Abstract
The DEAD-box RNA helicase family comprise enzymes that participate in every aspect of RNA metabolism, associated with a diverse range of cellular functions including response to abiotic stress. In the present study, we report on the identification of a new DEAD-box helicase ATP-binding protein (OsABP) from rice which is upregulated in response e to multiple abiotic stress treatments including NaCl, dehydration, ABA, blue and red light. It possesses an ORF of 2772 nt, encoding a protein of 923 aa, which contains the DEAD and helicase C-terminal domains, along with the nine conserved motifs specific to DEAD-box helicases. The in silico putative interaction with other proteins showed that OsABP interacts with proteins involved in RNA metabolism, signal transduction or stress response. These results imply that OsABP might perform important functions in the cellular response to specific abiotic stress.
Keywords: ABP, abiotic stress, expression profiles, helicases, rice
Plants, as sessile organisms, are continuously affected by abiotic stresses such as drought, high salinity, heavy metal, UV light or pollution, which can cause damage to lipids, proteins and DNA, reducing plant genome stability, growth and productivity.1,2 Plant adaptation to high stress habitats involves a combination of phenotypic plasticity and genetic adaptation. All plants perceive and transmit signals in response to different abiotic stresses, but few species are able to survive in high stress environments. Although there are numerous reports on the genetic, molecular and physiological bases of how plants respond to stress, the nature of plant adaptation to stress remains poorly understood.3
Among the major field-grown crops, rice (Oryza sativa L.) is one of the most important food crops in the world, since it feeds more than two billion people. However, rice plants are most sensitive to excess levels of salt, reduced or excess water supply and suboptimal temperature regimes.4 Rice also represents a model cereal system since it has a relatively small genome size as compared with other cereals, a vast germplasm collection, wide array of molecular genetic resources, and an efficient transformation system.5 Within this context, the study of new genes responsive to stress conditions is still required.
The DEAD-box RNA helicase family comprise enzymes that participate in every aspect of RNA metabolism. Although RNA helicases are associated with a diverse range of cellular functions, there have been relatively few reports of RNA helicases involvement in cellular response to abiotic stress. In the present study, we report on the identification of a new DEAD-box helicase ATP-binding protein, hereby entitled OsABP, responsive to abiotic stress, in rice plants.
The DEAD-box RNA helicase family has been defined by Linder et al.6 and named according to the highly conserved Asp-Glu-Ala-Asp (DEAD) residues present in motif II. RNA helicases are highly conserved enzymes that use ATP to bind or remodel RNA or ribonucleoprotein complexes (RNPs). They represent one of the largest protein classes in RNA metabolism and are found in all kingdoms of life.7 Despite their shared biochemical function (e.g., RNA unwinding) the DEAD-box helicases are involved in different molecular mechanisms such as RNA splicing, ribosome assembly, transcription initiation or nuclear export.8 They are also important cellular factors for regulatory events, in particular during organ maturation and cellular growth and differentiation.9 It has been suggested that RNA helicases can regulate gene silencing at nearly every level of the RNA interference (RNAi) pathways. For example, the Xeroderma pigmentosum group B (XPB) and group D (XPD) are DEAH box helicases which play specific and distinct roles in nucleotide excision repair (NER) pathway.10 Helicases are involved in almost every aspect of DNA and RNA metabolisms, which makes them very important molecules and thereby have several implications of general interest. The RNA helicases are divided mainly into two superfamilies: SF1 and SF2. The majority of RNA helicases belong to the superfamily 2 (SF2) and they are characterized by sequence homology within the helicase domain consisting of eight or nine conserved amino acid motifs.11
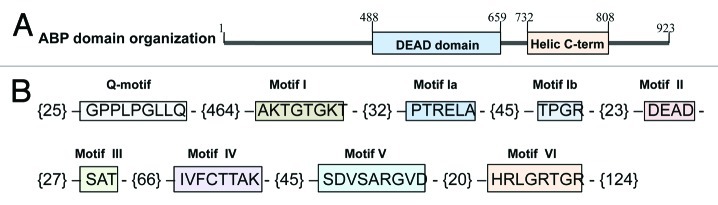
The genomic sequence of the OsABP helicase (LOC_Os06 g33520) was obtained from the Rice Genome Annotation Project funded by NSF (http://rice.plantbiology.msu.edu/). It contains an open reading frame (ORF) of 2772 nt, encoding for a protein of 923 aa. The present protein has a theoretical pI of the 8.7 and the molecular weight of 101.1 kDa. The protein domain search, performed in the NCBI Conserved Domain Database (NCBI-CDD; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) revealed the presence of the DEAD and helicase C-terminal domains (Fig. 1A). The DEAD domain contains several ATP-binding sites and it is involved in ATP-dependent RNA unwinding. The helicase superfamily C-terminal domain, associated with DEXD-, DEAD- and DEAH-box proteins, is found in a wide variety of helicases and helicase related proteins and it is characterized by the presence of a P-loop containing nucleoside triphosphate hydrolases. Members of the P-loop NTPase domain superfamily are defined by the conserved nucleotide phosphate-binding motif which binds the β-γ phosphate moiety of the bound nucleotides and the Mg2+ cation. These domains play functional roles in the regulation of protein activity and also confer structural and functional advantages.12 As all DEAD-box helicases, the OsABP protein is characterized by the presence of the nine conserved motifs: Q-motif (GPPLPGLLQ; aa 26–35), motif I (AKTGTGKT; aa 500–508), motif Ia (PTRELA; aa 541–547), motif Ib (TPGR; aa 593–597), motif II (DEAD; aa 611–615), motif III (SAT; aa 642–645), motif IV (IVFCTTAK; aa 711–719), motif V (SDVSARGVD; aa 754–763) and motif VI (HRLGRTGR; aa 801–809) (Fig. 1B). The Q motif is part of a cap-like structure on the classical RecA-like domain 1. It is essential for efficient binding of ssRNA as well as for the conformational changes that are driven by nucleotide binding and ATP hydrolysis. It was proposed that the Q motif could act as a sensor for the state of the bound nucleotide through the interactions with motif I and is important for both ATP and RNA binding.13 The hydrolysis of ATP through the A and B Walker motifs is necessary for the helicase nucleic acid unwinding activity. The motifs I and II are characteristic of NTP hydrolyzing proteins, while motif III is involved in coupling of ATP hydrolysis and unwinding. The motifs IV and V are involved in RNA binding, while motif VI is essential for the nucleic acid-dependent NTP hydrolysis.14
Figure 1. Schematic representation of the OsABP (LOC_Os06 g33520) domain organization (A) and the presence of the nine conserved motifs in the DEAD-box helicase family (B). The numbers within brackets represent the amino acids present between the motifs.
The comparison of amino acid sequences, performed by using the UniProt BlastP Service (www.uniprot.org/blast/), showed a 93 and 58% similarities with RNA helicase 31 from rice and Arabidopsis thaliana (Table 1). Other significant hits revealed a 62% and 64% sequence similarity with the RNA helicase 25, and 63% and 51% similarity with RNA helicase 26 form rice and A. thaliana. The RNA helicases 25, 26 and 31 are members of the DEAD-box family, and they possess ATP-dependent helicase activity and RNA-binding property.12 Although a large number of DEAD box proteins have been bioinformatically identified as putative predicted helicases, only for few of them the ATP-dependent RNA helicase activity has been demonstrated. The helicase activity of the OsABP has not been yet demonstrated.
Table 1. Percentage of similarity between OsABP and related RNA helicases form Arabidopsis thaliana and Oryza sativa.
| UniProt Acc. No. | Protein name | Organism | Length (aa) | Identity % |
|---|---|---|---|---|
|
Q0DBU5 |
DEAD-box ATP-dependent RNA helicase 31 |
Oryza sativa subsp japonica |
574 |
93 |
|
Q9FNM7 |
DEAD-box ATP-dependent RNA helicase 26 |
Arabidopsis thaliana |
850 |
51 |
|
Q0JL73 |
DEAD-box ATP-dependent RNA helicase 26 |
Oryza sativa subsp japonica |
536 |
63 |
|
B9SBJ5 |
Dead box ATP-dependent RNA helicase |
Ricinus communis |
751 |
59 |
|
Q9FFQ1 |
DEAD-box ATP-dependent RNA helicase 31 |
Arabidopsis thaliana |
761 |
58 |
|
Q94C75 |
DEAD-box ATP-dependent RNA helicase 25 |
Arabidopsis thaliana |
563 |
62 |
| Q5ZBH5 | DEAD-box ATP-dependent RNA helicase 25 | Oryza sativa subsp japonica | 594 | 64 |
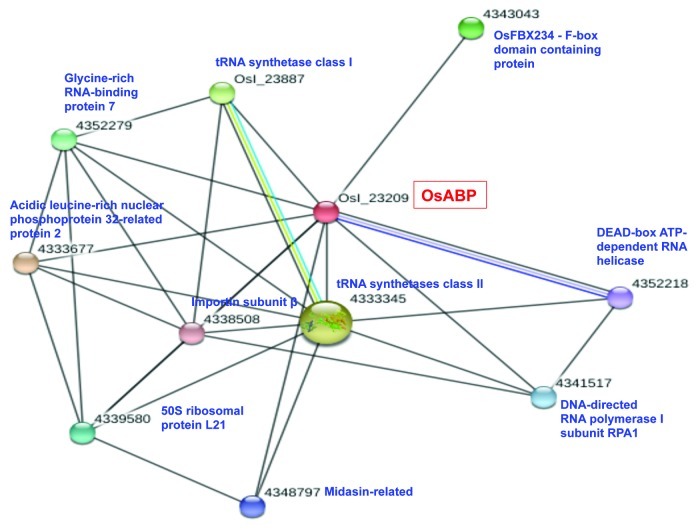
RNA helicases constantly interact with other proteins since they work as larger multi-component complexes. An increasing number of studies have focused on the identification of interacting proteins and the elucidation of their effects on RNA helicase activities.15 In the present study, the STRING computer service (http://string-db.org/) was used to determine the predicted protein-protein interaction for OsABP helicase. The results are graphically represented in Figure 2. The OsABP protein was predicted to interact with ten different proteins: a DEAD-box ATP-dependent RNA helicase, two tRNA synthetases form class I and II, the OsFBX234 which is an F-box domain containing protein, the RPA1 subunit of RNA polymerase I, the β subunit of importin, a midasin-related protein, the chloroplast precursor of the 50S ribosomal protein L21, an acidic leucine-rich nuclear phosphoprotein and the glycine-rich RNA-binding protein 7(GRRBP). The tRNA synthetases are enzymes that catalyze the esterification of a specific amino acid or its precursor to its compatible tRNAs to form aminoacyl-tRNA. The synthetase first binds ATP and the corresponding amino acid or its precursor to form an aminoacyl-adenylate and release inorganicpyrophosphate (PPi). The adenylate-aaRS complex then binds the appropriate tRNA molecule, and the amino acid is transferred from the aa-AMP to either the 2'- or the 3′-OH of the last tRNA nucleotide at the 3′-end. Some synthetases also mediate a proofreading reaction to ensure high fidelity of tRNA charging.16 The F-box proteins are characterized by the presence of the conserved F-box motif of approximately 40 amino acids and they represent one of the largest protein families, with about 700 members in Arabidopsis and rice. In plants, only a small portion of F-box proteins have been studied and they have been shown to play important roles in the regulation of various developmental processes and stress responses by integrating almost all phytohormone signaling pathways.17 In rice, two F-box proteins have been characterized: the gibberellin-insensitive dwarf2, which acts as a positive regulator of gibberellic acid signaling, and dwarf3 which controls tiller bud activity.18 It has been reported that some F-box containing genes are induced by abiotic stress and represents potential targets for microRNAs.19 Among the OsABP putative interacting partners, RNA polymerase I (Pol I) is an essential enzyme involved in transcription regulation and is the key convergence point that collects and integrates a vast array of information from cellular signaling cascades to regulate ribosome production that guides cell growth and proliferation. The DNA-dependent RNA polymerases catalyze the transcription of DNA into RNA using ribonucleoside triphosphates as substrates. The largest catalytic core component of RNA polymerase I, which synthesizes rRNA precursors, is the RPA1 subunit. It forms the polymerase active center together with the second largest subunit RPA2, and it promotes the translocation of Pol I by acting as a ratchet that moves the RNA-DNA hybrid through the active site.20 The importin is a specific protein involved in nuclear import-export. It transports other protein molecules into the nucleus by binding to their nuclear localization signals (NLS). It has two subunits denominated α and β. Members of the importin β family can bind and transport molecules in the nucleus, or can form heterodimers with importin α. As part of a heterodimer, importin β mediates interactions with the pore complex, while importin α acts as an adaptor protein to bind to NLS.21 Another putative interacting partner of OsABP, a member of the midasin-related proteins, is basically involved in the export of 60S ribosome subunits from the nucleus.22 As for the chloroplast ribosomal protein L21, recent studies showed that it is essentially required for chloroplast development and embryogenesis in plants.23 Members of the acidic leucine-rich nuclear phosphoprotein 32 family are implicated in intracellular signal transduction, transcription regulation, nucleocytoplasmic transport and mRNA metabolic processes.24 Finally, GRRBPs have been reported to be regulated by a number of external stimuli including cold, water stress, high salinity, wounding, and viral infection, demonstrating that these proteins might be involved in the responses of plants to changing environmental conditions.25 Recently, the pea GRRBP has been reported to be one of the interacting partner of pea Gβ subunit of heterotrimeric G-protein which is involved in stress signaling.26 Besides this, OsABP also interacts with a putative DEAD-box RNA helicase, belonging to the DDX helicase family with important roles in ribosome biogenesis.27 So, the bioinformatic predictions showed that OsABP protein interacts with several essential proteins involved in a wide variety of processes, especially RNA or DNA metabolism, nuclear transport, signal transduction and stress response. The experimental validation of these interactions is currently in progress.
Figure 2. Schematic representation of the putative interaction of OsABP with other proteins. STRING program (http://string-db.org/) was used for the bioinformatic prediction of protein-protein interaction.
All cells experience a range of stress conditions, either abiotic or biotic, that tend to decrease cellular fitness. RNA helicase activity may be required for cellular adaptation to the altered environmental conditions. Several reports have recently indicated that RNA helicase expression could be regulated in response to changes in specific environmental conditions, such as temperature, light, oxygen or osmolarity.28 Environmental regulation of RNA helicase gene expression is controlled through sensing and response pathways that are activated by stress. In consequence, RNA helicases play key roles in the regulation of gene expression. Their own expression can be environmentally regulated resulting in the induction of RNA helicase biochemical activity which provides the ability to regulate either the expression or the activity of downstream target mRNAs or functional RNAs.29
In the present study, the expression profiles of OsABP gene, performed by using the qRT-PCR technique, were analyzed in rice plants grown in greenhouse under normal conditions and submitted to a variety of stress conditions. Total RNA was extracted from rice cultivar IR64 wild type and stress challenged plants. For abiotic stress treatments, two weeks-old rice seedlings were transferred to salt solutions (200 mM NaCl) or abscisic acid (100 mM ABA) and kept at room temperature for 12 h. For cold and heat treatment, the plantlets were kept in incubators at 4°C and 42°C, respectively. Seedlings were kept on blotting paper for 12 h to mimic drought conditions. The light stress was induced by using red light (680 nm and 100 µM/m2/s2 intensity), blue light (430 nm and 30 µM/m2/s2 intensity) and white light. The pH stress was evaluated after exposure to simple solutions with low (pH 3.5) and high (pH 10.5) pH for 12 h. Plantlets grown at room temperature were taken as a control. The resulted data are graphically represented in Figure 3. The exposure to 200 mM NaCl resulted into 12.8-fold increase in the OsABP transcript level, while dehydration induced a 42-fold upregulation of the gene transcript. ABA treatments resulted into a 10-fold accumulation in the OsABP mRNA level (Fig. 3A). In the case of the light stress, OsABP gene was significantly upregulated in response to blue (37-fold compared with control) and red (22-fold compared with control) light, while downregulation (5.5-fold compared with control) was observed when the white light was used. Downregulation of OsABP transcript was evident also in response to cold and heat stresses. A 0.5-fold decrease in the amount of OsABP was showed under cold treatment, while the heat stress resulted into a 6.2-fold decrease in the transcript level (Fig. 3B). On the other hand, when low and high pH values were used, no significant differences between control and treatments were observed (Fig. 3A).

Figure 3. qRT-PCR analysis showing the expression levels of OsABP in two weeks-old rice plants grown in greenhouse and challenged with different abiotic stresses; (A) 200 mM NaCl, dehydration, 100 µM ABA, blue and red light, pH 3.5 and 10.5; (B) cold (4°C), heat (42°C) and while light; NT, non-treated. The relative expression is presented as fold change to control. Values are expressed as means ± SD of three independent replicated plants.
In plants, other DEAD-box-related helicases were proven to be responsive to different stress conditions. For example, the PDH47 (Pea DNA Helicase 47) and PDH45 were demonstrated to be induced by salinity, cold, heat and ABA treatments.30-33 PDH47 expression is differentially express in a tissue specific manner with induction by cold and salinity stress in shoots and roots and heat and ABA treatment in roots.29 PDH45 is a unique member of the RNA helicases DEAD-box family which transcript accumulates in response to general osmotic stress caused by salinity or desiccation.32,33 The single subunit of minichromosome maintenance 6 (MCM6) gene from pea was also recently reported to function as helicase and the overexpression of MCM6 promotes salinity stress tolerance without affecting yield.34-37 The Arabidopsis LOS4 RNA helicase was recently showed to be a temperature-regulated gene linked with other developmental processes such as flowering or vernalization.38,39 The study los4-1 and los4-2/ cryophyte mutants reveal that LOS4 is an early regulator of CBF transcription factor expression in response to plant chilling.38,39 The inactivation of LOS4 also affected the response to plant stress hormones. Germination studies indicated that LOS4 is required for the formation of a germination inhibitor, suggesting that helicase activity is necessary for the formation of a protein that inhibits the signal transduction pathways involved in germination.39 Genome-wide transcript analysis has identified other RNA helicases in Arabidopsis whose expression patterns are stress regulated. For example, the drh1 gene was showed to be moderately induced by cold stress.40 Similar analysis indicated that two putative Arabidopsis RNA helicase genes (At5g08610 and At1g59990) were downregulated in response to cold, salt and osmotic stress.28 In sorghum, a salt-responsive transcript HVD1 (Hordeum vulgare DEAD-box protein), encoding a putative ATP-dependent DEAD-box RNA helicase, was found to be induced under salt and cold stress.41 Liu et al. (2008)42 reported on a novel type of salt-responsive gene (AvDH1) from the halophyte dogbane plant that encodes a protein with high homology to DEAD-box helicases and is expressed in a salt-dependent manner. The AvDH1 gene was also strongly upregulated by low temperature, while no changes were observed in response to PEG-induced stress or ABA treatments, suggesting that AvDH1 follows in ABA-independent salt stress signaling networks. ABA plays crucial roles in growth regulation, seed dormancy and germination, stomatal movement, vegetative growth, and responses to biotic and abiotic stress. A recent study by He et al. (2012)43 reported on the identification of a DEXH box RNA helicase (ABO6), localized in mitochondria and required for gene splicing. The authors showed that in abo6 mutants, ROS accumulated more in mitochondria and the ABA signaling was impaired. Similar studies underline the importance of RNA helicases in chloroplasts. For example, in Arabidopsis plants it was demonstrated that RH3 helicase is directly involved in chloroplast intron splicing and in 50S ribosome biogenesis, while a knockdown of RH22 expression resulted in defective accumulation of chloroplast rRNA.44,45
In conclusion, the present results report on the identification of a new DEAD-box ATP-binding helicase responsive to abiotic stress conditions. The in silico analysis of putative interaction with other proteins showed that the OsABP interacts with essential proteins involved in tRNA and mRNA synthesis, transcription regulation, nuclear transport, phytohormone signaling pathways, signal transduction pathways or stress responsive proteins. The gene transcript is upregulated in response to salt, dehydration, ABA, blue and red light and downregulated under white light, cold and heat treatments. These results imply that OsABP might perform crucial functions directly involved in cellular response to specific abiotic stress, suggesting that it is a component of a general stress response mechanism. Nevertheless, further characterization of the protein functions is still needed and currently in progress. Although a relatively small percentage of the RNA helicase-related sequences in public databases have been identified as being stress-regulated, it is expected that abiotic-stress induced RNA helicase expression will be observed more frequently in the future as this scenario has the potential to open new perspectives related to the response to stress conditions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Short-Term Fellowship (Ref. No. F/ROM11–02) Program awarded through the International Center of Genetic Engineering and Biotechnology (ICGEB). Work on plant helicases, plant stress signaling and rice transformation in NT’s laboratory is partially supported by Department of Biotechnology (DBT), and Department of Science and Technology (DST), Government of India. We thank Dr. Shahinul Islam for his help in RNA isolation.
Glossary
Abbreviations:
- ABA
abcisic acid
- ABP
ATP-Binding Protein
- ATP
adenosine triphosphate
- DEAD
Aspartate-Glutamate-Alanine-Aspartate
- NaCl
sodium chloride
- Pol I
RNA polymerase I
- qRT-PCR
quantitative Real-Time Polymerase Chain Reaction
- SF1
helicase superfamily 1
- SF2
helicase superfamily 2
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21343
References
- 1.Schützendübel A, Polle A. Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J Exp Bot. 2002;53:1351–65. doi: 10.1093/jexbot/53.372.1351. [DOI] [PubMed] [Google Scholar]
- 2.Tuteja N, Ahmad P, Panda BB, Tuteja R. Genotoxic stress in plants: shedding light on DNA damage, repair and DNA repair helicases. Mutat Res. 2009;681:134–49. doi: 10.1016/j.mrrev.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Redman RS, Kim YO, Woodward CJDA, Greer C, Espino L, Doty SL, et al. Increased fitness of rice plants to abiotic stress via habitat adapted symbiosis: a strategy for mitigating impacts of climate change. PLoS One. 2011;6:e14823. doi: 10.1371/journal.pone.0014823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grover A, Minhas D. Towards the production of abiotic stress tolerant transgenic rice plants: issues, progress and future research needs. Proc Indian Nat Sci Acad. 2000;1:13–32. [Google Scholar]
- 5.Paterson AH, Freeling M, Sasaki T. Grains of knowledge: genomics of model cereals. Genome Res. 2005;15:1643–50. doi: 10.1101/gr.3725905. [DOI] [PubMed] [Google Scholar]
- 6.Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, et al. Birth of the D-E-A-D box. Nature. 1989;337:121–2. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 7.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–62. doi: 10.1016/S1097-2765(01)00329-X. [DOI] [PubMed] [Google Scholar]
- 8.Tuteja N, Tuteja R. Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery. Eur J Biochem. 2004;271:1835–48. doi: 10.1111/j.1432-1033.2004.04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuteja N, Tuteja R. Unraveling DNA helicases: motif, structure, mechanism and functions. Physica A. 2006;372:70–83. doi: 10.1016/j.physa.2006.05.014. [DOI] [Google Scholar]
- 10.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/S1097-2765(00)80177-X. [DOI] [PubMed] [Google Scholar]
- 11.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Aubourg S, Kreis M, Lecharny A. The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 1999;27:628–36. doi: 10.1093/nar/27.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanner NK. The newly identified Q motif of DEAD box helicases is involved in adenine recognition. Cell Cycle. 2003;2:18–9. doi: 10.4161/cc.2.1.296. [DOI] [PubMed] [Google Scholar]
- 14.Umate P, Tuteja R, Tuteja N. Architectures of the unique domains associated with the DEAD-box helicase motif. Cell Cycle. 2010;9:4228–35. doi: 10.4161/cc.9.20.13635. [DOI] [PubMed] [Google Scholar]
- 15.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 17.Zhan Y, Xu Y, Li Z, Deng X, Wu W, Xue Y. F-box protein DOR functions as a novel inhibitory factor for ABA-induced stomatal closure under drought stress in Arabidopsis thaliana. Plant Physiol. 2008;148:2121–33. doi: 10.1104/pp.108.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan YS, Chen XY, Yang K, Sun ZX, Fu YP, Zhan YM, et al. Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol Plant. 2010;8:1–8. doi: 10.1093/mp/ssq066. [DOI] [PubMed] [Google Scholar]
- 19.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, et al. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–72. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 21.Kose S, Imamoto N, Tachibana T, Yoshida M, Yoneda Y. β-subunit of nuclear pore-targeting complex (importin-β) can be exported from the nucleus in a Ran-independent manner. J Biol Chem. 1999;274:3946–52. doi: 10.1074/jbc.274.7.3946. [DOI] [PubMed] [Google Scholar]
- 22.Garbarino JE, Gibbons IR. Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics. 2002;3:18–24. doi: 10.1186/1471-2164-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin T, Pan G, Liu H, Wu J, Li Y, Zhao Z, et al. The chloroplast ribosomal protein L21 gene is essential for plastid development and embryogenesis in Arabidopsis. Planta. 2012;235:907–21. doi: 10.1007/s00425-011-1547-0. [DOI] [PubMed] [Google Scholar]
- 24.Matilla A, Radrizzani M. The Anp32 family of proteins containing leucine-rich repeats. Cerebellum. 2005;4:7–18. doi: 10.1080/14734220410019020. [DOI] [PubMed] [Google Scholar]
- 25.Kim JY, Kim WY, Kwak KJ, Oh SH, Han YS, Kang H. Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J Exp Bot. 2010;61:2317–25. doi: 10.1093/jxb/erq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhardwaj D, Lakhanpaul S, Tuteja N. Wide range of interacting partners of pea Gβ subunit of G-proteins suggests its multiple functions in cell signalling. Plant Physiol Biochem. 2012;58C:1–5. doi: 10.1016/j.plaphy.2012.06.005. [DOI] [PubMed] [Google Scholar]; DOI : .
- 27.Abdhaleem M, Maltais L, Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2008;81:618–22. doi: 10.1016/S0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 28.Owttrim GW. RNA helicases and abiotic stress. Nucleic Acids Res. 2006;34:3220–30. doi: 10.1093/nar/gkl408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–41. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vashisht AA, Pradhan A, Tuteja R, Tuteja N. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 2005;44:76–87. doi: 10.1111/j.1365-313X.2005.02511.x. [DOI] [PubMed] [Google Scholar]
- 31.Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci U S A. 2005;102:509–14. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amin M, Elias SM, Hossain A, Ferdousi A, Rahman MS, Tuteja N, et al. Over-expression of a Dead-box helicase, PDH45, confers both seedling and reproductive stage salinity tolerance to rice (Oryza sativa L.) Mol Breed. 2012;30:345–54. doi: 10.1007/s11032-011-9625-3. [DOI] [Google Scholar]
- 33.Sahoo RK, Gill SS, Tuteja N. Pea DNA helicase 45 promotes salinity stress tolerance in IR64 rice with improved yield. Plant Signal Behav. 2012;7 doi: 10.4161/psb.20915. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran NQ, Dang HQ, Tuteja R, Tuteja N. A single subunit MCM6 from pea forms homohexamer and functions as DNA helicase. Plant Mol Biol. 2010;74:327–36. doi: 10.1007/s11103-010-9675-7. [DOI] [PubMed] [Google Scholar]
- 35.Dang HQ, Tran NQ, Gill SS, Tuteja R, Tuteja N. A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol Biol. 2011;76:19–34. doi: 10.1007/s11103-011-9758-0. [DOI] [PubMed] [Google Scholar]
- 36.Tuteja N, Tran NQ, Dang HQ, Tuteja R. Plant MCM proteins: role in DNA replication and beyond. Plant Mol Biol. 2011;77:537–45. doi: 10.1007/s11103-011-9836-3. [DOI] [PubMed] [Google Scholar]
- 37.Vashisht AA, Tuteja N. Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobiol B. 2006;84:150–60. doi: 10.1016/j.jphotobiol.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci U S A. 2002;99:11507–12. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, et al. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005;17:256–67. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, et al. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura T, Muramoto Y, Yokota S, Ueda A, Takabe T. Structural and transcriptional characterization of a salt-responsive gene encoding putative ATP-dependent RNA helicase in barley. Plant Sci. 2004;168:63–70. doi: 10.1016/j.plantsci.2004.03.001. [DOI] [Google Scholar]
- 42.Liu HH, Liu J, Fan SL, Song MZ, Han XL, Liu F, et al. Molecular cloning and characterization of a salinity stress-induced gene encoding DEAD-box helicase from the halophyte Apocynum venetum. J Exp Bot. 2008;59:633–44. doi: 10.1093/jxb/erm355. [DOI] [PubMed] [Google Scholar]
- 43.He J, Duan Y, Hua D, Fan G, Wang L, Liu Y, et al. DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell. 2012;24:2–19. doi: 10.1105/tpc.112.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asakura Y, Galarneau E, Watkins KP, Barkan A, van Wijk KJ. Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 2012;159:961–74. doi: 10.1104/pp.112.197525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chi W, He B, Mao J, Li Q, Ma J, Ji D, et al. The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts. Plant Physiol. 2012;158:693–707. doi: 10.1104/pp.111.186775. [DOI] [PMC free article] [PubMed] [Google Scholar]