Abstract
The role of non-coding RNAs (ncRNAs), both short and long ncRNAs, in the regulation of gene expression has become evident in recent years. Non-coding RNA-based regulation is achieved through a variety of mechanisms; some are relatively well-characterized, while others are much less understood. MicroRNAs (miRNAs), a class of endogenous small RNAs, function as master regulators of gene expression in eukaryotic organisms. A notable, recently discovered role for long ncRNAs is that of miRNA decoys, also referred to as target mimics or sponges, in which long ncRNAs carry a short stretch of sequence sharing homology to miRNA-binding sites in endogenous targets. As a consequence, miRNA decoys are able to sequester and inactivate miRNA function. Engineered miRNA decoys are also efficacious and useful tools for studying gene function. We recently demonstrated that the potential of miRNA decoys to inactivate miRNAs in the model plants Arabidopsis thaliana and Nicotiana benthamiana is dependent on the level of sequence complementarity to miRNAs of interest. The flexibility of the miRNA decoy approach in sequence-dependent miRNA inactivation, backbone choice, ability to simultaneously inactivate multiple miRNAs, and more importantly, to achieve a desirable level of miRNA inactivation, makes it a potentially useful tool for crop improvement. This research addendum reports the functional extension of miRNA decoys from model plants to crops. Furthermore, endogenous miRNA decoys, first described in plants, have been proposed to play a significant role in regulating the transcriptome in eukaryotes. Using computational analysis, we have identified numerous endogenous sequences with potential miRNA decoy activity for conserved miRNAs in several plant species. Our data suggest that endogenous miRNA decoys can be widespread in plants and may be a component of the global gene expression regulatory network in plants.
Keywords: miRNA, decoy, target mimicry, gene regulation, miR171, miR160, miR319, miR156, miR172, soybean
Introduction
MicroRNAs (miRNAs), a class of small ncRNAs, are key regulators of gene expression controlling numerous functions from growth and development to stress responses in plants and animals.1-3 In plants, mature miRNAs are processed from long ncRNA precursors containing a stem-loop secondary structure by an RNase III-like enzyme, DCL1, in coordination with DRB1 (HYL1), and SE; and are further modified by HEN1 and exported by HST. The guide stand of the miRNA duplex is then loaded into the AGO1 protein, the major component of the RNA-induced silencing complex (RISC), and serves as the specificity determinant in target recognition (for review see reference 1). In plants miRNA-mediated gene repression occurs at the post-transcriptional level, triggering mRNA cleavage, as well as by inducing translational inhibition.4,5 In some instances, small RNAs derived from miRNA transcripts have been reported to induce DNA methylation pathways in animals and plants.6-8
MicroRNAs are found to interact with a wide range of target transcripts, both protein coding and non-coding, including non-coding transcripts serving as precursors for other types of small RNAs, such as trans-acting siRNAs (tasiRNAs).4,9,10 MicroRNA-based regulation of gene expression helps to achieve the spatial and temporal patterns necessary for proper genetic function of target transcripts.10-12 As a consequence, mis-regulation of miRNAs often leads to developmental abnormalities.4,13-15 Given their powerful influence, the system regulating miRNA activity must be equally robust. This system is a compilation of several layers of regulation, including pri-miRNA transcription rate16 and localization,17 processing,18 stability,19-21 and movement of both the precursor and mature miRNA.22,23
One layer of miRNA regulation recently identified in both plants and animals has been termed miRNA decoys, or target mimicry. MicroRNA decoys rely on the sequence-dependent interaction of miRNAs with other non-coding and/or protein coding RNAs. The first endogenous miRNA decoy example demonstrated, the interaction of mature miR399 in Arabidopsis with INDUCED BY PHOSPHATE STARVATION 1 (IPS1), a non-coding RNA containing a miR399 decoy, or mimic site, results in miR399 inactivation and subsequent upregulation of PHO2, the primary miR399 target transcript.24 Being the foremost example, the concept of ncRNA-mediated inactivation of miRNA function has been successfully established by overexpressing modified versions of the IPS1 transcript, sequestering a variety of miRNAs in Arabidopsis.24,25 It was also demonstrated that a modified ncRNA transcript endogenous to corn and a soybean miRNA precursor can serve as backbones for miRNA decoys.15 To date only one naturally-occurring miRNA decoy has been described in plants,24 however, bioinformatic analysis suggests the presence of orthologous miR399 decoys in other species,15,26 and that other miRNAs in plants may be regulated by endogenous decoys.15
Direct manipulation of miRNA antagonism through engineered decoy sequences affords the potential for practical applications for studying miRNA function and modification of plant characteristics. The range of functional plant miRNA decoys is not limited to the endogenous miR399 mimic example described,24,25 which contains a 3 nucleotide bulge structure. Rather, it has been demonstrated that a range of decoy configurations are efficacious when extended up to 5 nucleotide insertions and with as few as 1 mismatch at position 11, relative to the miRNA.15 In addition, we have shown that multiple decoy sites can be incorporated into a single transcript, downregulating the activity of multiple miRNAs simultaneously, and that functional decoys can exist as part of protein coding transcripts.15 In this addendum, we report that miRNA decoys are functional in plant species beyond Arabidopsis and N.benthamiana. Furthermore, computational analysis identified endogenous putative decoy transcripts in numerous plant species, suggesting the involvement of miRNA decoys in the global expression regulatory network of plants may be broad.
Results
miRNA decoys function in crops
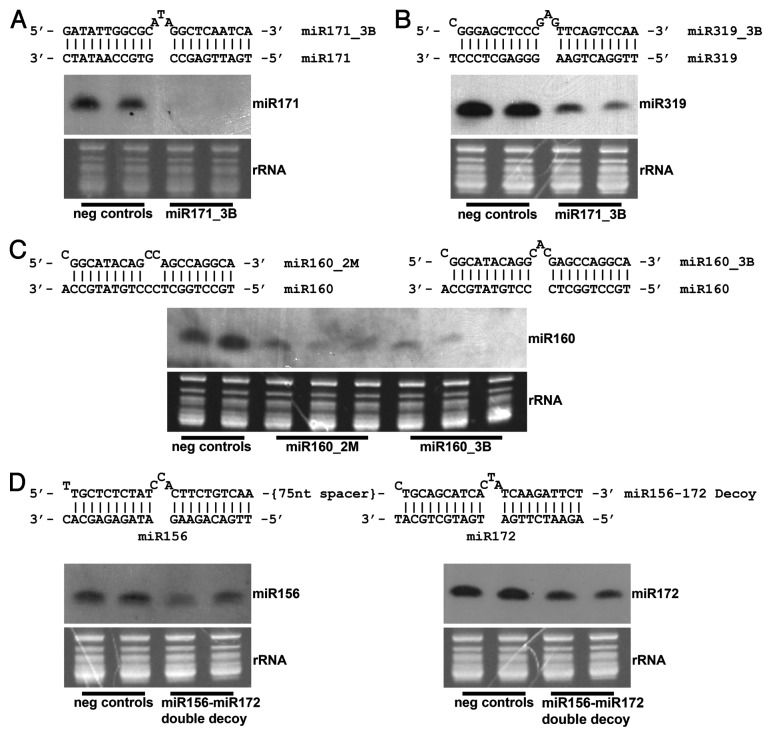
In order to demonstrate that miRNA decoy function is not limited to model plant species, such as Arabidopsis and N.benthamiana, decoy overexpression vectors with bulged or mismatched structures targeting several conserved miRNAs (miR160, miR171, miR172 and miR319) were constructed and transformed into soybean. All bulged decoys (miR160 and miR171) contained a 3 nucleotide insertion (3B) between the nucleotides interacting with 10 and 11 of the corresponding miRNA. Mismatched decoys (miR160, miR171, and miR319) were designed with two mismatches (2M) at the nucleotides interacting with 10 and 11 of the corresponding miRNA. All decoys were embedded in a long ncRNA transcript amplified from Z. mays. See Ivashuta, et al.15 for further details of general decoy design. A double decoy, containing two bulged decoy sites separated by a 75 nucleotide synthetic spacer, was designed to inhibit both miR156 and miR172. As in Arabidopsis, miR171, miR160 and miR319 in soybean are predicted to interact with the transcripts of GRAS (GIBBERELLIC ACID INSENSITIVE, REPRESSOR OF GA1, and SCARECROW) transcription factors, ARF (AUXIN RESPONSE FACTOR) transcription factors and TCP (TEOSINTE BRANCHED/CYCLOIDEA/PCF) transcription factors, respectively.4,13,14 The putative targets of miR156 and miR172 in soybean are also common with Arabidopsis, including orthologs of SPL (SQUAMOSA PROMOTER BINDING PROTEIN LIKE) transcription factors and AP2-like (APETALA2) transcription factors, which are involved in developmental timing.27-31
Our data indicate that expression of both bulged and mismatched decoys leads to degradation of mature miRNAs in soybean. The extent of miRNA degradation varied, depending on decoy type and miRNA targeted. The level of mature miRNA reduction observed ranged from very strong to mild; however, clearly visible phenotypes were apparent in all transformants. Plant height was reduced in miR171_3B transformants, leaf rigidity was decreased in transformants expressing miR319_3B, and altered leaf morphology was found in plants expressing miR160_3B, miR160_2M, as well as in miR156_3B-miR172_3B double decoy transformants (data not shown). A dramatic reduction in miRNA level was observed in the case of miR171_3B (Fig. 1A), miR319_3B (Fig. 1B) and miR160_3B (Fig. 1C) decoy transformants, wherein the decoys were embedded in the corn backbone with a bulge bearing three extra nucleotides. In the case of miR160_2M (Fig. 1C) and miR156_3B – miR172_3B (Fig. 1D) double decoy transformants, a relatively mild reduction in respective miRNAs was noted, but plants were accompanied with readily observed phenotypes in all transformants, indicating some functional thresholds had been met. Only in miR171_2M transformants, which show a small decrease in miR171 levels, were no dramatic phenotypic differences evident (data not shown). These results confirm previous observations made in model plants15,24,25 and suggest that decoy-dependent miRNA degradation, along with miRNA sequestration, are conserved mechanisms of miRNA inactivation in plants. While decoy dependent miRNA degradation was shown to be associated with activity of the proteins encoded by the SMALL RNA DEGRADING NUCLEASE (SDN) family, SDN1 and SDN2, in Arabidopsis,21 it is not clear how plants distinguish between miRNA/miRNA-target and miRNA/decoy interactions, as only the latter interaction leads to the destabilization of the mature miRNA in the majority of transformants tested.
Figure 1. Northern blot analysis of decoy-targeted miRNAs of interest. A bulged (3nt) decoy targeting (A) miR171 and (B) miR319 was embedded in a long non-coding transcript and constitutively overexpressed in soybean, leading to decreased levels of each mature miRNA. (C) A bulged and mismatched decoy was each singly overexpressed in the same long non-coding transcript, each targeting miR160. Levels of mature miR160 were reduced in transformants containing each type of decoy, including one bulge event showing almost total elimination of the miRNA. (D) Levels of mature miR156 and miR172 were simultaneously reduced by overexpressing a double decoy transcript, a long non-coding transcript containing a bulged decoy site targeting miR156, followed by a second bulged decoy site targeting miR172. The decoy sites were separated by a non-coding 75 nucleotide spacer.
Endogeneous miRNA decoys in plants
The scale of potential miRNA decoy-based regulation in plants was evaluated by conducting a computational identification of putative miRNA decoy sequences in various species. Selection of conserved miRNA families was based on Cuperus, et al.32 Mature miRNA sequences were downloaded from miRBase (www.mirbase.org, V17). Decoys were predicted as described previously15 in plant species for which genome sequences or transcriptome sequences were available and that represent important evolutionary lineages (Table 1). The predicted decoys were mapped to the species' ESTs from GenBank (as of 07/07/2011) to evaluate whether the decoys were expressed. The criteria to call a positive mapping included 95% identity and 95% coverage of the EST. The predicted decoys were then mapped and manually analyzed for homology to miRNA precursors found in miRbase. Any sequences with matches to the miRNA precursors were discarded. The remaining decoys were then classified as either coding or non-coding, by comparing them to the UniProt database (uniref. 90 from www.uniprot.org, as of February, 2011). Decoy sequences with an alignment length shorter than 100 amino acids were categorized as non-coding. Computational analysis (Table 1) indicates that putative decoy sites are present in various plant species. While the majority of decoy sites are found in protein coding transcripts, it must be noted that the bulk of sequence data sets used in this analysis are enriched for protein coding sequences due to sequencing and data processing methods.
Table 1. Computational prediction of miRNA decoys in plant species representing important lineages.
| species | decoy loci | decoys are ncRNA | Decoys with ESTs | miR156 | miR159 | miR319 | miR160 | miR166 | miR171 | miR408 | miR390 | miR395 | miR396 | miR397 | miR398 | miR162 | miR164 | miR167 | miR168 | miR169 | miR172 | miR393 | miR394 | miR399 | miR827 | version or release ID | Download link |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Physcomitrella patens |
23 |
5 |
16 |
1 |
|
1 |
3 |
3 |
2 |
4 |
5 |
3 |
|
|
|
|
|
1 |
|
|
|
|
|
|
|
152 |
www.phytozome.net |
| Pinus taeda |
47 |
17 |
* |
15 |
11 |
9 |
|
|
1 |
4 |
|
|
7 |
|
|
|
|
|
|
|
|
|
|
|
|
9 |
compbio.dfci.harvard.edu |
|
Arabidopsis thaliana |
58 |
4 |
48 |
20 |
1 |
2 |
|
|
3 |
|
2 |
|
8 |
2 |
|
|
2 |
4 |
2 |
2 |
6 |
|
1 |
2 |
1 |
10 |
www.arabidopsis.org |
| Glycine max |
80 |
10 |
58 |
14 |
1 |
10 |
2 |
|
4 |
|
10 |
|
10 |
|
|
3 |
|
4 |
|
7 |
7 |
1 |
7 |
|
|
1 |
in-house |
| Vitis vinifera |
56 |
1 |
33 |
4 |
6 |
2 |
|
|
5 |
|
2 |
1 |
6 |
|
1 |
1 |
3 |
1 |
|
5 |
4 |
|
8 |
4 |
|
145 |
www.phytozome.net |
| Populus trichocarpa |
137 |
7 |
47 |
8 |
4 |
6 |
6 |
2 |
13 |
4 |
17 |
4 |
9 |
15 |
1 |
|
1 |
3 |
|
22 |
6 |
1 |
7 |
5 |
3 |
156 |
www.phytozome.net |
| Brachypodium distachyon |
49 |
2 |
27 |
2 |
3 |
|
3 |
|
2 |
1 |
2 |
2 |
8 |
3 |
1 |
|
9 |
|
|
5 |
3 |
1 |
3 |
1 |
|
6.1 |
rice.plantbiology.msu.edu |
| Oryza sativa |
68 |
5 |
48 |
5 |
3 |
1 |
1 |
3 |
2 |
3 |
|
3 |
11 |
|
3 |
1 |
4 |
4 |
1 |
3 |
6 |
1 |
4 |
8 |
1 |
2.5a |
Maizesequence.org |
| Sorghum bicolor |
46 |
1 |
27 |
3 |
2 |
|
|
1 |
2 |
2 |
3 |
4 |
3 |
2 |
1 |
1 |
5 |
1 |
|
4 |
4 |
1 |
6 |
1 |
|
79 |
www.phytozome.net |
| Zea mays | 104 | 22 | 78 | 10 | 14 | 2 | 8 | 7 | 12 | 2 | 3 | 7 | 2 | 1 | 2 | 19 | 6 | 1 | 4 | 4 | 114 | www.phytozome.net |
Conclusion
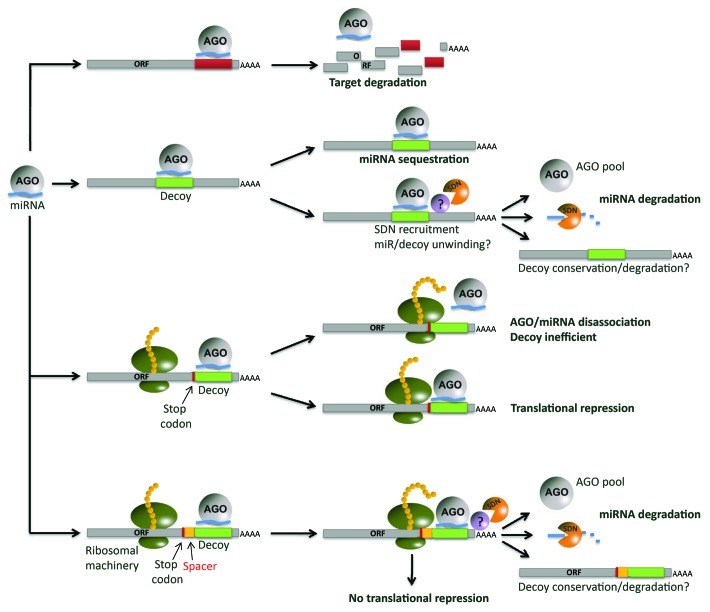
The discovery of miRNA regulation through target mimicry in plants and in animals reveals another level of complexity in controlling miRNA activity and gene regulation in eukaryotes. The new competing endogenous RNA (ceRNA) hypothesis describes cross-talk among mRNAs, transcribed pseudogenes and long non-coding RNAs via competition for shared microRNAs in humans33 and certainly mirrors the description of miRNA regulation put forth in plants.34 Previously published data,15,24,25 as well as the present study, demonstrate the ability of engineered decoys to modulate miRNA-regulated networks in various plants, including crops. This, combined with the computational prediction of endogenous miRNA decoys in multiple plant species, suggests that endogenous decoys may play a significant role in the control of gene expression in plants, consistent with the ceRNA hypothesis proposed in animals. Evidence that miRNA decoys can function when resident in a protein coding transcript and that a single transcript can harbor multiple miRNA decoy sites with up to 5 bulges or 3 mismatches15 allows for speculation that the scale of such miRNA decoy based regulation may be significant in plants. Similar principles of decoy-dependent regulation have shown pertinence to trans-acting small RNAs in plants,21 and may logically extend to other small RNA classes, including siRNAs and natural antisense siRNAs (nat-siRNAs). The interaction between miRNA and miRNA decoy and the molecular complex that may form during this interaction is not known, but it is likely different from the complex formed during miRNA/miRNA target interaction, as it leads to miRNA destabilization in plants and animals.15,25,35 In contrast, the interaction of a miRNA with its target has been shown to stabilize the miRNA in animals.36Figure 2 presents a model of miRNA interaction with different types of miRNA decoys and potential outcomes of such interactions. Mature miRNAs cause target degradation and are relatively stable in plants (top), while the interaction of a miRNA with a decoy results in miRNA degradation.15,25,35 In some instances miRNA decoys do not trigger obvious miRNA degradation. The extent of miRNA sequestration vs. degradation is difficult to predict and it is likely sequence-dependent. The presence and position of a miRNA decoy site within a protein coding transcript adds an additional level of complexity (third from top). Efficiency of translation can be affected by miRNA-miRNA decoy binding/association in some cases.15 At the same time, decoy efficiency decreases, possibly due to the interference of the decoy-bound AGO/miRNA complex with the translational machinery. However, inserting a spacer between the miRNA decoy site and open reading frame (ORF) may alleviate such interference and may increase both decoy and translational efficiency, as was shown to be the case in experiments in N. benthamiana.15 Further work is needed to clarify the mechanism underlying decoy-associated miRNA turnover and the contribution of decoys to global regulation of gene expression in plants.
Figure 2. Potential miRNA interactions. Mature miRNAs loaded into AGO bind to and inhibit target transcripts through cleavage or via translational inhibition (top). One must take into consideration the interactions of miRNAs with decoys embedded in long non-coding transcripts (second). The miRNAs are potentially sequestered by the decoy, or alternatively, degraded, a process that requires the presence of SDNs in Arabidopsis. Little is known about the stability of the non-coding transcript after interaction with the miRNA has taken place. A third path is the interaction with a decoy site found within a coding transcript. If the decoy site is located too closely to the ORF, putative steric interactions between the ribosomal machinery and the miRNA may be possible. Translational repression is another consequence. If the decoy is located in the 3′ UTR at an adequate distance from the stop codon (bottom), the decoy is efficient and translation will not be affected.
Acknowledgments
We would like to thank Vanitharani Ramachandran, Linda Rymarquis and Brent Brower-Toland for scientific conversation and review, as well as and Bala Karunanandaa, Ed Allen, Vijay Sharma, Ganesh Kumar, Anil Neelam, Miya Howell, Logan Huff and Nancy Brumley for support of this work.
Glossary
Abbreviations:
- ncRNA
non-coding RNA
- miRNA
microRNA
- siRNA
small interfering RNA
- tasiRNA
trans-acting small interfering RNA
- pri-miRNA
primary miRNA
- nat-siRNA
natural antisense small interfering RNA
- ceRNA
competing endogenous RNA
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21299
References
- 1.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 2.Shukla LI, Chinnusamy V, Sunkar R. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta. 2008;1779:743–8. doi: 10.1016/j.bbagrm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–8. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 4.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–6. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 5.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–90. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 6.Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, et al. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–22. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 7.Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, et al. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 2010;38:6883–94. doi: 10.1093/nar/gkq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, et al. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465–75. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–21. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–9. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 12.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutiérrez-Nava M, Poethig SR. -acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–81. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–75. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–63. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 15.Ivashuta S, Banks IR, Wiggins BE, Zhang Y, Ziegler TE, Roberts JK, et al. Regulation of gene expression in plants through miRNA inactivation. PLoS ONE. 2011;6:e21330. doi: 10.1371/journal.pone.0021330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–26. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 19.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–2. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, et al. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell. 2012;24:415–27. doi: 10.1105/tpc.111.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–4. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- 24.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–7. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 25.Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rymarquis LA, Kastenmayer JP, Hüttenhofer AG, Green PJ. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 2008;13:329–34. doi: 10.1016/j.tplants.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–20. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 28.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–41. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–5. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, et al. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–12. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 31.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–47. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011;23:431–42. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubio-Somoza I, Weigel D, Franco-Zorilla JM, García JA, Paz-Ares J. ceRNAs: miRNA target mimic mimics. Cell. 2011;147:1431–2. doi: 10.1016/j.cell.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–9. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]