Abstract
In this study, we present a 2nd generation genetic linkage map of a cross between the North American species Heterobasidion irregulare and H. occidentale, based on the alignment of the previously published 1st generation map to the parental genomes. We anchored 216 of the original 308 AFLP markers to their respective restriction sites using an in silico-approach. The map resolution was improved by adding 146 sequence-tagged microsatellite markers and 39 sequenced gene markers. The new markers confirmed the positions of the anchored AFLP markers, fused the original 39 linkage groups together into 17, and fully expanded 12 of these to single groups covering entire chromosomes. Map coverage of the genome increased from 55.3% to 92.8%, with 96.3% of 430 markers collinearly aligned with the genome sequence. The anchored map also improved the H. irregulare assembly considerably. It identified several errors in scaffold arrangements and assisted in reducing the total number of major scaffolds from 18 to 15. This denser, more comprehensive map allowed sequence-based mapping of three intersterility loci and one mating type locus. This demonstrates the possibility to utilize an in silico procedure to convert anonymous markers into sequence-tagged ones, as well as the power of a sequence-anchored linkage map and its usefulness in the assembly of a whole genome sequence.
Introduction
Genetic linkage maps have been used in genomic research for decades, both as a tool to identify candidate loci for phenotypic traits and as a way of studying chromosomal arrangement. Because of its potential for high yield of specific markers, a common fingerprinting method of choice has been amplified fragment length polymorphisms (AFLP) [1], in animals [2] and plants [2]–[3] as well as fungi [4]–[7]. As a consequence of recent improvements in and lowered costs for DNA sequencing technology, many organisms which previously have had their genome described with a genetic linkage map have since had their full genomes sequenced. These include a number of important fungi, such as Pleurotus ostreatus (http://genome.jgi.doe.gov/PleosPC15_2/PleosPC15_2.info.html), Coprinopsis cinerea [8], Gibberella zeae [9], Mycosphaerella graminicola [10], and the oomycete Phytophthora infestans [11]. A linkage map fully anchored to a whole genome sequence constitutes a very powerful tool, allowing immediate conversion of any mapped quantitative trait loci (QTL) to actual candidate genes. Without such a tool, the physical position of the QTLs will remain unknown, because the markers derived from random fingerprinting techniques like AFLP are anonymous and identification of candidate genes will require laborious map based cloning.
Anchoring and improving existing linkage maps assisted by genome sequence data has been done in various ways. In some cases the upgraded maps have been based only on new sequence-tagged markers without using the original map or sharing just a subset of the original markers to visualize the correlation between the maps [12]–[14]. Another approach has been to construct the new map complemented by markers derived from the known sequence [15]. Although these approaches can roughly link a map to its physical sequence, they don’t reveal the identity of every anonymous marker, which limits exploration of known QTLs. For organisms lacking known genome sequence, the process of converting anonymous markers into sequence-tagged sites has been even more cumbersome, usually involving repeated steps of selective AFLP with increasingly selective primers and manual excisions from polyacrylamide gels [16]–[17].
The root rot fungus Heterobasidion annosum sensu lato (s.l.) causes devastating damages to conifer forests all over the Northern Hemisphere. It is economically the world’s most important pathogen on conifers, causing yearly losses in Europe of at least €600 million [18]. The Heterobasidion species complex consists of phylogenetic distinctive, yet partially interfertile, species [19]–[20]. The interfertility phenomenon has been best described for the North American species H. occidentale and H. irregulare. It is controlled by at least five intersterility loci (S, P, V1, V2, V3) and compatible mating between individuals from the distinct species is possible only when both individuals in a pairing share a + allele for at least one of the intersterility loci [21]. However, the regular mating type system present in basidiomycetes is also present in H. annosum s.l. and is controlled by a bipolar mating system [22]–[23]. Albeit the system is controlled by a single MAT-A locus, there are probably more than a hundred mating type alleles in the species complex [24]–[25].
An AFLP markers-based linkage map of Heterobasidion species was originally published by Lind et al [26]. This 1st generation map was based on a mapping population of 102 single spore isolates originating from a compatible mating [27] between a H. occidentale isolate (TC-122-12) and a H. irregulare isolate (TC-32-1) [23]. The map has been used to locate several traits of interest, such as growth rate [28], virulence [29], resistance to Phlebiopsis gigantea overgrowth [30] and various intraspecific interactions [31]. The interfertility between TC-32-1 and TC-122-12 was mediated through a common + allele for the V3 intersterility locus, while the S, P and V2 loci were all polymorphic between the parents. This allowed for the mapping of these three intersterility loci, but also of a mating type locus, as each progeny isolate will carry the MAT-A [22] allele from either one of the parents. Previous efforts to identify these loci [26] provided a strong position for the S locus, a weaker one for P and no position at all for V2 and MAT-A [26].
The aim of this study was to upgrade the H. occidentale x H. irregulare linkage map [26] into a sequence-based, 2nd generation map covering the whole genome. We sought to achieve this by (i) de novo assemble the TC-122-12 parental strain genome and align it to the existing genome sequence of the TC-32-1 parental strain [23], (ii) anchor every AFLP marker of the original map to the genome sequences using an in silico approach; and (iii) to expand and fuse the anchored linkage groups into large groups covering whole individual chromosomes, by addition of new markers generated from the sequences of 146 SSRs and 39 known genes or ESTs. As a validation of the reliability of the final 2nd generation map, we wanted to use it to position three intersterility loci and one mating type locus. Finally, the correlation between physical and genetic map enabled assessment of recombination rates across the intersterility and mating type loci.
Materials and Methods
The parental strains TC-122-12 and TC-32-1 [32], the 102 progeny isolates used as a mapping population [27], the DNA extractions and the original linkage map [26] have been described elsewhere.
Sequence Data and Genome Assembly
Genome sequence data for the H. irregulare parental strain TC-32-1 were obtained from the JGI genome project (http://genome.jgi-psf.org/Hetan2/Hetan2.home.html). Version 2 of the assembly comprised 33.6 MB distributed over 14 large (1.3–3.6 MB) and one small (8 KB) scaffolds [23].
Heterobasidion occidentale (TC-122-12) mycelium was cultivated and submitted to DNA extraction according to Lind et al [26]. Library preparations and sequencing took place at the SNP&SEQ Technology Platform of Uppsala University Hospital, using Solexa/Illumina short-read ultra-high throughput DNA sequencing technology (version 1.6) according to the manufacturers’ instructions. Paired-end sequences were obtained from libraries with an insert length of 400 bp and subsequently screened for poor quality using the SolexaQA software, version 1.13 [33]. The sequenced reads were then assembled with the Velvet assembler version 1.1.02 using parameters obtained from VelvetOptimiser version 2.2.0 [34]. Scaffolds larger than 1000 bp were aligned to the H. irregulare genome using the NUCmer function of MUMmer version 3.0. Based on this alignment H. occidentale scaffolds were assigned corresponding H. irregulare positions.
Virtual AFLP
The principle of the virtual AFLP method, first described in 2003 [35], is to determine the genomic positions of AFLP markers obtained in vitro by an in silico simulation of the work of the restriction enzymes and the amplification steps. We used a DNA analysis software from AcaClone called pDRAW32 ver 1.1.110. Our markers to be anchored [26] were based on enzymatic cleavage with either Pst1-Mse1 or EcoR1-Mse1 pairs and amplified with two-nucleotide primer extensions. Thus, we scanned the TC-122-12 and TC-32-1 genomes for these pairs of restriction sites with the corresponding extensions, and selected candidates from pairs that flanked regions of the same size (in bp) as the sought markers.
Identification of Microsatellites and Primer Design
Microsatellite markers were designed to confirm the positions of the anchored AFLP markers and to further improve the linkage map. The genome sequence of TC-32-1 was scanned for SSR motifs using the Magellan software [36] and one hundred and forty-six candidates were selected. Motifs of large core units (hexa or penta nucleotides) and high number of repeats (from 5× to 33×) were primarily chosen to increase the possibility of polymorphisms between the parents. Primers for each microsatellite were designed using the Primer3 web browser tool (http://bioinfo.ebc.ee/mprimer3/), with primer lengths varying between 18 and 22 bases, melting temperatures set between 59 and 61°C and a GC clamp of one nucleotide used. The primers were picked to amplify regions varying ideally between 100 and 500 bases in length, with a few exceptions (Table S1). The markers were then amplified from the mapping population according to the following procedure: 0.2 ng of template DNA to a final volume of 5 µl per reaction, containing 0.5 mM dNTP (Fermentas), 1.8 mM MgCl2, 1 µl Taq buffer (Fermentas), 0.5 U DreamTaq Green DNA polymerase (Fermentas) and 0.4 µM of each primer (TAG Copenhagen). The reactions were run for 35 cycles of denaturation (50 s, 95°C), annealing (50 s, 60°C), and extension (80 s, 72°C). The PCR products were in a few cases analyzed on an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems) and the fragment lengths analyzed using the softwares GeneScan (PE Applied Biosystems) and Genotyper (PE Applied Biosystems). However, in most cases, the PCR products were analyzed by the Uppsala Genome Centre, using an ABI 3730 XL (PE Applied Biosystems).
Segregation Analysis of Sequenced Genes
A set of 16 genes previously described as associated with fungal-fungal interactions in Amylostereum areolatum, Neurospora crassa and H. irregulare [37]–[40] was selected, along with 23 genes from an EST library [41]. Specific primers for all 39 genes were designed using Primer3. Alleles were distinguished in the parental genomes TC-122-12 and TC-32-1 using PCR-RFLP (restriction fragment length polymorphism) [42] analysis (50 cycles, denaturation (30 s, 95°C), annealing (30 s, 60°C), extension (30 s, 72°C)) and agarose gel electrophoresis. The genes were mapped by scoring allele segregation in the mapping population [27] using the EvaGreen dye (Biotium Inc., Hayward, CA, USA) together with post-PCR DNA High Resolution Melting curve analysis [43].
Linkage Analysis
Segregation patterns for the 146 microsatellite markers and the 39 sequenced genes throughout the mapping population [27] were analyzed together with the existing set of 358 AFLP markers using the JoinMap 3.0 software [44]. The data set was coded as derived from haploid strains, originating from a diploid parent. Mapping groups were created of every cluster of markers joined together by a LOD score of 4 or higher. JoinMap suggested an internal marker order within each group. If the suggestion was in conflict with the physical position of the anchored markers, it was rejected. A manual suggestion was then created, with the markers arranged according to the anchored order. If this suggestion conflicted with the mapping data, the conflicting marker was moved within the suggested order, to a maximal extent of two markers away from its anchored positions (in one single case, three markers away). If the order was still not accepted, the conflicting marker was omitted. If the marker composition of a linkage group was supported by sequence based evidence, a LOD score of 2 was used as threshold for accepting the group. The sequence-anchored linkage map was visualized using MapChart 2.1 [45].
To compare the quality of sequence based and a non-sequence based genetic linkage map of the same genome, we estimated the genome coverage of the 1st generation map [26] prior to improvement by new markers or adjusted marker orders. The base pair coverage of each anchored linkage group was calculated based on the largest possible physical distance between any two markers in a linkage group and compared to the coverage of the 2nd generation map.
Mapping Mating Type Genes and Intersterility Genes
The scoring and mapping procedure for the mating type locus and intersteriliy loci have been previously described [26]. Mapping data for intersterility loci P and V2 were obtained by crossing the mapping population with tester strains known to carry a + allele for either the P locus (strain Sä 16-4) or the V2 locus (strain TC-111-4). The previous attempt at this was incomplete, as just 39 and 28 of the 102 progeny isolates expressed fertility with the respective tester strain (51 expected) [26]. We made new crosses to obtain a complete set of observations. To estimate and analyze local recombination rates across the mating type and intersterility loci, KB/cM-rates were calculated continuously across every linkage groups using a sliding window of two markers, moving one marker at a time.
Results
Assembling the H. occidentale (TC-122-12) Genome Sequence
The H. occidentale strain TC-122-12 was sequenced with an Illumina Genome Analyzer, resulting in 61 million quality screened paired-end reads from a 400-bp insert library. The Velvet assembler de novo-assembled the reads into 19 848 contigs with an N50 of 34 641 bp, covering in total 28.5 MB (internal gaps excluded). The single largest contig was 217 732 bp, while another 2 625 contigs were at least 1000 bp large. These were subsequently aligned to the assembly of H. irregulare strain TC-32-1 [23] using NUCmer. NUCmer identified 10 834 matches between the assemblies of TC-122-12 and TC-32-1, ranging in length from 66 bases up to 42 942 bases. The matches ranged in similarity from 65.4% to 100% identical. Median match similarity was 89.1%.
In Silico Anchoring AFLP Markers and Linkage Groups
A virtual AFLP approach was used to identify the pairs of restriction sites flanking each AFLP marker of the 1st generation linkage map [26]. To be considered as a candidate marker position, the distance between a pair of restriction sites had to be no more than 6 basepairs larger or smaller than the size given by the fragment analyzer (ABI Prism 310 Genetic Analyzer, PE Applied Biosystems) when the marker was originally scored. This frame was chosen to account for the potential misreads of the analyzing equipment and/or single nucleotide errors of the genome sequence. In the event that a tested marker had more than one putative genomic location, a closely linked marker was also tested, assuming that the markers would anchor to relatively proximate positions. Two linked markers rarely had more than one mutually putative position, and analyzing three linked markers was always enough to unambiguously locate them all. This was confirmed by microsatellite markers from the sequence covered by the anchored markers (see below and Figure 1).
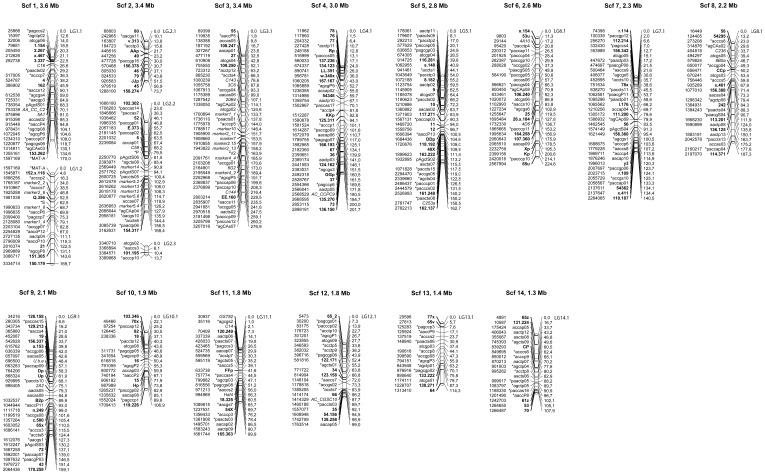
Figure 1. The 17 linkage groups of the 2nd generation map of the Heterobasidion annosum s.l. genome.
To the far left of each group bar is the position in basepairs on the H. irregulare or H. occidentale genome assembly. Next follow the marker names, where normal font indicates an AFLP marker or a mapped trait, bold stands for a microsatellite marker and italic names denotes a mapped gene of known sequence. To the right of the bar is the position in cM. Each group header tells the scaffold number associated with the group and its size in Mbp.
One hundred and twenty-seven TC-32-1 AFLP marker positions from the original map (80% of all) were successfully anchored to the H. irregulare genome assembly by use of the virtual AFLP-approach. The NUCmer-mediated alignment of the H. occidentale and the H. irregulare assemblies made it possible to also convert 89 TC-122-12 AFLP markers positions (59% of all) to the corresponding positions on the H. irregulare reference genome. Anchoring these 216 marker positions also situated thirty-four of the 39 linkage groups of the map [26] to the genome. The five remaining groups were small; four contained 2 markers and one contained 3 markers.
Creating a 2nd Generation Linkage Map
One hundred and forty-six microsatellite markers segregated according to a 1∶1 ratio and were mapped with strong significance to different linkage groups. One hundred and forty-one of these had sequence support for their mapped positions, while five microsatellite markers mapped to positions without sequence support. Thirty-nine sequenced genes could also be mapped to existing linkage groups. BLASTing their sequences against the genome confirmed 35 of these mapped positions. The remaining four did not map to expected positions according to the BLAST search. Collectively, these 185 new markers significantly increased the quality of the map (Table 1 and 2, Figure 1), bringing the total number of markers up to 430, concentrating the number of groups from 39 down to 17, increasing markers per group from on average 7.9 to 25.3 and lowering the average distance between two neighboring markers from 7.3 to 5.8 cM, as compared between the 1st and 2nd generation maps. The rearrangement of markers due to their discovered physical positions, in addition to the mapped 146 microsatellite markers and 39 sequenced genes, discovered and omitted 68 falsely linked AFLP markers. Four hundred and fourteen of the 430 markers (96.3%) were assigned genetic positions (in cM) fully aligned to their physical position (in bp). The other 16 had to be assigned a skewed genetic position in order to fit the map, but markers distorted by a distance of more than two mapped markers away from the expected position was not accepted (except on one occasion) (Figure 1).
Table 1. Compared mapping data between 1st and 2nd Heterobasidion annosum s.l. linkage maps.
| 2nd gen | 1st gen | |
| AFLP markers | 241 | 308 |
| SSR markers | 146 | – |
| Sequence markers | 39 | – |
| Traits | 4 | 2 |
| Total markers | 430 | 310 |
| Linkage groups | 17 | 39 |
| Avg markers per group | 25.3 | 7.9 |
| Mapped distance (cM) | 2476 | 2252 |
| cM/marker | 5.8 | 7.3 |
| KB/cM | 12.6 | 5.8 |
| Assembled size (KB) | 33649 | |
| Map coverage (KB) | 31233 | 18603a |
| Map coverage (%) | 92.8 | 55.3a |
The part of the assembly covered after anchoring the 1st generation map, calculated on the largest distance between the anchored markers of a linkage group, disregarding order among markers.
Table 2. Numeric data for each linkage group in the 2nd generation linkage map for Heterobasidion annosum s.l.
| Linkage group | Scaffold size b | Map covered (KB) c | Map covered (%)c | Size (cM) | KB/cM | Markers | AFLP | SSR | Gene | Traits | cM/marker |
| 1.1 | 3592 | 1571 | 92.1 | 170 | 9.2 | 27 | 16 | 7 | 3 | 1 a | 6.3 |
| 1.2 | 1738 | 169.7 | 10.2 | 20 | 9 | 5 | 4 | 2 a | 8.5 | ||
| 2.1 | 3445 | 1219 | 79.7 | 73.7 | 16.5 | 13 | 3 | 10 | 0 | 0 | 5.7 |
| 2.2 | 1477 | 168.4 | 8.8 | 27 | 14 | 5 | 7 | 1 | 6.2 | ||
| 2.3 | 49 | 13.7 | 3.6 | 4 | 3 | 1 | 0 | 0 | 3.4 | ||
| 3.1 | 3434 | 3118 | 90.8 | 276.9 | 11.3 | 41 | 22 | 5 | 13 | 1 | 6.8 |
| 4.1 | 2988 | 2886 | 96.6 | 201.7 | 14.3 | 37 | 15 | 21 | 1 | 0 | 5.5 |
| 5.1 | 2842 | 2604 | 91.6 | 162.7 | 16 | 37 | 22 | 14 | 1 | 0 | 4.4 |
| 6.1 | 2573 | 2558 | 99.4 | 224.6 | 11.4 | 28 | 16 | 10 | 2 | 0 | 8 |
| 7.1 | 2304 | 2190 | 95.1 | 140.5 | 15.6 | 35 | 22 | 13 | 0 | 0 | 4 |
| 8.1 | 2239 | 2180 | 97.4 | 187.2 | 11.6 | 26 | 15 | 9 | 2 | 0 | 7.2 |
| 9.1 | 2115 | 2030 | 96.0 | 159.1 | 12.8 | 32 | 16 | 14 | 2 | 0 | 5 |
| 10.1 | 1875 | 1669 | 89.0 | 106.9 | 15.2 | 19 | 11 | 8 | 0 | 0 | 5.3 |
| 11.1 | 1817 | 1631 | 89.8 | 99.9 | 16.3 | 25 | 17 | 5 | 3 | 0 | 4 |
| 12.1 | 1764 | 1758 | 99.7 | 99.0 | 17.8 | 23 | 14 | 8 | 1 | 0 | 4.3 |
| 13.1 | 1386 | 1293 | 93.3 | 114.3 | 11.3 | 16 | 11 | 5 | 0 | 0 | 7.1 |
| 14.1 | 1267 | 1262 | 99.6 | 107.9 | 11.7 | 20 | 14 | 6 | 0 | 0 | 5.4 |
| Total | 33641 | 31233 | 92.8 | 2476.3 | 12.6 | 430 | 240 | 146 | 39 | 5 | 5.7 |
Both groups contain mating type marker MAT-A.
Size of the assembled scaffold or chromosome corresponding to the respective linkage group.
Part of the assembled scaffold or chromosome covered by the respective linkage groupSupporting Information Legends.
After anchoring the 1st generation map to the physical genome sequences of the parental strains, it covered 55.3% of the genome. After the addition of the 185 new markers, the entire 2nd generation map covered 92.8%, which also increased the map-wide KB/cM level from 5.8 to 12.6 KB/cM (Table 2).
Locating Mating Type and Intersterility Loci
New data increased the observed number of progeny isolates capable of expressing fertility with the intersterility tester strain TC-111-4 (carrying the V2+ allele) from 28 to 42, and those expressing fertility with the tester Sä 16-4 (carrying the P + allele) from 39 to 54. This allowed us to use the 2nd generation linkage map to locate the P-locus to scaffold 2, approximately between positions 2 239 084 and 2 250 770 bp in the H. irregulare genome (version 2, http://genome.jgi-psf.org/Hetan2/Hetan2.home.html). We were also able to position the V2-locus between 1 943 822 and 2 091 761 bp on scaffold 3. The genetic position of the previously mapped S-locus [26], was confirmed and anchored to scaffold 1 between positions 1 981 038 and 1 990 633 bp. The position of mating type locus MAT-A is known from the H. irregulare genome analysis to be located at 1 597 169 bp on scaffold 1 [23], in a poorly mapped area between the linkage groups 1.1 and 1.2. When mapped together with the markers from these groups, the mating type marker was positioned close to this locus in both groups, although the linkage data was insufficient to fuse the entire groups together.
The genome-wide recombination rate, calculated as the mean across all 17 linkage groups, was 12.6 KB/cM (Table 2). The mean for an individual linkage group (LG) (if disregarding the very small LG 2.3) spanned from 8.8 KB/cM (LG 2.2) to 17.8 (LG 12.1). Compared to this, recombination rates around the intersterility loci and mating type locus were in most cases elevated compared to the rest of genome (Figure 1). The putative position for intersterility locus S, on linkage group 1.2, had a recombination rate of 0.6 KB/cM, compared to a mean of 10.2 KB/cM for the linkage group as a whole. The suggested position for intersterility locus P has a recombination rate of 0.8 KB/cM, compared to the mean value of 8.8 for LG 2.2 in general. The mating type locus on LG 1.1, with a group mean of 9.2 KB/cM, has a local recombination rate of 6.8 KB/cM. The locus for intersterility locus V2 has a lower recombination rate than the rest of LG 3.1, 19.2 KB/cM compared to the group mean of 11.3 KB/cM. We also observed another local recombination hot spot close to the mapped position of V2 (8 cM away, between positions 1 908 660 and 1 910 855), with a recombination rate of 0.5 KB/cM. Elevated recombination rates of 1 KB/cM or less occurred between 29 neighbouring markers pairs throughout the linkage map. They were most frequent on LG 2.2 (5 hotspots) and did not occur at all on LGs 2.3, 8.1, 10.1 and 11.1. On LG 1.2, 2.2, and 3.1, containing the intersterility loci S, P and V2, we found 1, 5 and 3 hotspots, respectively (including the hotspots across the intersterility loci).
Discussion
Using an in silico approach, 127 H. irregulare (TC-32-1) and 89 H. occidentale (TC-122-12) AFLP markers were anchored to the genome sequences of both species and incorporated into a 2nd generation linkage map. This total of 240 is considerably less than the 308 AFLP markers present in the 1st generation map, a decrease based on the exposure and omittance of falsely linked markers. Scoring errors and statistical ambiguities are known to cause a certain degree of false positive linkages during compilation of large sets of markers [46]. Data on physical positions is a powerful way to discern a false link from a correct one since the randomly occurring false linkages can easily be identified when a correct, sequence-based order of markers is enforced.
Although the 1st generation map contained an equal amount of markers from each parent, more H. irregulare than H. occidentale markers remain in the 2nd generation map (Table 2). The reason for this is that the H. occidentale genome by necessity was aligned using the H. irregulare genome as reference. The restriction sites of the 20 unanchored TC-122-12 markers probably exist among the 2 625 H. occidentale scaffolds, but some scaffolds might be too different from the H. irregulare genome to be aligned and positioned. Some of the missing markers might also be located in unassembled parts of the H. occidentale genome. Such are likely to exist since the 28.5 MB of the assembled H. occidentale genome represents only 85% of the 33.6 MB of the H. irregulare genome; a discrepancy probably accounted for by transposable elements, which were estimated to comprise 16.2% of the H. irregulare genome [23] and are probably also frequent in H. occidentale.
The 2nd generation map covers 92.8% of the assembled H. irregulare sequence, compared to the 55.3% covered by the 1st generation map. This massively improved coverage originates from the use of microsatellite markers to expand and merge formerly individual groups into larger ones. However, despite their physical closeness, linkage groups 1.1 and 1.2, and 2.1, 2.2 and 2.3, could not be joined. The areas between these groups contain a lot of transposable elements [23], which complicates the acquisition of unique microsatellite markers. If these unmapped areas contain recombinational hotspots or otherwise have high recombination rates, the groups would also be genetically unlinked.
The anchoring of AFLP markers revealed that four of the 39 linkage groups in the 1st generation map [26] contained markers from two separate scaffolds. The other 35 solely consisted of markers from individual scaffolds, but the genetic position (in cM) of the markers never fully correlated to their physical positions (in bp). Incorrect marker orders impair the significance of QTL effects and complicate estimation of QTL sizes, which introduces uncertainty when identifying candidate genes. Our 2nd generation map rectifies this problem by including only 16 markers with a genetic position in conflict with the physical position, and of these, only one is misplaced by a distance of more than two markers (three). The other 414 markers are mapped in concordance with sequence data which enables sharp and accurate QTL mapping. Furthermore, these results advice caution when interpreting QTL data from linkage maps without sequence-anchored markers, in terms of confidence in genome coverage, linkage group composition and marker order.
Of the 17 linkage groups (LG) remaining after the improvement of the map, twelve (LG 3.1–14.1) span almost the entire lengths of their respective scaffolds (Table 2), covering between 89% (LG 10.1) and 99.6% (LG 14.1) of the assembled sequence. None of the markers of these LGs showed any linkage to those of any other group in the 2nd generation map, which suggests that the corresponding scaffolds represent individual chromosomes. This is also validated by the fact that six of the scaffolds have both their telomeric regions sequenced, and that the other six have one telomeric region sequenced [23]. Scaffold 1 may well also represent a chromosome, since (i) it has one sequenced telomeric region, (ii) LG 1.1 and 1.2 together cover 92.1% of its sequence, and (iii) the MAT-A locus links to the downstream end of LG 1.1 and upstream start of LG 1.2. Scaffold 2 is more obscure. Its three linkage groups only cover 79.7% of the sequence and it does not have any telomeric region sequenced. Since groups 2.1–2.3 does not link to any other groups, however, nothing contradicts that the scaffold represents a distinct chromosome. A fourteen chromosome arrangement was also suggested after the whole genome assembly of H. irregulare [23] and corresponds well to previous karyotyping data [47].
Anchoring this linkage map to sequence information was greatly beneficial for the assembling process of the H. irregulare genome. Version 1 of the assembly was constructed without consulting the map and consisted of 38 scaffolds, 18 of which were larger than 300 KB (http://genome.jgi-psf.org/Hetan1/Hetan1.home.html). In version 2 (http://genome.jgi-psf.org/Hetan2/Hetan2.home.html), map-based linkage information improved the assembly considerably. Scaffolds 16 and 12 of version 1 were joined into scaffold 9 of version 2, while scaffolds 11, 15 and 17 were joined into scaffold 4. Similarly, scaffolds 7 and 3 were split and joined into scaffolds 5 and 6. Map-based evidence thus decreased the total number of large scaffolds from 18 to 15. This highlights the usefulness of a high quality linkage map; it is not only a potent tool to target genomic regions governing phenotypic traits, but can also be used to fine-tune genome assemblies and increase our understanding of chromosomal arrangement.
Although the in silico-anchoring of AFLP markers is easiest with assemblies consisting of few, large scaffolds, it is potentially more beneficial for fragmented assemblies since it allows for joining of multiple scaffolds carrying linked markers. Although the number of candidate restriction sites could be more manageable in smaller genomes, the approach is also useful for larger ones; if the AFLP adapters and nucleotide extensions are specific enough to produce a unique marker in vitro, its original locus could also be determined in silico.
In the 1st generation map [26], the loci for MAT-A and V2 could not be positioned, but it was postulated that this was due to insufficient phenotype mapping data or to insufficient map coverage. Remedying these aspects enabled mapping of both loci, and the position for MAT-A (Figure 1) was confirmed by its physical position in the H. irregulare genome version 2 [23]. The molecular mechanism behind the intersterility concept is yet unknown, but since the occurrence of certain intersterility + alleles strictly follow the affinity for certain hosts [22], these traits might either be located physically close to each other or indeed be controlled by the same genes [21]. The putative loci are currently being examined for candidate genes, which will be investigated further using expression studies and gene deletions.
The most common approach to study genome-wide recombination rates is the so-called Marey mapping, i.e. comparing genomic and physical maps [48]. A simplified version of this was applied by running a sliding window across the scaffolds, comparing the distance in cM between pairwise neighbouring markers to their physical positions. Recombination frequencies measured in this way fluctuated a lot across the scaffolds, with in total 29 regional hotspots with a recombination rate below 1 KB/cM distributed over 13 of the 17 scaffolds. Interestingly, some of these hotspots coincided with the position for the intersterility loci S and P, and to a lesser extent with MAT-A. The mapped position for the V2 locus had no increased recombination rate, but just 8 cM away there was a position with a very similar recombination rate to S and P. Since only 42 of the assumed 51 existing isolates carrying the V2+ allele were identified, the current position is not necessary absolute and might in reality correlate to the neighbouring hot spot. The degree of recombination elevation were similar across all intersterility loci compared to the genome average of 12.6 KB/cM; for S, P and V2 respectively 0.6, 0.8 and 0.5 KB/cM, given that the local hot spot close to V2 is relevant. This is comparable to the increased recombination rate around the mating type locus measured in Cryptococcus neoformans, from 13.2 to 0.27 KB/cM [49].
The recombination rate in the region of MAT-A also increased (6.8 KB/cM), but not to the same extent as for the intersterility loci. This could be partially explained by the difficulties in mapping the mating type locus, physically situated in an area containing transposable elements. Affected recombination rates have been described previously for sex-determining regions of fungal genomes. In Cryptococcus, the increased recombination rates were accredited to flanking recombination hot spots on either side of the MAT-A locus, and it was suggested that this served to suppress recombination within the locus itself, as has also been described in Neurospora tetrasperma [50]. Hsueh et al. (2006) further argued the reason for this suppression to be the preservation of MAT as a single unit, as recombination within the locus could generate sterile or self-fertile progeny. The same argument could be made regarding the intersterility loci; if the increased recombination rate stems from flanking hot spots, the locus itself would be inherited as an intact entity. As individuals with disrupted intersterility loci would not carry any + alleles, they would invariably be sterile.
In this study, a virtual AFLP-approach was used to anchor AFLP markers from a previously published linkage map to the physical map. This provided the opportunity to determine the exact sequence for already identified and future QTLs. The reliability of this method was validated by microsatellite markers that confirmed the positions of the anchored AFLP markers. The genome coverage of the map was hence increased from 55.3% to 92.8% of the assembled sequence. This allowed the sharpened positioning of loci for intersterility and mating type, and the discovery that recombination rates were elevated around intersterility loci in particular. The described improvements to the linkage map also suggest that maps derived solely from random fingerprinting data might be fragmented, incomplete and assembled incorrectly, but of great use in genome assemblage following whole genome sequencing efforts.
Supporting Information
Primer combinations for every microsatellite marker or mapped gene in the 2nd generation linkage map of Heterobasidion annosum s.l.
(DOCX)
Acknowledgments
Thanks to Kajsa Himmelstrand, Kerstin Dalman, Rena Gadjieva and Joakim Halldin Stenlid for their assisting lab work in obtaining marker data for sequenced genes.
Funding Statement
This project was funded by the Swedish Foundation for Strategic Research (grant RBb08-0011), Carl Tryggers Foundation (grant CTS 07:271), the Nilsson-Ehle donations (grant 2008-11-12), Anna and Nils Håkanssons Stiftelse (grant 2007-04-12) and CF Lundströms Stiftelse (grant 1347). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, et al. (1995) AFLP: A new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Petro D, Onyeka TJ, Etienne S, Rubens S (2011) An intraspecific genetic map of water yam (Dioscorea alata L.) based on AFLP markers and QTL analysis for anthracnose resistance. Euphytica 179: 405–416. [Google Scholar]
- 3. Lopez-Lavalle LAB, Matheson B, Brubaker CL (2011) A genetic map of an Australian wild Gossypium C genome and assignment of homoeologies with tetraploid cultivated cotton. Genome 54: 779–794. [DOI] [PubMed] [Google Scholar]
- 4. Terashima K, Matsumoto T, Hayashi E, Fukumasa-Nakai Y (2002) A genetic linkage map of Lenintula edodes (shiitake) based on AFLP markers. Mycological Research 106: 911–917. [Google Scholar]
- 5. Ueng P, Zhao Y (2009) Genetic linkage map of Phaeosphaeria nodorum, the causal agent of stagonospora nodorum blotch disease of wheat. Phytopathology 99: S132–S132. [Google Scholar]
- 6. Chang SW, Jung G (2008) The first linkage map of the plant-pathogenic basidiomycete Typhula ishikariensis. Genome 51: 128–136. [DOI] [PubMed] [Google Scholar]
- 7. Kema GHJ, Goodwin SB, Hamza S, Verstappen ECP, Cavaletto JR, et al. (2002) A Combined Amplified Fragment Length Polymorphism and Randomly Amplified Polymorphism DNA Genetic Linkage Map of Mycosphaerella graminicola, the Septoria Triciti Leaf Blotch Pathogen of Wheat. Genetics 161: 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stajich JE, Wilke SK, Ahren D, Au CH, Birren BW, et al. (2010) Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proceedings of the National Academy of Sciences of the United States of America 107: 11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cuomo CA, Gueldener U, Xu JR, Trail F, Turgeon BG, et al. (2007) The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317: 1400–1402. [DOI] [PubMed] [Google Scholar]
- 10.Goodwin SB, Ben M’Barek S, Dhillon B, Wittenberg AHJ, Crane CF, et al. (2011) Finished Genome of the Fungal Wheat Pathogen Mycosphaerella graminicola Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis. Plos Genetics 7. [DOI] [PMC free article] [PubMed]
- 11. Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- 12. Miao H, Zhang SP, Wang XW, Zhang ZH, Li M, et al. (2011) A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 182: 167–176. [Google Scholar]
- 13. Foulongne-Oriol M, Dufourcq R, Spataro C, Devesse C, Broly A, et al. (2011) Comparative linkage mapping in the white button mushroom Agaricus bisporus provides foundation for breeding management. Current Genetics 57: 39–50. [DOI] [PubMed] [Google Scholar]
- 14. Lee J, Jurgenson JE, Leslie JF, Bowden RL (2008) Alignment of Genetic and Physical Maps of Gibberella zeae. Applied and Environmental Microbiology 74: 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Labbé J, Zhang X, Yin T, Schmutz J, Grimwood J, et al. (2008) A genetic linkage map for the ectomycorrhizal fungus Laccaria bicolor and its alignment to the whole-genome sequence assemblies. New Phytologist 180: 316–328. [DOI] [PubMed] [Google Scholar]
- 16. Xu DH, Ban T (2004) Conversion of AFLP markers associated with FHB resistance in wheat into STS markers with an extension-AFLP method. Genome National Research Council Canada 47: 660–665. [DOI] [PubMed] [Google Scholar]
- 17. Brugmans B, van der Hulst RG, Visser RG, Lindhout P, van Eck HJ (2003) A new and versatile method for the successful conversion of AFLP markers into simple single locus markers. Nucleic Acids Research 31: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodward S, Stenlid J, Karjalainen R, Hûttermann A, editors. (1998) Heterobasidion annosum. Biology, Ecology, Impact and Control. Cambridge, UK: CAB International.
- 19.Niemelä T, Korhonen K (1998) Taxonomy of the Genus Heterobasidion In: Woodward S, Stenlid J, Karjalainen R, Hüttermann A, editors. Heterobasidion annosum Biology, Ecology, Impact and Control. Wallingford, UK: CAB International. 27–33.
- 20. Otrosina WJ, Garbelotto M (2010) Heterobasidion occidentale sp. nov. and Heterobasidion irregulare nom. nov. : a disposition of North American Heterobasidion biological species. Fungal Biology 114: 16–25. [DOI] [PubMed] [Google Scholar]
- 21. Chase TE, Ullrich RC (1990) Five genes determining intersterility in Heterobasidion annosum . Mycologia 82: 73–81. [Google Scholar]
- 22.Korhonen K (1978) Intersterility groups of Heterobasidion annosum. Communicationes Instituti Forestalis Fenniae 94: 25 pp.
- 23.Olson Å, Aerts A, Asiegbu FO, Belbahri L, Bouzid O, et al. (2012) Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytologist. [DOI] [PubMed]
- 24. Chase TE, Ullrich RC (1983) Sexuality, distribution, and dispersal of Heterobasidion annosum . Mycologia 75: 825–831. [Google Scholar]
- 25. Stenlid J (1985) Population structure of Heterobasidion annosum as determined by somatic incompatibility, sexual incompatibility and isoenzyme patterns. Canadian Journal of Botany 63: 2268–2273. [Google Scholar]
- 26. Lind M, Olson Å, Stenlid J (2005) An AFLP-marker Based Genetic Linkage Map of Heterobasidion annosum Locating Intersterility Genes. Fungal Genetics and Biology 42: 519–527. [DOI] [PubMed] [Google Scholar]
- 27. Olson Å, Lind M, Stenlid J (2005) In vitro test of virulence in the progeny of a Heterobasidion interspecific cross. Forest Pathology 35: 321–331. [Google Scholar]
- 28. Olson Å (2006) Genetic linkage between growth rate and the intersterility genes S and P in the basidiomycete Heterobasidion annosum s.lat. . Mycological Research 110: 979–984. [DOI] [PubMed] [Google Scholar]
- 29. Lind M, Dalman K, Stenlid J, Karlsson B, Olson Å (2007) Identification of quantitative trait loci affecting virulence in the basidiomycete Heterobasidion annosum s.l. . Current Genetics 52: 35–44. [DOI] [PubMed] [Google Scholar]
- 30. Samils N, Olson Å, Stenlid J (2008) The capacity in Heterobasidion annosum s.l. to resist overgrowth by the biocontrol agent Phlebiopsis gigantea is a heritable trait. Biological Control 45: 419–426. [Google Scholar]
- 31. Lind M, Stenlid J, Olson Å (2007) Genetics and QTL mapping of somatic incompatibility and intraspecific interactions in the basidiomycete Heterobasidion annosum s.l. . Fungal Genetics and Biology 44: 1242–1251. [DOI] [PubMed] [Google Scholar]
- 32. Olson Å, Stenlid J (2001) Mitochondrial control of fungal hybrid virulence. Nature 411: 438. [DOI] [PubMed] [Google Scholar]
- 33. Cox M, Peterson D, Biggs P (2010) SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zerbino D, Briney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rombauts S, van de Peer Y, Rouzé P (2003) AFLPinSilico, simulating AFLP fingerprints. Bioinformatics (Oxford) 19: 776–777. [DOI] [PubMed] [Google Scholar]
- 36. Lim S, Notley-McRobb L, Lim M, Carter D (2004) A comparison of the nature and abundance of microsatellites in 14 fungal genomes. Fungal Genetics and Biology 41: 1025–1036. [DOI] [PubMed] [Google Scholar]
- 37.Gardes M, Bruns TD (1996) ITS-RFLP matching for identification of fungi. In: Clapps JP, editor. Species Diagnostics Protocols, PCR and Other Nucleic Acid Metods. Totowa, New Jersey, USA: Humana press.
- 38. Adomas A, Eklund M, Johansson M, Asiegbu FO (2006) Identification and analysis of differently expressed cDNAs during nonself-competitive interaction between Phlebiopsis gigantea and Heterobasidion parviporum. FEMS Microbiology Ecology 57: 26–39. [DOI] [PubMed] [Google Scholar]
- 39. Hutchison E, Brown S, Tian C, Glass NL (2009) Transcriptional profiling and functional analysis of heterokaryon in Neurospora crassa reveals that ROS, but not metacaspases, are associated with programmed cell death. Microbiology 155: 3957–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iakovlev A, Olson Å, Elfstrand M, Stenlid J (2004) Differential gene expression during interactions between Heterobasidion annosum and Physisporinus sanguinolentus . FEMS Microbiology Letters 241: 79–85. [DOI] [PubMed] [Google Scholar]
- 41. Karlsson M, Olson Å, Stenlid J (2003) Expressed sequences from the basidiomycetous tree pathogen Heterobasidion annosum during early infection of scots pine. Fungal Genetics and Biology 39: 51–59. [DOI] [PubMed] [Google Scholar]
- 42. van der Nest MA, Steenkamp E, Slippers B, Mongae A, van Zyl K, et al. (2011) Gene expression associated with vegetative incompatibility in Amylostereum areolatum . Fungal Genetics and Biology 48: 1034–1043. [DOI] [PubMed] [Google Scholar]
- 43. Li Y, Chu Z, Liu X, Hao D (2010) A Cost-effective High-resolution Melting Approach using the EvaGreen Dye for DNA Polymorphism Detection and Genotyping in Plants. Journal of Integrative Plant Biology 52: 1036–1042. [DOI] [PubMed] [Google Scholar]
- 44.Van Ooijen J, Voorrips R (2001) JoinMap 3.0, Software for the calculation of genetic linkage maps. Wageningen, the Netherlands: Plant Research International.
- 45. Voorrips R (2002) MapChart: Software for the graphical presentation of linkage maps and QTLs The Journal of Heredity. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- 46.Anderson M, Kasuga T, Mitchelson K (1993) A partial physical karyotype of Heterobasidion annosum. In: Johansson M, Stenlid J, editors. Proceedings of the Eighth International Conference on Root and Butt Rots. Wik and Haikko: International Union of Forestry Research Organisations (IUFRO). 303–313.
- 47. Gale LR, Bryant J, Calvo S, Giese H, Katan T, et al. (2005) Chromosome Complement of the Fungal Plant Pathogen Fusarium graminearum Based on Genetic and Physical Mapping and Cytological Observations. Genetics 171: 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chakravarti A (1991) A graphical representation of genetic and physical maps: the Marey map. Genomics 11: 219–222. [DOI] [PubMed] [Google Scholar]
- 49. Hsueh Y-P, Idnurm A, Heitman J (2006) Recombination Hotspots Flank the Cryptococcus Mating-Type Locus: Implications for the Evolution of a Fungal Sex Chromosome. PLoS Genet 2: e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gallegos A, Jacobson D, Raju N, Skupski M, Natvig DO (2000) Suppressed recombination and pairing anomaly on the mating-type chromosome of Neurospora tetrasperma . Genetics 154: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer combinations for every microsatellite marker or mapped gene in the 2nd generation linkage map of Heterobasidion annosum s.l.
(DOCX)