Abstract
Cytokinins are plant hormones with profound roles in growth and development. Cytokinin signaling is mediated through a 'two-component' signaling system composed of histidine kinases, histidine-containing phosphotransfer proteins, and response regulators. Phylogenetic analysis of two-component signaling elements from the monocot rice and the dicot Arabidopsis reveals lineage-specific expansions of the type-B response regulators, transcription factors that act as positive regulators for the cytokinin signal. We recently reported in Plant Physiology on a functional analysis of rice type-B response regulators. A type-B response regulator from a subfamily comprised of both monocot and dicot type-B response regulators complemented an Arabidopsis type-B response regulator mutant, but a type-B response regulator from a monocot-specific subfamily generally did not. Here, we extend this analysis to demonstrate that the promoter of an Arabidopsis cytokinin primary response gene is induced by type-B response regulators from a shared subfamily, but not by one from a lineage-specific subfamily. These results support a model in which the type-B response regulators of monocots and dicots share conserved roles in the cytokinin signaling pathway but have also diverged to take on lineage-specific roles.
Keywords: Arabidopsis, cytokinin, response regulator, rice, two-component signaling
Cytokinins control multiple aspects of plant growth and development, including regulation of cell division and metabolism, stimulation of chloroplast development, modulation of shoot and root development, and delay of leaf senescence.1-4 Cytokinin signal transduction is mediated through a multistep histidine-to-aspartate phosphorelay system, evolutionarily related to the two-component signaling systems of prokaryotes.5-7 Signaling is initiated when cytokinin binds to the histidine kinase-linked receptors with extracellular CHASE domains. Binding of cytokinin induces autophosphorylation on His, and subsequently Asp residues within the intracellular signaling domains. Phosphoryl groups are then transferred to nucleocytoplasmic histidine-containing phosphotransfer proteins, which convey the signal to the type-B response regulators (RRs) in the nucleus. The type-B RRs act as transcription factors and control the expression of primary cytokinin-responsive genes, including type-A RRs, which act as negative regulators of the signal transduction pathway.8-10
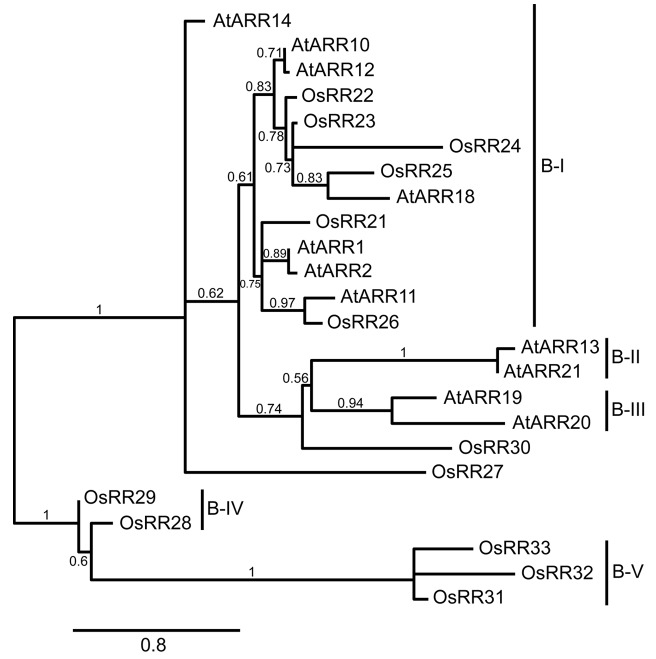
The current model of cytokinin signal transduction in plants has been established largely based on studies in Arabidopsis. However, similar two-component signaling elements have been identified in the monocot rice supporting a common mechanism for transmission of the cytokinin signal.11-14 In our paper published in Plant Physiology, we characterized the rice signaling elements based on several complementary approaches with a particular focus on their role in cytokinin signaling.15 To gain insight into the conserved and unique features of the two-component signaling elements of plants, we performed a phylogenetic analysis. Based on this analysis, rice and Arabidopsis have a similar cohort of histidine kinases and phosphotransfer proteins. Interestingly, although both rice and Arabidopsis contain the same types of RRs (types A, B, and C), substantial lineage-specific expansions of the RRs have occurred, such that there are few 1:1 orthologous relationships. Lineage specific expansion is defined in relative terms as the proliferation of a protein family in a particular lineage, relative to the sister lineage, with which it is compared.16 It is believed to be of critical importance to the evolution of genome plasticity as such expansions provided opportunities for functional overlap, which in turn can lead to the emergence of new functions. For example, rice contains 13 type-B RRs. Six of these are found in subfamily-I, which also contains the type-B RRs from Arabidopsis that participate in cytokinin signaling,15 but the other seven type-B RRs from rice fall into monocot-specific subfamilies (Fig. 1).
Figure 1. Phylogenetic relationship of type-B RRs of Arabidopsis and rice. A phylogeny based on the receiver domains of type-B RRs was constructed using the phylogeny.fr pipeline.22 Sequences from rice are designated with the prefix Os; sequences from Arabidopsis are designated with the prefix At. Nomenclature of rice elements is based on Schaller et al.23 and of Arabidopsis elements on Schaller et al.24 Subfamily designations are from Tsai et al.15
We used the NanoString nCounter system to gain information on the expression profile of rice two-component signaling elements, in which the cytokinin-regulated expression of rice genes was analyzed over a time course in the shoot and root.15 Cytokinin did not regulate expression of most type-B OsRRs, consistent with what has been found in Arabidopsis, and only five of the 13 type-B OsRRs (OsRR21, OsRR22, OsRR23, OsRR24, and OsRR26) were expressed at relatively high levels. Significantly, these five genes are members of the subfamily-I type-B OsRRs implicated in carrying out the majority of cytokinin signaling.
To elucidate the degree of functional overlap among type-B response regulators from rice and Arabidopsis, we performed a functional complementation assay based on the cytokinin insensitivity exhibited by the Arabidopsis type-B response regulator mutant, arr1–3;arr12–1.15 A rice subfamily-I type-B RR (OsRR22) restored cytokinin sensitivity of arr1–3;arr12–1 to wild-type level based on hypocotyl and root growth response assays. However, transgenic expression of a member of the monocot-specific subfamily IV (OsRR29) did not. Interestingly, both OsRR22 and OsRR29 could rescue the seed size phenotype of the arr1–3;arr12–1 mutant, suggesting that OsRR29 is functional but within a more limited developmental context than OsRR22.
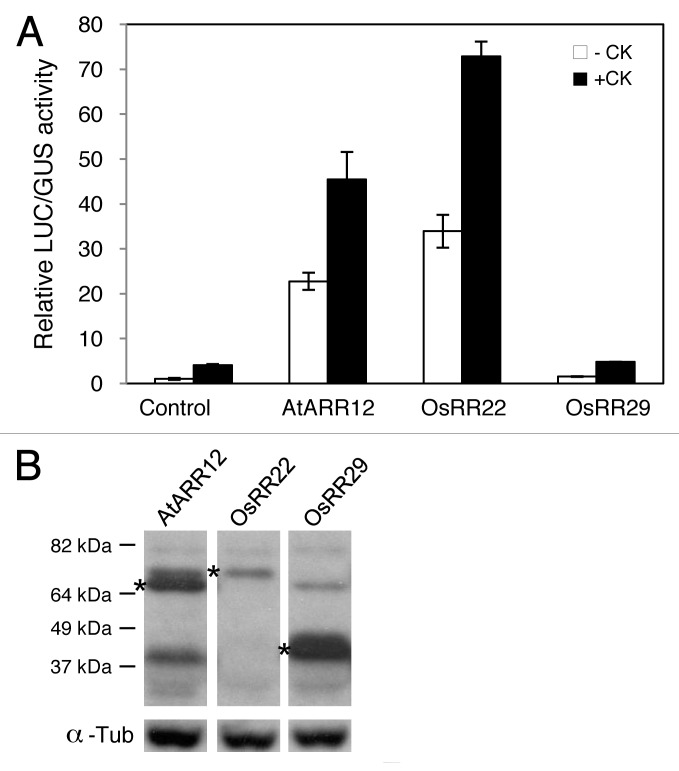
To test transactivation activity of OsRR22 and OsRR29 on a cytokinin primary response gene, AtARR6, we performed a transient expression assay using mesophyll protoplasts of Arabidopsis leaves.17,18 In transfected protoplasts, the activity of the AtARR6 promoter fused to the firefly luciferase (LUC) reporter gene was induced by cytokinin treatment about 4-fold (Fig. 2). Ectopic expression of AtARR12 and OsRR22 tagged with the Myc epitope was sufficient to activate AtARR6-LUC expression in the absence of exogenous cytokinin; AtARR12 and OsRR22 activated ARR6-LUC expression about 20- and 30-fold, respectively. Cytokinin treatment further enhanced the effect of AtARR12 and OsRR22 on the ARR6-LUC activity, inducing expression to over 45- and 70-fold, respectively, compared with basal levels. In contrast, OsRR29 did not transactivate AtARR6-LUC expression either in the absence or presence of cytokinin, even though OsRR29 was present at higher protein levels than OsRR22 (Fig. 2).
Figure 2. Functional analysis of AtARR12, OsRR22, and OsRR29 in the Arabidopsis protoplast transient expression system. (A) Transactivation activity of AtARR12, OsRR22, and OsRR29 on a cytokinin primary response gene, AtARR6. Arabidopsis mesophyll protoplasts were isolated from mature leaves of the wild-type plants, and cotransfected with the ARR6-LUC reporter and an effector plasmid expressing Myc-tagged AtARR12, OsRR22, or OsRR29 protein.17 Transfected protoplasts were treated with (+CK) or without (-CK) 100 nM t-zeatin for 3h. The UBQ-GUS construct was used as an internal control to normalize the variations of each transfection. (B) Proteins levels of AtARR12, OsRR22, and OsRR29 based on immunoblot analysis using the Myc epitope. Total proteins are extracted from transfected protoplasts according to the method by Kim et al.25 α-Tubulin was detected as a loading control.
The differing abilities of OsRR22 and OsRR29 to stimulate expression of a known cytokinin primary response gene provides a mechanistic explanation for their differing abilities to functionally complement the arr1–3;arr12–1 mutant. Subfamily-I type-B RRs, such as AtARR12 and OsRR29, share substantial sequence homology within their Myb-like DNA binding domains and are potentially capable of targeting a similar cohort of genes for regulation.15 In contrast, OsRR29 differs from the subfamily-I type-B RRs in its Myb-like DNA-binding motif, suggesting that it may target divergent promoter elements for regulation. In addition, based on sequence divergence in other parts of the type-B RRs, differences may exist in their response to upstream signaling elements, in their transciptional activation domains, and/or in their interaction with other proteins, all of which could affect their degree of functional overlap. Nevertheless, within the developmental context of seed growth, OsRR29 was able to rescue a subfamily-I mutant phenotype. This suggests that a shared target set of genes exist relevant to seed growth, which can be regulated by both OsRR22 and OsRR29. Alternatively, seed-specific factors may modulate OsRR29 so that it is able to recognize a broader range of promoter elements, thereby increasing its functional overlap with OsRR22.
Completion of the rice genome sequence has facilitated the comprehensive identification of genes involved in cytokinin signaling and metabolism, but functional analysis of these genes is still in its infancy. Of particular interest is the finding that OsCKX2, a cytokinin oxidase/dehydrogenase, was identified as a quantitative trait locus (QTL) responsible for increased grain productivity in rice.19 Reduced expression of OsCKX2 in an indica variety of rice results in cytokinin accumulation in the inflorescence meristems and a consequent increase in the number of reproductive organs, thereby enhancing the grain yield. Consistent with the ability of OsRR29 to rescue the seed size phenotype of the Arabidopsis arr1–3;arr12–1 double mutant, cytokinins may have other critical roles in rice grain development. Our results suggest that a large set of conserved orthologs of type-B response regulators in rice and Arabidopsis play general but essential roles in cytokinin signaling in multiple developmental contexts. On the other hand, lineage-specific type-B response regulators might have evolved for new biological functions (e.g., the role of OsRR30/Ehd1 in short-day promotion of flowering20), response to diverse environmental pressures (e.g., pathogen responses21), and organizational complexity (e.g., panicle architecture19). The in vivo functions of lineage-specific type-B response regulators remain to be fully elucidated.
Acknowledgments
This work was supported by the United States Department of Agriculture (grant no. 2011–67013–30069 to JJK and no. 2007–35304–18323 to GES), and by the Human Frontier Science Program (LT000757/2009-L to HJK)
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21293
References
- 1.Mok DW, Mok MC. Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- 2.Mok DWS, Martin RC. Cytokinin metabolic enzymes. In: Mok DWS and Mok MC, eds. in Cytokinins: Chemistry, Activity and Function. Boca Raton, FL: CRC Press, 1994:129-37. [Google Scholar]
- 3.Werner T, Motyka V, Strnad M, Schmülling T. Regulation of plant growth by cytokinin. Proc Natl Acad Sci U S A. 2001;98:10487–92. doi: 10.1073/pnas.171304098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perilli S, Moubayidin L, Sabatini S. The molecular basis of cytokinin function. Curr Opin Plant Biol. 2010;13:21–6. doi: 10.1016/j.pbi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 6.To JP, Kieber JJ. Cytokinin signaling: two-components and more. Trends Plant Sci. 2008;13:85–92. doi: 10.1016/j.tplants.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Goulian M. Two-component signaling circuit structure and properties. Curr Opin Microbiol. 2010;13:184–9. doi: 10.1016/j.mib.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–18. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Argyros RD, Mathews DE, Chiang Y-H, Palmer CM, Thibault DM, Etheridge N, et al. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 2008;20:2102–16. doi: 10.1105/tpc.108.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishida K, Yamashino T, Yokoyama A, Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:47–57. doi: 10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- 11.Pareek A, Singh A, Kumar M, Kushwaha HR, Lynn AM, Singla-Pareek SL. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006;142:380–97. doi: 10.1104/pp.106.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du L, Jiao F, Chu J, Jin G, Chen M, Wu P. The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics. 2007;89:697–707. doi: 10.1016/j.ygeno.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 2007;48:523–39. doi: 10.1093/pcp/pcm022. [DOI] [PubMed] [Google Scholar]
- 14.Cheng X, Jiang H, Zhang J, Qian Y, Zhu S, Cheng B. Overexpression of type-A rice response regulators, OsRR3 and OsRR5, results in lower sensitivity to cytokinins. Genet Mol Res. 2010;9:348–59. doi: 10.4238/vol9-1gmr739. [DOI] [PubMed] [Google Scholar]
- 15.Tsai YC, Weir NR, Hill K, Zhang W, Kim HJ, Shiu SH, et al. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012;158:1666–84. doi: 10.1104/pp.111.192765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–59. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–9. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 18.Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, et al. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:814–9. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–5. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 20.Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–36. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell. 2010;19:284–95. doi: 10.1016/j.devcel.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465-9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP, Kurata N, et al. Nomenclature for two-component signaling elements of rice. Plant Physiol. 2007;143:555–7. doi: 10.1104/pp.106.093666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaller GE, Kieber JJ, Shiu S-H. Two-Component Signaling Elements and Histidyl-Aspartyl Phosphorelays. In: Somerville C and Meyerowitz EM, eds. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists. 2008:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, et al. Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature. 2008;455:1134–7. doi: 10.1038/nature07289. [DOI] [PubMed] [Google Scholar]