Abstract
SEPALLATA3 (SEP3) is important in determining flowering time as well as floral organ identity. Although much is known about the regulation of floral organ identity by SEP3, its role as a downstream gene of FLOWERING LOCUS T (FT) for the regulation of ambient temperature-responsive flowering is poorly understood. Here, we show that SEP3 as a downstream gene of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 (SPL3) and FT modulates the flowering time in response to different ambient temperatures. SEP3 overexpression showed temperature-insensitive flowering at 23°C and 16°C. This suggests that altered SEP3 activity affects ambient temperature-responsive flowering. However, a lesion in SEP3 did not obviously affect ambient temperature-responsive flowering. SEP3 expression was affected by altered SPL3 and FT activities in the leaf and shoot apical regions at different temperatures. These results suggest that the miR156-SPL3-FT circuitry directly or indirectly regulates SEP3 expression for the regulation of ambient temperature-responsive flowering in Arabidopsis.
Keywords: SEP3, miR156-SPL3-FT circuitry, ambient temperature, flowering time
Flowering, which is the transition from the vegetative phase to the reproductive phase, is very important for plant reproduction and survival in continuously changing environments.1 This requires the precise perception and processing of various environmental factors and endogenous developmental cues. Thus, an extremely complicated network and a finely-tuned crosstalk control floral transition in plants. Molecular genetic analyses in Arabidopsis thaliana have revealed that flowering time is regulated by five major floral pathways: photoperiod, vernalization, autonomous, gibberellic acid (GA), and thermosensory.2-4 Although some studies reported several components and mechanisms for the regulation of ambient temperature-responsive flowering in plants, the molecular mechanisms underlying the responses of plants to changes in ambient temperature are still limited.
The regulatory modules consisting of microRNAs (miRNAs) and their targets principally affect diverse growth and development in plants.5,6 In Arabidopsis and other plant species, miR156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) regulatory module is a well-known example of such modules. miR156-SPL regulatory modules, which are highly conserved in plant species,7,8 play essential roles in the regulation of a variety of developmental processes.9-15 A variety of genes, such as MADS box and MYB family genes, have been identified as downstream targets of this module. A recent report showed that the miR156-SPL3 module controls ambient temperature-responsive flowering by negatively regulating FLOWERING LOCUS T (FT) expression.16
A set of flowering time genes called floral integrators that include FT17,18 and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1),19,20 are pivotal in the integration of environmental and endogenous signals to induce flowering in Arabidopsis.4,21 FT is a major output for ambient temperature-responsive flowering.22,23 However, weakly temperature-sensitive flowering of ft-10 mutants, an RNA null allele of FT, suggests that other flowering time genes, such as SOC1 and TWIN SISTER OF FT (TSF),24 redundantly function as outputs in the thermosensory pathway. Among the target genes of FT in the leaf,25 we have shown that various loss- and gain-of-function alleles of FRUITFULL (FUL) normally respond to ambient temperature changes,16 suggesting that FUL plays a limited role in the ambient temperature-responsive flowering.
Here, we show that SEPALLATA3 (SEP3), a downstream gene of SPL3 and FT, regulates flowering time in response to different ambient temperatures. Increased SEP3 activity resulted in the ambient temperature-insensitive flowering at 23°C and 16°C. Also, SEP3 expression was affected by altered SPL3 and FT activities. Our results suggest that a model in which the miR156-SPL3-FT circuitry directly or indirectly regulates SEP3 expression in the leaf and shoot apical regions to control ambient temperature-responsive flowering in Arabidopsis.
SEP3 is Involved in the Ambient Temperature-Responsive Flowering
Because overexpression of SEP3 showed an early flowering and curled leaf phenotype, as seen in 35S::SPL3(-) [a miR156-resistant version with the miR156 binding element mutated] and 35S::FT plants25 (Fig. 1), we investigated the phenotype of loss- and gain-of function mutants of SEP3 (sep3–2 and 35S::SEP3 plants, respectively) at 23°C and 16°C under long-day (LD) condition. 35S::SEP3 plants flowered with a similar number of leaves at both temperatures (3.9 and 4.9 leaves at 23°C and 16°C, respectively) (Fig. 1). This indicated that the flowering of 35S::SEP3 plants was almost insensitive to the changes in ambient temperature. This further suggested that altered SEP3 activity can affect the ambient temperature-responsive flowering. Also, the curled leaf phenotype seen in 35S::SEP3 plants grown at 23°C was attenuated at the lower temperature (data not shown). This phenotypic attenuation at 16°C was also found in 35S::SPL3(-) and 35S::FT plants.16,25 This suggests that low temperature influences leaf curling in these transgenic plants overexpressing SEP3, SPL3(-), or FT. In contrast to 35S::SEP3 plants, sep3–2 mutants showed slightly late flowering at 23°C and 16°C (15.5 and 35.4 leaves, respectively). This indicated that sep3–2 mutants normally responded to ambient temperature changes like wild-type plants.
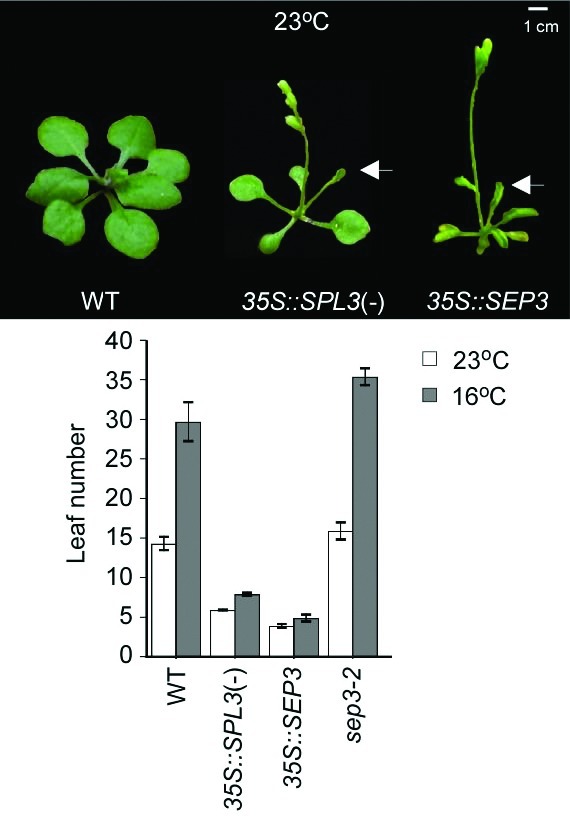
Figure 1. Flowering phenotypes of wild-type, 35S::SPL3(-), 35S::SEP3, and sep3–2 plants. Photographs were taken when 35S::SPL3(-) and 35S::SEP3 plants flowered at 23°C. 35S::SPL3(-) and 35S::SEP3 plants are early flowering and small with curled leaves (denoted by arrows). Total leaf numbers of each plant grown at 23°C and 16°C under long-day (LD) conditions are presented. Error bars indicate the standard deviation (SD).
SPL3 and FT Positively Regulate SEP3 Expression in the Leaf and the Shoot Apical Regions
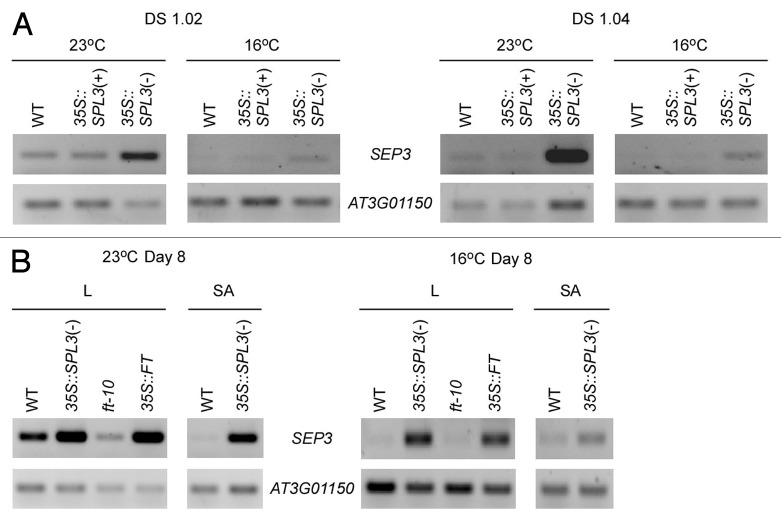
Because 35S::SEP3 plants showed temperature-insensitive flowering as similarly seen in 35S::SPL3(-) plants (Fig. 1), we further investigated the SEP3 expression in 35S::SPL3(+) plants [35S::SPL3(+) plants had an intact miR156 binding element in its 3′-untranslated region (UTR)] and 35S::SPL3(-) plants at defined growth stages (DS)1.02 and DS1.04.26 We used a developmentally synchronized stage to compare SEP3 expression levels due to the altered plastochron length of SPL3-overexpressing plants.16 In 35S::SPL3(-) plants at growth stage DS1.02, SEP3 expression was significantly increased at 23°C and 16°C (Fig. 2A). However, the levels of SEP3 expression were not altered in 35S::SPL3(+) plants at both temperatures, compared with those in wild-type plants. This suggested that the miR156-sensitive SPL3 version does not affect SEP3 expression. Moreover, at DS1.04, the upregulation of SEP3 was more apparent in 35S::SPL3(-) plants. This indicated that increased SPL3 activity upregulated SEP3 expression at different temperatures.
Figure 2.SEP3 expression in 35S::SPL3(+), 35S::SPL3(-), 35S::FT, and ft-10 plants. (A) RT-PCR analyses of SEP3 expression at defined growth stages (DS)1.02 and DS1.0426 of wild-type (WT), 35S::SPL3(+), and 35S::SPL3(-) plants grown at 23°C and 16°C under LD conditions. (B) RT-PCR analyses of SEP3 expression in the leaves (L) and in the shoot apical regions (SA) of 8-d-old seedlings in WT, 35S::SPL3(-), ft-10, and 35S::FT plants grown at 23°C and 16°C under LD conditions. AT3G01150 was used as an internal control.29
We further analyzed SEP3 expression in the leaf and the shoot apical region of 35S::SPL3(-), ft-10, and 35S::FT plants, because SEP3 is expressed in the leaf and the shoot apex,27 and SEP3 is a downstream gene of FT in the leaf.25 In the leaf of 8-d-old 35S::FT plants, SEP3 expression was strongly upregulated at 23°C and 16°C (Fig. 2B), consistent with the results reported previously.25 However, the downregulation of SEP3 were more apparent in ft-10 plants grown at 23°C than at 16°C. Furthermore, the upregulation of SEP3 was observed in the leaf of 8-d-old 35S::SPL3(-) plants. This suggested that SEP3 is a downstream gene of SPL3 and FT in the leaf. In the shoot apical region of 35S::SPL3(-) plants, SEP3 expression was highly increased at both temperatures (Fig. 2B). Collectively, these results suggested that SEP3 is regulated by SPL3 and FT in the leaf, whereas SPL3 regulates SEP3 in the shoot apical region.
Here, we have demonstrated that increased SEP3 activity affects flowering time by the changes in ambient temperature. Also, we showed that SEP3 acts downstream of SPL3 and FT in the leaf and the shoot apical regions. Based on these results, we propose that SEP3 acts as a downstream gene of SPL3 and FT in the leaf and the shoot apical regions to modulate ambient temperature-responsive flowering (Fig. 3). In the leaf, changes in miR156 activity by different ambient temperatures affect transcriptional regulation of FT by directly binding of SPL3 protein to FT genomic region.16 Because FT protein requires partner-dependent transcriptional regulation, the regulation of SEP3 through the FT-X (unknown factor) module controls flowering time by the changes in different ambient temperatures. It is also possible that SPL3 protein could directly bind to SEP3 genomic loci to regulate SEP3 expression. Because the misexpressed version of rSPL3 (miR156-resistant version) and FT in the shoot apex (FD::rSPL3 and FD::FT plants) still showed ambient temperature-responsive flowering,16 the miR156-SPL3 module directly regulates SEP3 expression for age-dependent flowering in the shoot apical region.9 However, we cannot exclude the possibility that the regulation of SPL3 via FT-FD module affects SEP3 activity to modulate photoperiodic flowering in the shoot apical region, because FD protein binds to the G-box motifs in the SPL genomic region.16,28 Thus, further investigation is required to elucidate the mechanism of interaction between the FT-X module and SEP3 in the leaf and between SPL3 and SEP3 in the shoot apical region. Furthermore, because loss-of-function mutants of SEP3 showed ambient temperature-responsive flowering, the temperature response of ft sep3 double mutants will shed light on the question whether SEP3 redundantly acts as an output gene with FT in the control of flowering time by the changes in different ambient temperatures.
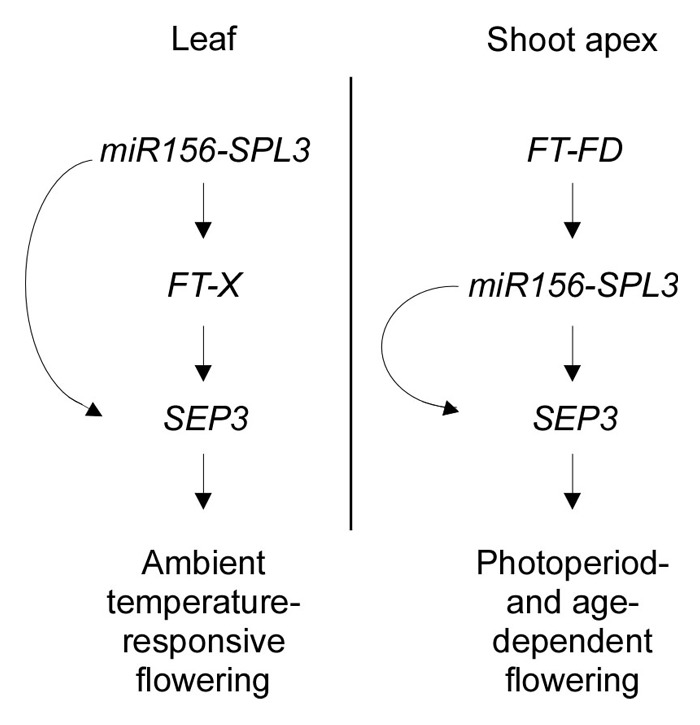
Figure 3. A proposed molecular interaction between the miR156-SPL3-FT circuitry and SEP3 for ambient temperature-responsive flowering. In the leaves, changes in ambient temperature affect the miR156-SPL3 module, which regulates the FT expression by directly SPL3 binding to the FT genomic region.16 Because FT functions through partner-dependent transcriptional activation of unknown factors, the FT-X module controls SEP3 expression to regulate ambient temperature-responsive flowering. However, we cannot dismiss the possibility that SPL3 protein directly binds to SEP3 loci. In the shoot apical region, the miR156-SPL3 module regulates SEP3 expression to modulate age-dependent flowering.9 Furthermore, the FT-FD module positively regulates SPL3 expression, which affects SEP3 expression for the photoperiodic flowering.28 Arrows represent promotion effects.
Glossary
Abbreviations:
- GA
Gibberellic acid
- miRNAs
microRNAs
- SPL
SQUAMOSA PROMOTER BINDING PROTEIN-LIKE
- FT
FLOWERING LOCUS T
- SOC1
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1
- TSF
TWIN SISTER OF FT
- FUL
FRUITFULL
- SEP3
SEPALLATA3
- LD
Long-day
- UTR
Untranslated region
- DS
Developmental stage
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21366
References
- 1.Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 2004; 16 Suppl: S18-31; PMID: 15037730; 10.1105/tpc. 015958. [DOI] [PMC free article] [PubMed]
- 2.Lee JH, Lee JS, Ahn JH. Ambient temperature signaling in plants: an emerging field in the regulation of flowering time. J Plant Biol. 2008;51:321–6. doi: 10.1007/BF03036133. [DOI] [Google Scholar]
- 3.Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. Trends Genet. 2010;26:519–27. doi: 10.1016/j.tig.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Fornara F, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell. 2010;141:550–, 550, e1-2. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 5.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–8. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 6.Dugas DV, Bartel B. MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol. 2004;7:512–20. doi: 10.1016/j.pbi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–4. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 8.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–9. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 9.Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008; 20: 1231-43; PMID:18492871. [DOI] [PMC free article] [PubMed]
- 10.Yu N, Cai WJ, Wang S, Shan CM, Wang LJ, Chen XY. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell. 2010;22:2322–35. doi: 10.1105/tpc.109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–47. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–95. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–49. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–9. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010;24:2678–92. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH. The miR156-SPL3 module regulates ambient temperature-responsive flowering via FT in Arabidopsis thaliana. Plant Physiol 2012; PMID:22427344; http:/ /dx.doi.org/10.1104/pp.111.192369. [DOI] [PMC free article] [PubMed]
- 17.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–2. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 18.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, et al. Activation tagging of the floral inducer FT. Science. 1999;286:1962–5. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 19.Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–76. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky M, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis Science 2000; 288: 1613-6; PMID: 10834834; 10/1126/science.1219644. [DOI] [PubMed]
- 21.Lee JH, Hong SM, Yoo SJ, Park OK, Lee JS, Ahn JH. Integration of floral inductive signals by FLOWERING LOCUS T and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1. Physiol Plant. 2006;126:475–83. [Google Scholar]
- 22.Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007;21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blázquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003;33:168–71. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–89. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 25.Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell. 2005;17:2661–75. doi: 10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, et al. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF. APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 2001;26:385–94. doi: 10.1046/j.1365-313X.2001.2641042.x. [DOI] [PubMed] [Google Scholar]
- 28.Jung JH, Ju Y, Seo PJ, Lee JH, Park CM. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012;69:577–88. doi: 10.1111/j.1365-313X.2011.04813.x. [DOI] [PubMed] [Google Scholar]
- 29.Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH. Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 2010;51:1694–706. doi: 10.1093/pcp/pcq128. [DOI] [PubMed] [Google Scholar]