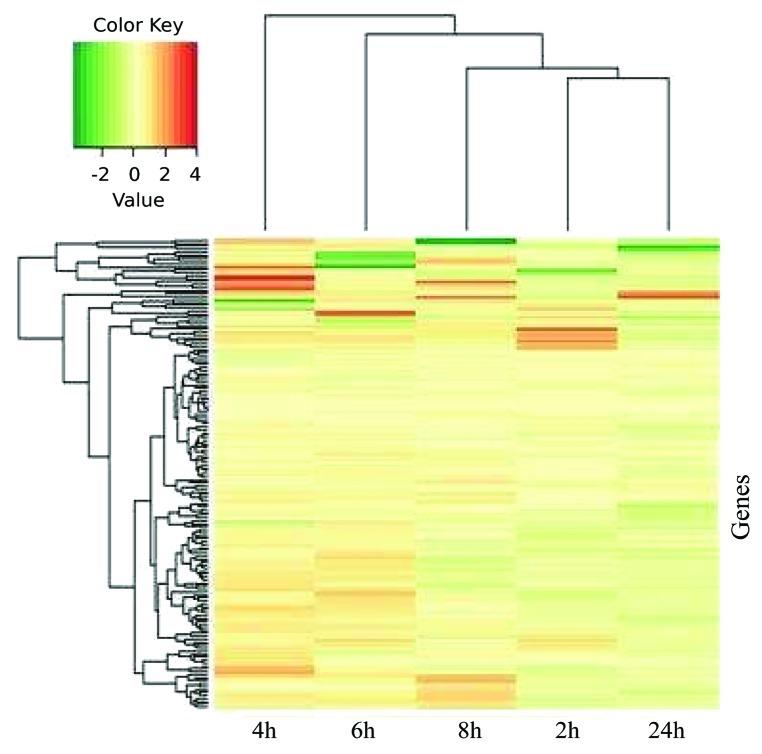
The fold-change of gene expression from the control and treatment data for each of the five analyzed time points is illustrated in Figure 2, which demonstrates that the majority of the genes was downregulated after 2 h of salt stress, while at 4 h and 6 h expression of several genes was increased relative to the control. However, after longer stress treatments, at 8 h and 24 h, the expression of most genes was reduced compared with the control. Since transcript abundance of many genes was reduced at the 2 h and 24 h time points, compared with control, salinity stress appeared to induce a transient response at the gene expression level. We note that the patterns of up- and downregulation match the clustering based on log-fold changes shown in Figure 2. One of the genes analyzed, At1g22110 (ribosomal protein L30e) was repressed compared with the control treatment in three of the five analyzed time points. None of the other genes was significantly downregulated by salinity stress. However, two and six genes showed upregulation upon salt stress in three and two time points, respectively (Supplementary file 1, Table S1). One of the genes upregulated at three time points was AT3G08010, encoding ATAB2, a chloroplast-located A/U-rich RNA-binding protein presumably functioning as an activator of translation. Inactivation of ATAB2 leads to a strong impairment in Arabidopsis and thylakoid membrane biogenesis, finally resulting in albino plants.24 Activation of ATAB2 expression by salt stress, as observed here, may be an important factor contributing to counteract the negative effect of salt stress on plant performance. The other gene showing enhanced expression in three time points was AT4G11175, encoding chloroplast translation initiation factor IF-1. Although the biological role of the gene has not been analyzed yet in Arabidopsis, it can be assumed to play an important role in translational control. The fact that AT4G11175 shows elevated expression during salinity stress may point to its involvement in the control of translation of transcripts important for establishing salt tolerance.
Figure 2. Fold-change of gene expression data from the control and treatment conditions. Heat map of salt-regulated transcripts in Col-0 plants subjected to 150 mM NaCl stress. Arabidopsis plants were grown on Murashige-Skoog (MS) medium. Seedlings were collected 16 d after germination and transferred to liquid medium, and salt stress was applied. Expression levels were determined by calculating the log-fold changes between salt treated and non-treated samples. The translation-related genes in different time points after salt treatment were subjected to hierarchical clustering with the average method by using R. Red and green indicate higher and lower expression values, respectively. Intensity of the colors is proportional to the absolute value of log2 of the fold difference in expression. White indicates no change in gene expression.
Among the genes upregulated by salt stress is AT1G33140 (60S ribosomal protein L9–1, also called PIGGYBACK 2), a gene previously found to affect leaf patterning.25 AT5G14660, also upregulated by salt stress, encodes a plant-specific peptide deformylase 1B (PDF1B) that catalyzes the removal of the N-formyl group from the N-terminus of nascent proteins synthesized in chloroplasts.26,27 A knockout of Arabidopsis PDF1B led to an albino phenotype.28 Similarly, in rice, PDF1B has been shown to be essential for chloroplast development.29 Thus, activation of PDF1B expression by salt stress as observed here may be important for a transient activation of translational capacity during the initial phase of salinity stress. AT3G02660 encodes a dual-targeted (chloroplast and mitochondria) tyrosyl-tRNA synthetase30; its mutation leads to an embryo-defective morphological phenotype during seed development31 (www.seedgenes.org). The other genes upregulated by salt stress, namely AT3G13580, AT3G20230 and AT5G16710, have to our knowledge not been functionally characterized yet.
Taken together, several genes encoding elements of the translational machinery are transiently activated at the transcript level upon salinity stress, which may influence the translation capacity of the plant. In fact, salinity stress has been reported to cause a transient suppression of de novo protein synthesis in Arabidopsis suspension cultures from which the cells recover within a time frame of 4 h.13 In accordance with this we found that most of the salt-affected translation-associated genes were upregulated at 4 h of salt treatment, but not at 2 h (Supplementary file 1, Table S1). In addition, although we observed salinity-triggered expression changes, a further possibility is that translation of these altered transcripts is affected as well. Of note, protein abundance of chloroplast elongation factor EF-Tu (AT4G20360) increased upon salt, but not sorbitol treatment in Arabidopsis cell suspension cultures13; its expression increased in our data set at the 2 h time point (Supplementary file 1, Table S1).
Cluster-based approach to identify global effectors
Since only few genes showed significant changes over time in our initial analysis (see above), we tried to explore the data set through a descriptive cluster-based analysis. To identify key players in this experimental set-up we employed a clustering-based method by using dynamic time warping (DTW).32,33 As the genes were pre-selected by means of their proposed function in relation to translation and due to similar expression profiles (see Methods), we could not find clusters of genes which could be separated based on their sub-cellular localization, biological process or molecular function. Therefore, we used a novel way to determine the global effectors which is based on the concepts of entropy and variation of information (VI)34 for the clusterings of data from control and treatment conditions.
We clustered all genes for the treatment and the control separately, and calculated the difference between the clusters from the control and the clusters from the treatment data by calculating the respective VI. We then iteratively removed a gene xi, and repeated the clustering for the set of genes without xi at iteration i. Similarly, we calculated the differences between the clusters of control data and the clusters of treatment data. This resulted in 61 genes of the 170 genes tested, as listed in Supplementary file 1, Table S4. Almost none of the 61 genes have so far not been tested with respect to salt physiology. We aligned the treatment and control clusterings in the following way: for each cluster in the treatment, we determined the most similar cluster from the control data with respect to the genes they share. The five genes contributing to the difference between the most similar clusters are shown in Table 1. Three out of four global regulators (see Supplementary file 1, Table S2 and S3), determined in the inferential analysis (differential expression), also appeared in the 61 genes.
Table 1. Genes were located in different clusters by aligning the control and treatment clusterings. The five genes encode ribosomal proteins and show maximum absolute value of log-fold changes over the entire time domain in the range from 1.60 to 3.63.
| AGI code | Annotation | Maximum absolute values of fold change |
|---|---|---|
| AT2G47610 |
60S ribosomal protein L7a-1 |
3.63 |
| AT1G07320 |
50S ribosomal protein L4, chloroplastic (CL4) (R-protein L4) |
1.60 |
| AT3G63490 |
50S ribosomal protein L1, chloroplastic (CL1) |
1.60 |
| AT4G17560 |
50S ribosomal protein L19–1, chloroplastic |
2.09 |
| AT3G04770 | 40S ribosomal protein Sa-2 (p40 protein homolog) | 2.07 |
As outlined, the five genes from Table 1 contributed to the difference between the salt-treated and the untreated samples following the descriptive analysis approach described in the previous paragraphs. The specific functions of AT2G47610 (60S ribosomal protein L7a), AT1G07320 (chloroplast 50S ribosomal protein L4, RPL4) and AT3G63490 (chloroplast ribosomal protein L1) were not studied in detail yet, although the latter was reported to cause an embryo-defective phenotype when mutated (www.seedgenes.org). With respect to AT4G17560 (chloroplast ribosomal protein L19), a truncated cDNA of the salt cress homolog to this gene led to increased salt tolerance in Arabidopsis, suggesting that it might have caused the silencing of the homologous Arabidopsis gene, which may play a negative role in salt tolerance. However, it has to be noted that the truncated cDNA had a complete open reading frame encoding a small protein of around 100 amino acids. The possibility that the truncated peptide functions in salt tolerance cannot therefore be ruled out.35 Finally, the expression of AT3G04770 was found to be upregulated in a T-DNA insertion mutant of APUM23 that encodes a nucleolar Puf domain protein involved in pre-rRNA processing; the apum23 mutant showed slow growth.36
AT1G32990 is among the genes detected by the descriptive approach; it encodes plastid ribosomal protein L11 (PRPL11) and was previously found to be downregulated in transgenic Arabidopsis plant overexpressing the ethylene receptor NTHK1 from tobacco after 12 h of salt stress.37 Such plants showed enhanced sensitivity to salt stress.38 A knockout mutant deficient for PRPL11 was previously reported; although neither the abundance of plastid ribosomes nor their assembly into polysomes was affected, the mutant showed significantly reduced translation of the large subunit of Rubisco (RbcL), the CO2-fixing enzyme located in the plastid stroma.39 The severe phenotype of the prpl11 mutant, a drastic growth depression and pale leaves, are consistent with an important function of PRPL11 for protein synthesis in plastids and chloroplast function. Thus, the salt stress-dependent upregulation of PRPL11 expression as observed here may contribute to sustain chloroplast functionality at least during the initial period under salt stress. It may be interesting to overexpress PRPL11 transgenic plants to determine the effect on salt tolerance in the future. Two other genes found to be affected by salt stress in the descriptive approach are AT3G08010 (encoding ATAB2) and AT5G14660 (PDF1B), as outlined above.
Salinity stress has a major effect on plant growth and performance and in particular is known to affect chloroplast function and hence photosynthesis. By using qRT-PCR-based expression profiling, we have shown here that a small number of genes coding for the plastidial translation machinery are rapidly altered by salt stress, although the majority of the translational genes tested did not significantly change their expression over the analyzed time course. The responsive genes are determined by using the proposed method following a comparison of clusterings based on the variation of information index. Most importantly, several of the salt-responsive genes are known to encode key functions in chloroplast biology, including PRPL11, ATAB2 and PDF1B. These genes thus may represent prime targets for the biotechnological optimization of plant and crop growth under saline conditions.
Materials and Methods
Plants cultivation, NaCl treatment and sample preparation
Arabidopsis thaliana (L.) Heynh. (Col-0) seeds were surface sterilized and placed on Murashige and Skoog (MS) semisolid agar plates (with 1% sucrose, 1.5% agar, pH 6.8)40 in a growth chamber at 8 h light (150 µE m−2 s−1, 20°C, 60% relative humidity)/16 h dark (16°C, 75% relative humidity) cycle. Uniformly developed 16-d-old seedlings from plates were transferred into liquid mediums41, and kept on a rotary shaker (100 rpm, continuous light) for 20 h. Thereafter, NaCl (150 mM final concentration) was added to the medium. Seedlings were harvested 2, 4, 6, 8 and 24 h after application of the salt, and control samples (not treated with NaCl) were harvested in parallel. The experiment was performed in three biological replicates.
RNA extraction, gDNA digestion and cDNA synthesis
RNA was extracted from eight to ten seedlings from each biological replicate, using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany, 74904) according to the manufacturer’s instructions. Genomic DNA (gDNA) contamination in extracted RNA was eliminated using TURBO DNA-free Kit (Ambion, Darmstadt, Germany, AM1907). Elimination of gDNA contamination was subsequently confirmed by qRT-PCR using intron-specific primers for MAF5 (At5g65080; FP 5′-TTTTTTGCCCCCTTCGAATC-3′ and RP 5′-ATCTTCCGCCACCACATTGTAC-3′). RNA integrity was checked on 1% (w/v) agarose gels, and RNA concentration was checked before and after gDNA digestion using a Nanodrop-2000 spectrophotometer (Peqlab, Erlangen, Germany). Thereafter, cDNA was synthesized from 2 µg of total RNA using RevertAid H minus First-strand cDNA Synthesis Kit (Fermentas, Sankt Leon-Rot, Germany, K1632). Oligo-dT primers were used to initiate cDNA synthesis. The synthesized cDNA was checked by qRT-PCR using ACTIN2 (At3g18780, FP 5′-TCCCTCAGCACATTCCAGCAGAT-3′ and RP 5′-AACGATTCCTGGACCTGCCTCATC-3′) as a reference gene; cDNA integrity was checked using GAPDH 5′ and 3′ end primer pairs (At1g13440; 3′ end: FP 5′-TTGGTGACAACAGGTCAAGCA-3′, RP 5′-AAACTTGTCGCTCAATGCAATC-3′; 5′ end: FP 5′-TCTCGATCTCAATTTCGCAAAA-3′, RP 5′-CGAAACCGTTGATTCCGATTC-3′). qRT-PCR analysis was done using SYBR Green PCR Master Mix (Applied Biosystems, Darmstadt, Germany, 4309155).
Gene selection and design of qRT-PCR primers
More than 95% of chloroplast proteins are encoded by the nucleus and enter the chloroplast after translation.42 In this study, genes related to the chloroplast translational machinery were chosen based on a previously established set of genes encoding plastid-localized proteins.43 Genes of this set were screened for high expression in leaves using the Arabidopsis eFP browser [http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi] and Genevestigator [https://www.genevestigator.com/gv/] online tools. The selected genes were then used to identify highly co-expressed genes (r-value 0.95 - 1.0, Pearson correlation coefficient; AtGenExpress tissue data set), using the expression angler tool [http://bar.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi], and only genes highly expressed in leaf tissue were selected and added to the gene list. This process was repeated several rounds until no new genes were found. The final list included 548 genes, which again were cross-checked with the set of 2090 genes to remove duplicates. Finally, 170 genes proposed to have a role in leaf translation were selected (see Supplementary file 2 for gene names and annotations). qRT-PCR primers were designed using QuantPrime (http://www.quantprime.de/) and are given in Supplementary file 2.
Data Analysis
Preprocessing
Data preprocessing was performed with the “HTqPCR” package in R,44 which is frequently used for analysis of qRT-PCR data. For each biological replicate, the mean of the two technical replicates was determined, and the data were normalized based on the reference gene (At3g18780, ACTIN2). The genes were first categorized as undetermined and determined, based on whether or not at least one NA value was present in the replicates, respectively; moreover, the genes were also divided into unreliable and reliable, based on whether or not the relative Ct values were outside the chosen confidence interval (here, 90%), respectively. The genes which were categorized as undetermined or unreliable in at least three replicates over the five time pointes were filtered out and were not considered in the subsequent analysis.
Inferential statistical analysis
Analysis of differential expression was performed on the data remaining after preprocessing and normalization. Differential expression was investigated: between time points in one condition, between consecutive time points across conditions, and over all time points, as summarized below:
1. Differential expression based on linear models: By employing the limma package,45 we performed two types of analysis: we attempted to first determine the genes which are differentially expressed at each time point relative to the control, and second, to find the differentially expressed genes between consecutive time points across conditions.
2. Differentially expressed genes over all time points (global regulators): Different statistical procedures were applied to find the genes which are differentially expressed over the entire period of the experiment, termed global regulators. To this end, time course,46 limma45 and samr47 packages were used.
Descriptive analysis-a cluster-based approach for global effectors
We used a novel way of quantifying the distance between the gene expression profiles, called Dynamic Time Warping (DTW).32,33 The distance measure is based on the idea of aligning a pair of time-resolved profiles over the entire (or partial) time span of the experiment by stretching and/or compressing time points, and subsequently, calculating the distance based on the found alignment. The resulting distance matrix, when used in clustering, guarantees that the genes assigned to different clusters cannot be aligned to each other even after stretching or compressing of their profiles along their expression curves.
Due to the small number of time points in the experimental setting, cubic spline interpolation was initially performed between first time point (2 h) and the fourth (corresponding to 8 h) with 60 equidistant time points in between. Finally, after the interpolation, time point 24 h was added to the resulting profile. Interpolation over the entire time span was not performed in order to reduce artifacts arising from the non-uniform sampling over time.
We then used DTW (R package dtw48) by applying Global alignment, ‘itakura’ windowing and a symmetric step pattern which has been defined by the Rabiner-Juang with the weighting type “d”49, as shown in Figure 3.
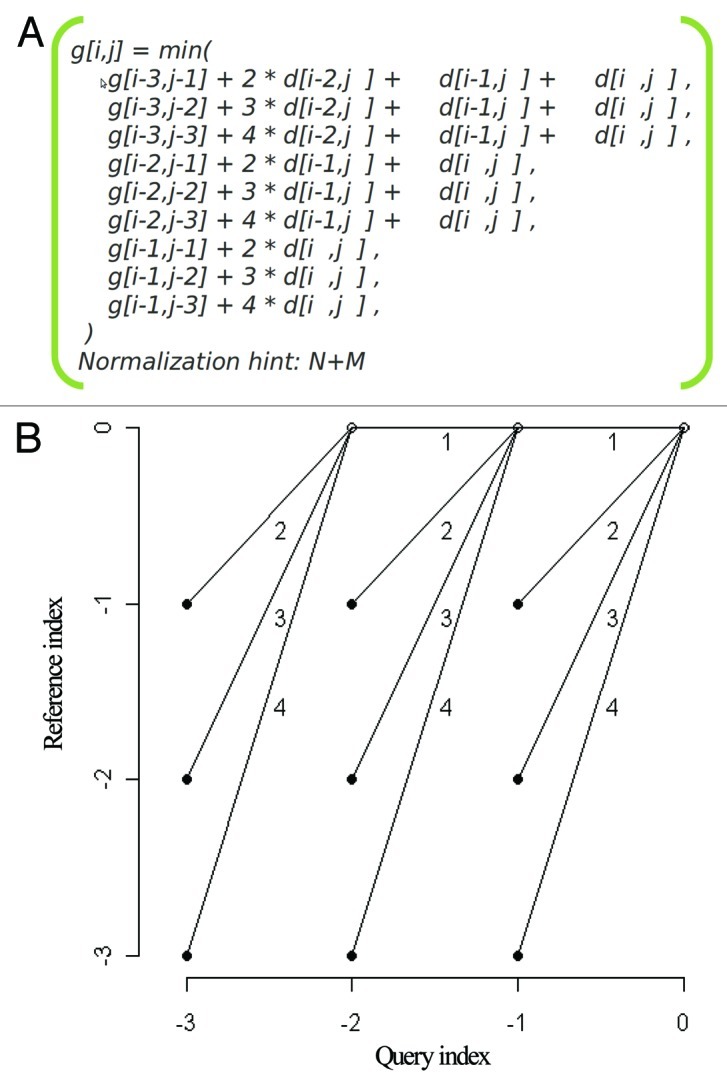
Figure 3. Path constraints. a) The step pattern used in calculating the distances between time-resolved expression profiles. b) Visualization of the global path constrain with the slope weighting on every possible path.
We used DTW to align the expression profile of genes over the entire time period and calculate the distances for all gene pairs. Thereafter, a K-medoids clustering algorithm was used with the distance matrix, resulting from DTW, as input. Clustering was performed for number of clusters ranging from 2 to 40 (for each 1000 repetitions are conducted) to determine the best number of clusters by using silhouette index.50
We employed the clustering algorithm on the control and the treatment separately, and then determined the genes which change their clustering membership between the two conditions, as described in the following section.
Determining global effectors; genes contributing to the difference between clustering of control and treatment based on variation of information
Variation of Information (VI)34 is a distance measure which is usually used to quantify the differences between two clusterings. Variation of information for two clusterings, quantifies the information in each of the clusterings in addition to the amount of information that one of the clustering gives about the other.
The entropy of a clustering is calculated as:
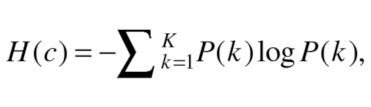 |
(1) |
where 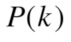 is the probability of selecting all
is the probability of selecting all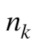 members of cluster number
members of cluster number 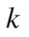 among all
among all 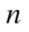 elements, so
elements, so 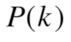 is
is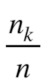 Entropy is a non-negative value and it becomes zero only if there exists no uncertainty, i.e., when all points group in a single cluster.34 Given two clusterings,
Entropy is a non-negative value and it becomes zero only if there exists no uncertainty, i.e., when all points group in a single cluster.34 Given two clusterings,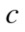 and
and 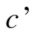 , with
, with 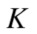 and
and 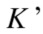 clusters, respectively, the information that one has about the other is called mutual information and defined as follows:
clusters, respectively, the information that one has about the other is called mutual information and defined as follows:
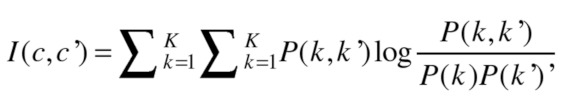 |
(2) |
where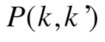 is the probability that a point belongs to
is the probability that a point belongs to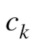 in clustering
in clustering 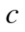 and to
and to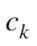 in
in 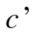 The variation of information, used as a comparison criterion for two clustering
The variation of information, used as a comparison criterion for two clustering 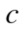 and
and 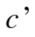 is then:
is then:
| (3) |
In our analysis, with the help of clustering and variation of information, we attempt to determine a set of global effectors, i.e., genes which contribute to large differences between clusterings of control and treatment data.
Thereafter, we removed the genes one by one and again calculate the difference between control and treatment without the chosen gene. If increases upon removal of gene
increases upon removal of gene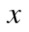 from the control data, i.e.,
from the control data, i.e.,
| (4) |
this gene can be considered as potentially informative. The same holds for the treatment data, and if
| (5) |
we can say that x also plays an important role in the treatment data since its removal increases the entropy in clustering of the treatment data.
By using Eqs. (4) and (5), we arrive at the Eq. (6)
| (6) |
On the other hand, 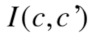 quantifies how much each of the variables reduces the uncertainty (the average over all variables). Hence, reducing the uncertainty can be encapsulated in the following inequalities:
quantifies how much each of the variables reduces the uncertainty (the average over all variables). Hence, reducing the uncertainty can be encapsulated in the following inequalities:
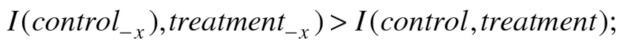 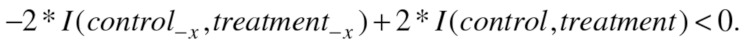
|
(7) |
By combining the inequalities from Eqs. (6) and (7), we obtain Eq. (8):
| (8) |
We can substitute some parts in Eq. (8) with the variation of information, so the final inequality used for selection of the genes acting as global effectors is given in Eq. (9), below.
| (9) |
Therefore, if the removal of a gene satisfies the inequality in Eq. (9), the gene can be considered as a global effector as it contributes to the difference between control and treatment clusterings.
Supplementary Material
Authors’ contributions
M.A.O, F.S.A. and B.M.R. designed the study. M.A.O. did all the experiments and primary data analysis. N.O. and Z.N. analyzed the data. All authors wrote the paper and interpreted the results.
Acknowledgments
We thank Dr. Salma Balazadeh for her comments and scientific discussions, Dr. Samuel Arvidsson for his helps in primary data analysis and scientific discussions and Dr. Armin Schlereth for his helps in robot programming for high-throughput qRT-PCR. M. A. O. thanks the University of Potsdam and the Ministry of Science, Research and Technology, Iran, for financial support.
Glossary
Abbreviations:
- PCR
Polymerase Chain Reaction
- PRPL11
Plastid Ribosomal Protein L11
- ATAB2
Arabidopsis TAB2
- PDF1B
Peptide Deformylase 1B
- eIF3A
eukaryotic Initiation Factor 3 A
- qRT-PCR
quantitative Real-Time PCR
- IF-1
Initiation Factor - 1
- DTW
Dynamic Time Warping
- VI
Variation of Information
- APUM23
ARABIDOPSIS PUMILIO 23
- NTHK1
Nicotiana tabacum Histidine Kinase-l
- RbcL
Rubisco large subunit
- Col-0
Colombia-0
- MS
Murashige and Skoog
- gDNA
genomic DNA
- cDNA
Complementary DNA
- FP
Forward primer
- RP
Reverse primer
- Ct
Cycle threshold
- lfc
Log-fold change
- Eq
Equation
Supplemental Material
Supplemental materials may be found at www.landesbioscience.com/journals/psb/article/21218.
Footnotes
These authors contributed equally to the work and should be considered as joint first authors.
Previously published online: www.landesbioscience.com/journals/psb/article/21218
References
- 1.Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot. 2009;103:551–60. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–81. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 3.Parida AK, Das AB. Salt tolerance and salinity effects on plants: a review. Ecotoxicol Environ Saf. 2005;60:324–49. doi: 10.1016/j.ecoenv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Shao Hong-bo cL-y, Zhao Chang-xing, Guo Qing-jie, Liu Xian-an, Ribaut Jean-Marcel. Plant Gene Regulatory Network Szstem Under Abiotic Stress. Acta Biologica Szegediensis. 2006;50:9. [Google Scholar]
- 5.Mehta P, Kraslavsky V, Bharti S, Allakhverdiev SI, Jajoo A. Analysis of salt stress induced changes in Photosystem II heterogeneity by prompt fluorescence and delayed fluorescence in wheat (Triticum aestivum) leaves. J Photochem Photobiol B. 2011;104:308–13. doi: 10.1016/j.jphotobiol.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–42. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–7. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 8.Rausell A, Kanhonou R, Yenush L, Serrano R, Ros R. The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J. 2003;34:257–67. doi: 10.1046/j.1365-313X.2003.01719.x. [DOI] [PubMed] [Google Scholar]
- 9.Brinker M, Brosché M, Vinocur B, Abo-Ogiala A, Fayyaz P, Janz D, et al. Linking the salt transcriptome with physiological responses of a salt-resistant Populus species as a strategy to identify genes important for stress acclimation. Plant Physiol. 2010;154:1697–709. doi: 10.1104/pp.110.164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosová K, Vítámvás P, Prášil IT, Renaut J. Plant proteome changes under abiotic stress--contribution of proteomics studies to understanding plant stress response. J Proteomics. 2011;74:1301–22. doi: 10.1016/j.jprot.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Pang Q, Chen S, Dai S, Chen Y, Wang Y, Yan X. Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J Proteome Res. 2010;9:2584–99. doi: 10.1021/pr100034f. [DOI] [PubMed] [Google Scholar]
- 12.Singh BN, Mishra RN, Agarwal PK, Goswami M, Nair S, Sopory SK, et al. A pea chloroplast translation elongation factor that is regulated by abiotic factors. Biochem Biophys Res Commun. 2004;320:523–30. doi: 10.1016/j.bbrc.2004.05.192. [DOI] [PubMed] [Google Scholar]
- 13.Ndimba BK, Chivasa S, Simon WJ, Slabas AR. Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2005;5:4185–96. doi: 10.1002/pmic.200401282. [DOI] [PubMed] [Google Scholar]
- 14.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/S0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 15.P.Palečková KMa. Activity of ribosomes and tmRNA of Streptomyces aureofaciens during development and stress conditions induced by changes in temperature and the presence of antibiotics. Communicating Current Research and Educational Topics and Trends in Applied Microbiology 2007; Formatex:11-8. [Google Scholar]
- 16.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–40. doi: 10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 17.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta 2010; 1803:673-83. [DOI] [PubMed]
- 18.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3:RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 19.Mukhopadhyay P, Reddy MK, Singla-Pareek SL, Sopory SK. Transcriptional downregulation of rice rpL32 gene under abiotic stress is associated with removal of transcription factors within the promoter region. PLoS One. 2011;6:e28058. doi: 10.1371/journal.pone.0028058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogalski M, Schöttler MA, Thiele W, Schulze WX, Bock R. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell. 2008;20:2221–37. doi: 10.1105/tpc.108.060392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, et al. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zorb C, Schmitt S, Neeb A, Karl S, Linder M, Schubert S. The biochemical reaction of maize (Zea mays L.) to salt stress is characterized by a mitigation of symptoms and not by a specific adaptation. Plant Sci. 2004;167:91–100. doi: 10.1016/j.plantsci.2004.03.004. [DOI] [Google Scholar]
- 23.Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, et al. Comparative genomics in salt tolerance between Arabidopsis and aRabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barneche F, Winter V, Crèvecoeur M, Rochaix JD. ATAB2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. EMBO J. 2006;25:5907–18. doi: 10.1038/sj.emboj.7601472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, et al. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development. 2008;135:1315–24. doi: 10.1242/dev.016469. [DOI] [PubMed] [Google Scholar]
- 26.Dirk LM, Williams MA, Houtz RL. Eukaryotic peptide deformylases. Nuclear-encoded and chloroplast-targeted enzymes in Arabidopsis. Plant Physiol. 2001;127:97–107. doi: 10.1104/pp.127.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dirk LMA, Schmidt JJ, Cai Y, Barnes JC, Hanger KM, Nayak NR, et al. Insights into the substrate specificity of plant peptide deformylase, an essential enzyme with potential for the development of novel biotechnology applications in agriculture. Biochem J. 2008;413:417–27. doi: 10.1042/BJ20071641. [DOI] [PubMed] [Google Scholar]
- 28.Giglione C, Vallon O, Meinnel T. Control of protein life-span by N-terminal methionine excision. EMBO J. 2003;22:13–23. doi: 10.1093/emboj/cdg007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon S, Giglione C, Lee DY, An S, Jeong DH, Meinnel T, et al. Rice peptide deformylase PDF1B is crucial for development of chloroplasts. Plant Cell Physiol. 2008;49:1536–46. doi: 10.1093/pcp/pcn121. [DOI] [PubMed] [Google Scholar]
- 30.Duchêne AM, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters NM, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2005;102:16484–9. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg M, Rogers R, Muralla R, Meinke D. Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 2005;44:866–78. doi: 10.1111/j.1365-313X.2005.02580.x. [DOI] [PubMed] [Google Scholar]
- 32.Sakoe H, Chiba S. Dynamic-Programming Algorithm Optimization for Spoken Word Recognition. Ieee T Acoust Speech. 1978;26:43–9. doi: 10.1109/TASSP.1978.1163055. [DOI] [Google Scholar]
- 33.Hempel S, Koseska A, Nikoloski Z, Kurths J. Unraveling gene regulatory networks from time-resolved gene expression data - a measures comparison study. BMC Bioinformatics. 2011;•••:12. doi: 10.1186/1471-2105-12-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meila M. Comparing clusterings - an information based distance. J Multivariate Anal. 2007;98:873–95. doi: 10.1016/j.jmva.2006.11.013. [DOI] [Google Scholar]
- 35.Du J, Huang YP, Xi J, Cao MJ, Ni WS, Chen X, et al. Functional gene-mining for salt-tolerance genes with the power of Arabidopsis. Plant J. 2008;56:653–64. doi: 10.1111/j.1365-313X.2008.03602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, et al. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010;64:960–76. doi: 10.1111/j.1365-313X.2010.04393.x. [DOI] [PubMed] [Google Scholar]
- 37.He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005;44:903–16. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 38.Cao WH. Ethylene receptor regulates plant response to salt stress. Institute of Genetics and Developmental Biology. Beijing: Chinese Academy of Sciences, 2004. [Google Scholar]
- 39.Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D. Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J. 2001;27:179–89. doi: 10.1046/j.1365-313x.2001.01076.x. [DOI] [PubMed] [Google Scholar]
- 40.Ljung K, Bhalerao RP, Sandberg G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001;28:465–74. doi: 10.1046/j.1365-313X.2001.01173.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Zhu QG, Meng QW, Wu CA. Transcript profiling during salt stress of young cotton (Gossypium hirsutum) seedlings via Solexa sequencing. Acta Physiol Plant. 2012;34:107–15. doi: 10.1007/s11738-011-0809-6. [DOI] [Google Scholar]
- 42.Soll J, Schleiff E. Protein import into chloroplasts. Nat Rev Mol Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- 43.Richly E, Leister D. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene. 2004;329:11–6. doi: 10.1016/j.gene.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Dvinge H, Bertone P. HTqPCR: high-throughput analysis and visualization of quantitative real-time PCR data in R. Bioinformatics. 2009;25:3325–6. doi: 10.1093/bioinformatics/btp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smyth GK. Limma: linear models for microarray data. New York: Springer, 2005:397–420. [Google Scholar]
- 46.Tai YC. timecourse: statistical analysis for developmental microarray time course data. 2007.
- 47.Li RTaGCaBNaJ. samr: SAM: Significance Analysis of Microarrays. 2011.
- 48.Giorgino T. Computing and Visualizing Dynamic Time Warping Alignments in R: The dtw Package. J Stat Softw. 2009;31:1–24. [Google Scholar]
- 49.Itakura F. Minimum prediction residual principle applied to speech recognition. Ieee T Acoust Speech 1975; As23:67-72.
- 50.Rousseeuw PJ. Silhouettes - a Graphical Aid to the Interpretation and Validation of Cluster-Analysis. J Comput Appl Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.