Abstract
Previously, we showed that OsGLU3, a β-1,4-endoglucanase, can affect the cellulose synthesis for root elongation in rice. And the phosphate starvation induced root elongation in rice depends on the function of OsGLU3. Here, we further showed that OsGLU3 is also dispensable for nitrogen starvation induced root elongation in rice.
Keywords: cellulose, nitrogen starvation, root elongation
Root is an important plant organ for anchoring the plant and acquiring minerals and water from the soil. Roots move through the rhizosphere by elongating.1 Root elongation is determined by the proliferation and elongation of cells, primarily within the elongation zone.2 Different from the animal cells, the plant cells were covered with a lay of cell wall, which can support the shape of the cell. Cellulose is a major component of the cell wall.3 Therefore, the synthesis and remodeling of cellulose plays an important role in plant development and also the root elongation.
It is well known that rice can alter its root architecture to adapt to environmental stresses, such as phosphate starvation. To check whether nitrogen starvation also affects cellulose content to modulate rice root architecture in rice, we observed rice root architecture under the stress and measured the cellulose content.
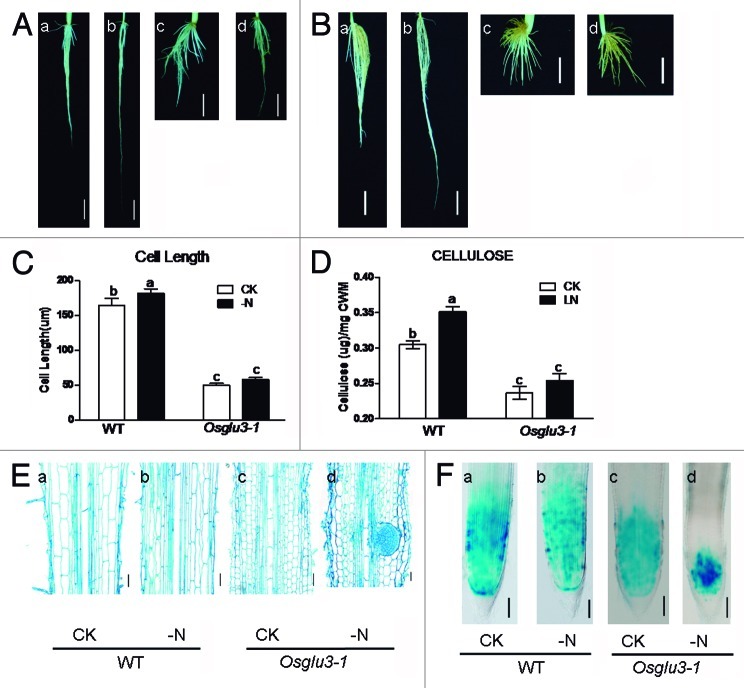
The Wild type (WT, SSBM) were grown for 10 d in media without nitrogen and transferred to low nitrogen media (2 mg/L nitrogen) or control media (40 mg/L nitrogen) for another 20 d respectively. The nitrogen starvation stress leads to an around 20% increase of primary root elongation in WT as compared with that grown under the control condition, Figure 1A). To determine whether the root elongation under nitrogen starvation is due to the induction of cell elongation or enhanced mitotic activity, longitudinal sections of the primary root maturation zone and the expression patterns of OsCYCB1;1:GUS under the treatment were analyzed. It showed that nitrogen starvation can lead to an increase of approximately 13% in rice root cell elongation (Fig. 1C and E). Also, the starvation activates root mitotic activity as visualized by a broader GUS staining in the root tip (Fig. 1F). Together, this indicates that similar to phosphate starvation, the nitrogen starvation also stimulates primary root elongation by inducing root cell elongation and activating root mitotic activity.
Figure 1. Nitrogen starvation induces root elongation in an OsGLU3 dependant way in rice. (A) The phenotype of WT (SSBM) and Osglu3–1 under nitrogen-starvation. a, WT under control condition; b, WT under nitrogen starvation; c, Osglu3–1 under control condition; d, Osglu3–1 under nitrogen starvation. Bar = 2cm. (B) The phenotype of WT (Dongjin) and the T-DNA insertion mutant Osglu3–2 under nitrogen starvation. a, WT under control condition; b, WT under nitrogen starvation; c, Osglu3–2 under control condition; d, Osglu3–2 under nitrogen starvation. a-b, Bar = 2cm; c-d, Bar = 1cm. (C) Nitrogen-starvation could not induce root cell elongation in Osglu3–1. (D) The root crystalline cellulose (TFA-insoluble) content of Osglu3–1 was not obviously changed under nitrogen starvation. Data are mean values from three independent experiments, each performed in triplicate. Error bars represent standard error (SE). Different letters are used to indicate means that differ significantly (p < 0.05). CWM: cell wall material. (E) Nitrogen starvation could not induce the cell elongation in Osglu3–1.Bar = 50µm. (F) Restricted OsCYCB1;1:GUS expression in Osglu3–1 under nitrogen starvation. Bar = 100µm
Furthermore, we also found that the nitrogen starvation can lead to an approximately 15% increase in root cellulose content (Fig. 1D). This indicates that the nitrogen starvation, can also affect cellulose synthesis to modulate root architecture. To test this hypothesis, we checked the root architecture and cellulose content of the Osglu3–1 mutant under nitrogen starvation stress. It showed that nitrogen starvation could not obviously induce root and root cell elongation in Osglu3–1 (Fig. 1A, C and E). Under the stress, the defect of root cell division of Osglu3–1 was more severe as visualized by a decrease of OsCYCB1;1:GUS expression in Osglu3–1 (Fig. 1F). Also the root cellulose content of Osglu3–1 was not obviously increased under the stress (Fig. 1D). Consistent with these observations, the nitrogen starvation induced root elongation is also abolished in the OsGLU3 loss function mutant, Osglu3–2 (Fig. 1B). Therefore, it suggests that nitrogen starvation induced primary root elongation depends on the activity of OsGLU3.
Altogether, these data with our previous work indicate that both the phosphate and nitrogen starvation can affect root cell wall cellulose synthesis to modulate root architecture in an OsGLU3 dependant way in rice. It was well known that under the phosphate or nitrogen starvation, plants showed an increased root: shoot biomass ratio. These responses might be resulted from the alteration of carbon partitioning to favor root growth, therefore improve the ability to acquire the mineral elements.4 Since, the exogenous application of glucose can elevate the cellulose content in rice root,5 we proposed that both the nitrogen and phosphate starvation may induce the carbon partitioning to root tissue, which leads to an induction of cellulose synthesis and favors the root elongation in rice. Furthermore, OsGLU3 is a key player in this system, as loss of function of it can abolish the response. However, how the plants sensing the external starvation signals to alter the carbon partitioning and how the plants integrate the internal carbon signal and the external starvation signals to modulate the root mitotic activity are still need to be addressed in future.
Acknowledgments
This work was supported by Zhejiang Provincial Natural Science Foundation of China (LR12C15001) and National Natural Science Foundation of China (30900098, 30971851).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21334
References
- 1.Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- 2.Beemster GT, Fiorani F, Inzé D. Cell cycle: the key to plant growth control? Trends Plant Sci. 2003;8:154–8. doi: 10.1016/S1360-1385(03)00046-3. [DOI] [PubMed] [Google Scholar]
- 3.Sandhu AP, Randhawa GS, Dhugga KS. Plant cell wall matrix polysaccharide biosynthesis. Mol Plant. 2009;2:840–50. doi: 10.1093/mp/ssp056. [DOI] [PubMed] [Google Scholar]
- 4.Hermans C, Hammond JP, White PJ, Verbruggen N. How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci. 2006;11:610–7. doi: 10.1016/j.tplants.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhang JW, Xu L, Wu YR, Chen XA, Liu Y, Zhu SH, et al. OsGLU3, a putative membrane-bound endo-1,4-beta-glucanase, is required for root cell elongation and division in rice (Oryza sativa L.) Mol Plant. 2012;5:176–86. doi: 10.1093/mp/ssr084. [DOI] [PubMed] [Google Scholar]