Abstract
Remote Ischemic Preconditioning (RIPC) induced by brief episodes of ischemia of the limb protects against multi-organ damage by ischemia-reperfusion (IR). Although it has been demonstrated that RIPC affects gene expression, the proteomic response to RIPC has not been determined. This study aimed to examine RIPC induced changes in the plasma proteome. Five healthy adult volunteers had 4 cycles of 5 min ischemia alternating with 5 min reperfusion of the forearm. Blood samples were taken from the ipsilateral arm prior to first ischaemia, immediately after each episode of ischemia as well as, at 15 min and 24 h after the last episode of ischemia. Plasma samples from five individuals were analysed using two complementary techniques. Individual samples were analysed using 2Dimensional Difference in gel electrophoresis (2D DIGE) and mass spectrometry (MS). Pooled samples for each of the time-points underwent trypsin digestion and peptides generated were analysed in triplicate using Liquid Chromatography and MS (LC-MS). Six proteins changed in response to RIPC using 2D DIGE analysis, while 48 proteins were found to be differentially regulated using LC-MS. The proteins of interest were involved in acute phase response signalling, and physiological molecular and cellular functions. The RIPC stimulus modifies the plasma protein content in blood taken from the ischemic arm in a cumulative fashion and evokes a proteomic response in peripheral blood.
Introduction
Ischemic preconditioning is a potent innate mechanism observed in many species whereby cells develop tolerance to ischemia-reperfusion (IR) injury when exposed to controlled periods of transient, sub-lethal ischemia prior to a prolonged ischaemia [1], [2] . However, local ischemic preconditioning is not clinically applicable to most patients. During the past decade, a simple technique of preconditioning has been developed with the potential for rapid translation into clinical practice [3].
Remote ischemic preconditioning (RIPC) is a phenomenon where brief episodes of ischemia of one tissue (e.g., skeletal muscle) protect against IR injury in an organ at a remote location [4]. RIPC has great potential for clinical application as it can be applied non-invasively using a standard blood pressure cuff to induce cycles of IR to skeletal muscle [3], [5]. We have previously demonstrated that brief episodes of limb ischemia protected the donor heart after transplantation [6], providing multi-organ protection against cardiopulmonary bypass-induced tissue injury [7] and effective protection during evolving myocardial infarction [8]. We have also demonstrated that IR and RIPC induced a genomic response in the myocardium and circulating leukocytes of experimental animals and in humans [9]–[11]. Additionally, we observed that RIPC decreased expression of kinin receptors [12], neutrophil adhesion and also modified the functional responses of human neutrophils [13]. We have also applied RIPC to clinical practice and demonstrated, in a randomized controlled trial, the benefits of the RIPC in children undergoing cardiac surgery [14]. A recent large randomized controlled trial further demonstrated a beneficial effect of the RIPC, as a complement to angioplasty, on myocardial salvage in patients with acute myocardial infarction [5].
Although the clinical benefits of RIPC are apparent, the mechanism underlying this protection remains unknown. Others and we have previously suggested the existence of a blood-borne effector of the RIPC stimulus that is transferred from the transiently ischemic limb to remote organs rendering them resistant to prolonged ischemia [6], [15]. Furthermore, it appears that transient limb ischemia not only remotely preconditions through a humoral mechanism, but also that plasma transfer from the ischemic limb of one species may protect against IR injury in other species [15].
It is intuitive to believe that the observed changes in gene expression in response to the RIPC [9], [10] will result in changes protein expression. However, the proteomic response to RIPC has not been studied to date. The purpose of this study was to determine 1) if the plasma from the transiently ischemic limb has a modified proteomic profile, 2) if the proteomic changes are cumulative with each subsequent episode of transient ischemia, and 3) if the RIPC stimulus evokes a global proteomic response early and late after the induction of the transient limb ischemia.
Materials and Methods
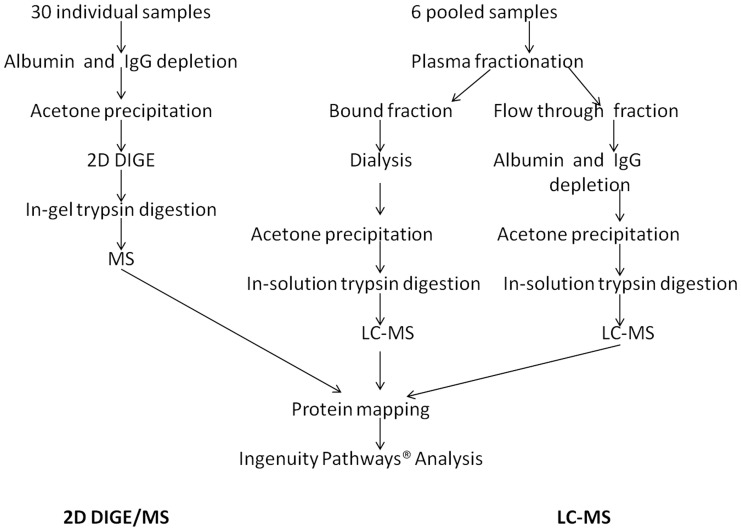
This study was approved by the Royal Children’s Hospital Ethics in Human Research Committee (#29007) and written informed consent was obtained from the participants. Five healthy adult male volunteers 36.2±6.3 (mean ± SD), not on any medications were fasted overnight and underwent the RIPC protocol. The protocol consisted of 4 cycles of 5 minutes of ischemia alternating with 5 minutes of reperfusion. Ischemia was induced by inflating a standard blood pressure cuff to a pressure exceeding systolic, as previously described [14]. Venous blood samples were collected from the same arm at 6 time-points: baseline, at the beginning of each period of re-perfusion and then at 15 minutes and 24 hours following application of the RIPC stimulus. Blood samples were collected in S-Monovette® tubes (Sarstedt, Australia), containing 1 volume of citrate per 9 volumes of blood. The samples were centrifuged at 3000 rpm for 10 min at 10°C (Megafuge 1.0R, Heraeus), the plasma was collected and stored at −80°C. The samples were then analysed using two methods described below ( Figure 1 ).
Figure 1. Two complementary proteomic methods used to assess RIPC induced changes in the plasma proteome.
2D DIGE- 2Dimensional Difference in gel electrophoresis, LC– Liquid chromatography, -MS-mass spectrometry.
Two-Dimensional Difference in Gel Electrophoresis (2D-DIGE) and Mass Spectrometry
The analysis was conducted on 30 individual samples (6 samples from 5 individuals).
Albumin and IgG depletion was performed using the albumin IgG depletion kit (GE Healthcare, Australia). The remaining proteins were precipitated using acetone precipitation, as specified in the depletion kit and resuspended in buffer containing 7 mol/L urea, 2 mol/L thiourea, 4% 3-[3-cholaamidopropyl]-1-propane-sulfonate and 30 mmol/L Tris. The protein content of each sample was quantified using the Bradford assay (Bio-Rad, Hercules, CA, USA) and bovine serum albumin standards [16].
The internal standard, consisting of an equal amount of each of the 30 samples, was labelled with Cyanine 2 (Cy2) fluorescent dye (GE Healthcare, Australia) and run on each gel to control for gel-to-gel variation. Each sample was randomised to be labelled with either Cy 3 or Cy5 dye and then randomized to 15 gels. The Cy2, Cy3 and Cy5 samples (50 mg of sample/400 pmol of Cy dye) for each gel were pooled and loaded onto the Immobilized pH Gradient (IPG) strip. One 24 cm, pH 3–11 strip per gel was rehydrated with 15 ml IPG buffer and 3 ml DeStreak solution (GE Healthcare, Australia). Proteins were separated based on isoelectric point (first dimension) and molecular weight (second dimension) using previously published methodology [16]. Gels were scanned using the Typhoon Trio variable mode imager (GE Healthcare, Australia) [16]. Data obtained from the scanning were quantified using DeCyder software version 6.5 (GE Healthcare, Australia). The Differential In-gel analysis (DIA) was used to optimize spot detection. The Biological Variation Analysis (BVA) module was used for analysis of each sample according to the corresponding time point. The filtering parameters were set to determine protein spots that had a p-value of <0.05 and a 1.5-fold change in abundance between the time points.
Proteins of interest were excised from the gels robotically using the Ettan Spot-picker (GE Healthcare, Australia) and prepared for in-gel trypsinolysis as previously described [16]. Gel plugs were consecutively washed with 25 mM NH3HCO3 followed by 50% v/v acetonitrile for 15 min each. Following dehydration by incubation at 37°C for 30 min, gel plugs were incubated with modified porcine trypsin in 25 mM NH3HCO3 (Promega) (pH 9, 37°C, 15 h). Trifluoroacetic acid (0.5% w/v) was subsequently added to neutralise the trypsin. The digested proteins were concentrated directly onto a thin layer affinity matrix solution of α-cyano-4-hydroxycinnamic acid for analysis by MALDI-TOF MS. The MS reflector mode was used to generate a protein mass fingerprint for the identification of each protein (4700 Proteomics Analyzer, Applied Biosystems, USA), operating at a resolution of 10,000–15,000 FWHM (Full Width at Half Maximum). Reordered in positive reflector mode at a laser intensity of 2950, spectra were acquired at 200 Hz using a YAG laser (335 nm). A mass filter that excluded matrix cluster ions and trypsin autolysis peaks was applied. Ten of the most intense peptide ions were selected for further MS analysis (MS/MS). All MS/MS data from the TOF-TOF was acquired using a default 1 kV method at laser energy 3000–3500. The PMF and MS/MS data were combined and submitted for database searching as described in the protein mapping section below. Protein identity was listed for samples that gave a significant (P<0.05). A peptide mass tolerance of 100 ppm and up to 1 missed cleavage allowed when searching against all databases.
Liquid Chromotography (LC) and Mass Spectrometry
This method was applied to better assess the heparin-bound proteins. The analysis was conducted on 6 pooled samples from 5 individuals taken at 6 time points. Plasma fractionation was performed using the ÄKTA™ Fast Protein Liquid Chromatography (Amersham Pharmacia Biotech AB, Uppsala, Sweden). The plasma proteins were separated into two fractions, based on their affinity to heparin [17]. One fraction contained proteins that bind heparin (bound fraction) and the other fraction contained those that do not bind heparin (flow through fraction) [17]. Plasma samples were diluted 1∶3 in 50 mMTris-HCl, 0.1 M NaCl pH 7.5 (Kjellberg 2006) and passed through a 0.22 µM spin filter (Agilent Technologies, Australia) by centrifugation for 5 min. The samples were then fractionated in duplicate runs by injecting 400 µL of the sample into the AKTA system, through a 1 mL Hi-Trap Heparin column (GE healthcare, Australia) at a flow rate of 1 ml/min for 5 mins to collect the flow through fraction. The bound fraction was then eluted off the column under high salt conditions with 50 mM Tris-HCl, 3.0 M NaCl pH 7.5 for 13 mins (Kiellberg 2006). Between each sample run, the column was re-equilibrated with 50 mMTris-HCl, 0.1 M NaCl pH 7.5 for 7 minutes. Samples from the bound fraction were dialysed against phosphate buffered saline to reduce the salt concentration in preparation for acetone precipitation. Dialysis was performed for 48 hours with a change of buffer at 24 hours, with 25 mm×16 mm cellulose dialysis tubing (Sigma Aldrich, St Louis, USA) [18].
Albumin and IgG were depleted from the flow through fraction using the Albumin and IgG removal kit (GE Healthcare, Australia). This was performed to increase the probability of detecting low abundance proteins that are not bound to heparin. The bound fraction was not subjected to this depletion protocol as albumin and IgG do not bind to heparin and are therefore not present in this fraction.
Both the bound fraction and the flow through fraction underwent acetone precipitation and quantification [16]. In-solution trypsin digestion was performed and samples prepared for MS using a standard protocol where four volumes of ice cold acetone were added to the samples and precipitation was carried out overnight at −20°C. Protein pellets were obtained by centrifugation at 13 000 g for 20 mins at 8°C and were resuspended in 6M Urea, 100 mM Tris buffer. The protein content of each sample was quantified using the Bradford assay (Bio-Rad, Hercules, CA, USA) by comparing against a standard curve of bovine serum albumin concentration [19]. In-solution trypsin digestion was performed on 50 µg of protein from each sample. The samples were reduced with 10 mM dithiothreitol for one hour, followed by alkylation with 55 mM iodoacetamide for one hour. The concentration of urea was reduced to <1M by diluting the sample with 0.4M Tris buffer at pH 7.8. Sequencing grade porcine trypsin (Promega, Madison, WI, USA) was added at a ratio of 1∶20 and trypsin digestion then carried out overnight at 37°C. The reaction was stopped by titration with concentrated acetic acid until the pH was lower than pH 6.
Following trypsin digestion, samples were passed through Oasis HLB extraction cartridges (Waters, Ireland) preconditioned with methanol and equilibrated with 2% acetonitrile and 0.1% Trifluroacetic acid (TFA). Bound peptides were first eluted with 80% acetonitrile containing 0.1% TFA, followed by 100% acetonitrile and 0.1% TFA. The combined eluant was lyophilised by freeze drying, after which each was reconstituted in 200 µL of 0.1% formic acid in preparation for mass spectrometry.
LC MS/MS was carried out on a LTQ Orbitrap Velos (Thermo Scientific, West Palm Beach, FL, USA) equipped with a nanoelectrospray interface coupled to an Ultimate 3000 RSLC nanosystem (Dionex, Sunnyvale, CA, USA). The nanoLC system used an Acclaim Pepmap nano-trap column (Dionex – C18, 100 Å, 75 µm×2 cm) and an Acclaim Pepmap RSLC analytical column (Dionex - C18, 100 Å, 75 µ m×15 cm). Typically for each LCMSMS experiment 1 µl of each peptide preparation, equating to 250 ng total peptide, was loaded onto the enrichment (trap) column followed by separation and elution of peptides from the analytical column employing a gradient from 3% to 45% acetonitrile over 90 minutes. The LTQ-Orbitrap Velos mass spectrometer was operated in the data dependent mode with nano ESI spray voltage of +1.6 kv, capillary temperature of 250°C and S-lens RF value of 60%. All spectra were acquired in positive mode with full scan MS spectra scanning from m/z 300–2000 in the flight time mode at 60,000 resolution after accumulating to a target value of 1.00e6 with maximum accumulation of 500 ms. The 8 most intense peptide ions with charge states ≥2 were isolated at a minimum threshold value of 2000 and fragmented by low energy collision induced dissociation (CID) with normalized collision energy of 35, activation Q of 0.25 and activation time of 10 ms. A dynamic exclusion of 1 repeat over 10 sec with exclusion duration of 15 sec was set. At all times, monoisotopic precursor selection was enabled. Each sample was run in triplicate with 2 blank injections between each triplicate set to minimize the effect of sample carryover [20]. Data processing was carried out using Expressionist Refiner MS (Genedata, Basel, Switzerland) to align MS data, carry out noise reduction, and for peak extraction (clustering). Clustering of MS/MS spectra was employed to identify the spectra of the same peptide (from triplicate runs) and to replace them with a single representative spectrum. Once clustered, peak area intensity measurements of precursor ions were extracted and analysed for statistical relevance using Genedata Analyst to compare each of the 3 samples collected post ischemia and 15 minutes and 24 hours thereafter with the baseline sample. Subsequently, peptides showing up or down-regulated expression (p<0.001 above a computer generated false discovery rate) across the time-points were collated into lists for identification using a targeted MS/MS approach. For biomarker discovery using targeted mass spectrometry analysis, the mass spectrometer was operated in the data-dependant mode as described above with the following modifications. The 10 most intense peptide ions with charge states ≥2 were isolated at a minimum threshold value of 2000 from an assigned parent list. A dynamic exclusion of 4 repeats over 30 sec with exclusion duration of 15 sec was set.
The MS data was loaded onto Proteome Discoverer 1.2 software suite (Thermo Scientific, West Palm Beach, FL, USA) and submitted to Mascot v.2.2.04 (Matrix Science, London, UK) www.matrixscience.com) to match against the National Centre for Biotechnology Information (NCBInr), Bethesda, US database. An initial filter of precursor mass was set between 300 to 6000 Da. The peptide mass tolerance was set to 20 ppm and 0.8 Da for MS/MS fragmentation ions Searches were carried out on the latest version of the NCBInr human database (National Centre for Biotechnology Information, Bethesda, US). Enzyme specificity was trypsin with a maximum of 2 missed cleavages. Cysteine carbaidomethylation (+57.0215 Da) and methionine oxidation (+15.9949 Da) were set as the fixed and variable modification respectively for all searches. ESI-FTICR was set as the default instrument search setting. All the spectra were searched against the decoy database to achieve a targeted false discovery rate of 1%. Only those peptides that matched the database with medium (FDR <0.05) or high confidence (FDR <0.01), ie protein score greater than 40, with spectra that matched the original data analysis for fragmentation pattern, retention time and mass to charge ratio were considered when assigning a positive match. Individual MS/MS spectra from the targeted runs within a precursor tolerance of 2 ppm and maximum R/T difference of 1.5minutes were merged (clustered) into single representative spectrum.
Results
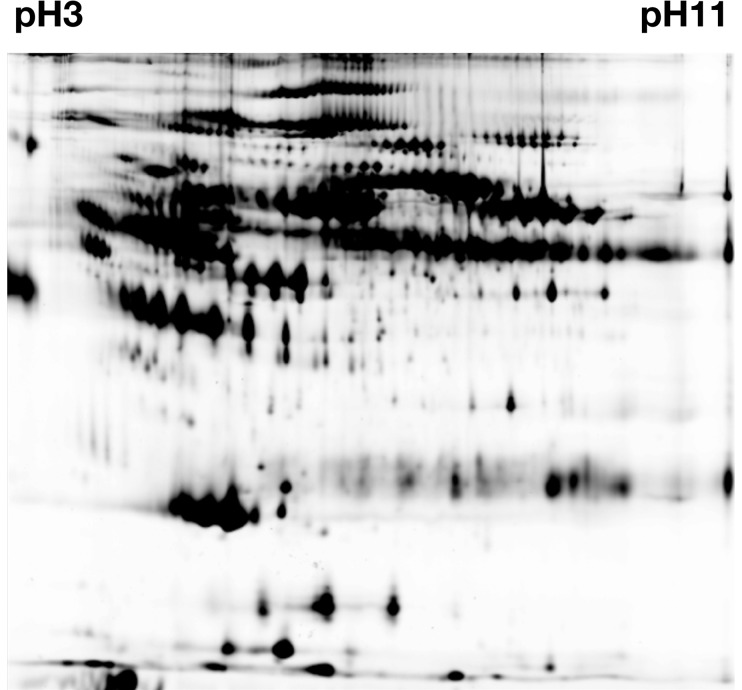
Using the 2D DIGE ( Figure 2 ) with individual plasma samples, 33 spots were determined to have changed significantly in response to RIPC, p<0.05. From these protein spots, 6 proteins were successfully identified by MS and are presented in Table 1 .
Figure 2. The protein pattern from a representative 2D-DIGE gel of human plasma proteins. pH 3 to pH 11 - left to right.
Table 1. Significantly changed proteins 2D DIGE/MS.
| Accession number | Protein | Protein score□ | p-value(t-test) | Average ratio | Main function |
| gi 178751 | α2-antiplasmin precursor | 64 | 0.046 | 1.37 | Serine protease inhibitor |
| gi 4557321 | Apolipoprotein A-1* | 197 | 0.045 | −1.21 | Lipid transport |
| gi 8101268 | Complement C3 | 393 | 0.0047 | 1.46 | Immune response |
| gi 223002 | Fibrin beta* | 294 | 0.028 | 1.18 | Haemostasis |
| gi 223170 | Fibrinogen gamma* | 198 | 0.018 | 1.24 | Haemostasis |
| gi 8853069 | Vitronectin precursor | 120 | 0.014 | −1.32 | Cell adhesion |
Proteins that were also found to change significantly using LC-MS.
□The protein score indicates the confidence with which the proteins identified match the NCBInr human protein database. Only scores greater than 40 were considered to match with sufficient confidence. Average ratio indicates the degree of difference in the abundance between two protein spot groups. Values below zero indicate a down-regulation, whereas, values greater than zero indicate up-regulation.
Using LC-MS analysis, 806 peptides were differentially expressed compared with the baseline sample (p<0.001), and of these, 133 (16.5%) peptides were successfully mapped to 48 known proteins in the NCBInr database ( Table 2 ). The remaining peptides could not be matched to proteins currently available in the database.
Table 2. Significantly changed proteins using LC-MS.
| Protein | Cluster number* | Accession number | Protein | Protein score | Main function |
| 1 | 03064; 03114 | gi152207506 | Alpha-1-antitrypsin | 50.52 | Major plasma protease inhibitor |
| 2 | 0355; 01053; 01172; 01516; 01772; 02364; 02525; 02607; 02613; 02623; 02632; 03082; 03101; 03169; 03213 | gi3212456 | Albumin | 1409.65 | Maintenance of osmotic pressure (carrier protein) |
| 3 | 03271; 03316; 03379 | gi4502027 | Albumin pre-proprotein | 3915.51 | Albumin synthesis |
| 4 | 0851 | gi4502067 | Alpha-1-microglobulin/Bikunin precursor | 760.52 | Trypsin inhibitor |
| 5 | 02542; 02625 | gi2098275 | Amyloidogenic TransthyretinVariants | 717.28 | Molecular Transport |
| 6 | 1075 | gi999513 | Antithrombin Iii ComplexChain A, | 1280.26 | Protease inhibition |
| 7 | 00345; 00421; 00991; 01738; 02127; 02426; 03248; 03288; 03368 | gi90108664 | Apolipoprotein A-I | 221.85 | Lipid Transport |
| 8 | 01759; 00319; 01798; 02803; 03173 | gi24987503 | Apolipoprotein A-Ii | 605.36 | Lipid Transport |
| 9 | 03249 | gi619383 | Apolipoprotein D | 476.73 | Lipid Transport |
| 10 | 0822 | gi6573461 | Apolipoprotein H | 1950.32 | Lipid Transport |
| 11 | 03243 | gi4262120 | Beta-globin | 119.95 | Haemoglobin synthesis |
| 12 | 0393 | gi218511956 | Complement C1r | 591.36 | Immune response |
| 13 | 1014; 0920 | gi81175238; gi1314244 | Complement C4B | 2498.95 | Immune response |
| 14 | 0868 | gi21730336 | Complement C8 gamma | 564.78 | Immune response |
| 15 | 1056; 0797; 0524; 0480; 0412; 0230; 0102; 0681 | gi119625338 | Fibrin beta | 321.18 | Haemostasis |
| 16 | 0781; 0400 | gi223170 | Fibrinogen gamma | 395.17 | Haemostasis |
| 17 | 1015; 0927; 0853 | gi109658664 | Fibronectin 1 | 2796.95 | Endothelial cell activation |
| 18 | 1072; 0237 | gi4504165 | Gelsolin precursor | 1365.75 | Actin binding |
| 19 | 01892; 02104 | gi169791771 | Haemoglobin | 470.28 | Oxygen binding |
| 20 | 01942 | gi63080988 | Haemoglobin alpha-2globin mutant | 470.28 | Oxygen binding |
| 21 | 00935 | gi47679339 | Haemoglobin beta | 110.75 | Oxygen binding |
| 22 | 03201; 02348; 02396 | gi4826762; gi229323; gi296653 | Haptoglobin | 653.78 | Haemoglobin binding |
| 23 | 02059 | gi45580723 | Haptoglobin 2-alpha | 400.55 | Acute phase response |
| 24 | 01237 | gi119589124 | Hemopexin, isoform | 504.33 | Heme binding |
| 25 | 0900; 0793 | gi4504489 | Histidine-rich glycoproteinprecursor | 1076.52 | Protein binding |
| 26 | 01381 | gi229536 | Immunoglobulin A Light chain | 439.93 | Acute phase response |
| 27 | 02489; 03175 | gi8569502 | Immunoglobulin G-1 (Fc Fragment) | 1139.00 | Acute phase response |
| 28 | 01081; 01369 | gi184747 | Immunoglobulin G-1 heavy chain constant region | 353.79 | Acute phase response |
| 29 | 02730 | gi25987833 | Immunoglobulin G-2 heavy chain constant region | 427.39 | Acute phase response |
| 30 | 02372 | gi311771988 | Immunoglobulin G-Aptamer Complex | 570.83 | Acute phase response |
| 31 | 0653; 00247 | gi2414492 | Immunoglobulin heavy chainconstant region | 275.38 | Acute phase response |
| 32 | 0937 | gi553485 | Immunoglobulin kappa chainvariable region | 117.71 | Acute phase response |
| 33 | 0777 | gi3328006 | Immunoglobulin light chainvariable region | 92.82 | Acute phase response |
| 34 | 1049; 0875; 0770; 03057 | gi166007160 | Immunoglobulin M | 840.10 | Acute phase response |
| 35 | 0884; 0779; 0218 | gi4467842 | Immunoglobulin M heavy chain | 105.89 | Acute phase response |
| 36 | 0942; 0925; 0421; 0356; 0344 | gi55958063 | Inter-alpha (globulin) inhibitor H2 | 1816.20 | Protease inhibition |
| 37 | 1024; 0892; 0871; 0801; 0762; 0662 | gi225311 | Lipoprotein B100 | 5942.27 | Lipid transport |
| 38 | 0374 | gi156616294 | N-acetylmuramoyl-L-alanineamidase precursor | 705.96 | Peptidoglycan biosynthesis |
| 39 | 0465 | gi160877748 | Neuropilin-1 B1 Domain In Complex With A Vegf-Blocking Fab, Chain L | 909.96 | Protein signalling |
| 40 | 0994; 0889; 0632; 0348; 0168 | gi8569387 | P14-Fluorescein-N135q-S380c-Antithrombin-Iii, Chain I | 1280.26 | Protease inhibition |
| 41 | 0519 | gi229528 | Protein Len, Bence-Jones | 687.73 | Immune response |
| 42 | 03436 | gi229526 | Protein Rei, Bence-Jones | 558.14 | Immune response |
| 43 | 01545 | gi223069 | Protein Tro alpha 1 H | 707.61 | Immune response |
| 44 | 00132; 00479; 01819; 00088; 00270; 00395; 00603; 00910; 00978; 01032; 01972; 01602; 01127; 01975; 02244; 03107; 3362 | gi110590597; gi194383506; gi110590599 | Transferrin | 3130.44 | Iron binding |
| 45 | 01415 | gi1881852 | Sry-related HMG box gene | 101.3 | DNA binding |
| 46 | 02051; 02692 | gi18655424 | Vitamin D Binding Protein | 994.05 | Vitamin D sterol transport |
| 47 | 03398 | gi139641 | Vitamin D-binding protein precursor | 1023.55 | Vitamin D sterol transport |
| 48 | 02516 | gi4699583 | Zinc-Alpha-2-Glycoprotein | 145.06 | Lipid transport |
Cluster number refers to the number alocated to each peptide fragment in the Genedata software. Accession number refers to the corresponding protein from the NCBInr human protein database. Protein score is a score assigned by the Proteome Discoverer software to indicate the confidence with which the proteins identified match the NCBInr human protein database. Only Protein scores greater than 40 were considered to match with sufficient confidence.
- Each cluster number equals a unique piptide identified for the particular protein.
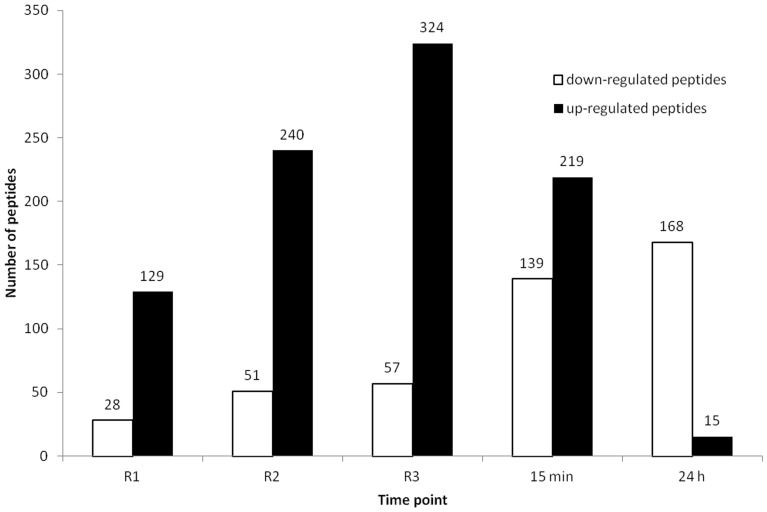
The number of up-regulated peptides increased with reperfusion ( Figure 3 ), and the number of up-regulated peptides peaking at 324 peptides following the third cycle of ischemia. Similarly, the number of down-regulated peptides increased steadily throughout the RIPC protocol, with the highest number of down-regulated peptides observed 24 hours following application of the RIPC stimulus.
Figure 3. The number of peptides that changed significantly compared to the baseline.
R1, R2 and R3 refer to the blood samples taken immediately after the first, second and third period of transient limb ischemia catching the blood coming from the ischemic arm at the beginning of each cycle of reperfusion (R), demonstrating a cumulative proteomic response to the RIPC stimulus. The remaining two samples were taken from the same arm at 15 minutes and 24 hours after completion of the RIPC stimulus, demonstrating an early and late global proteomic response to the RIPC stimulus.
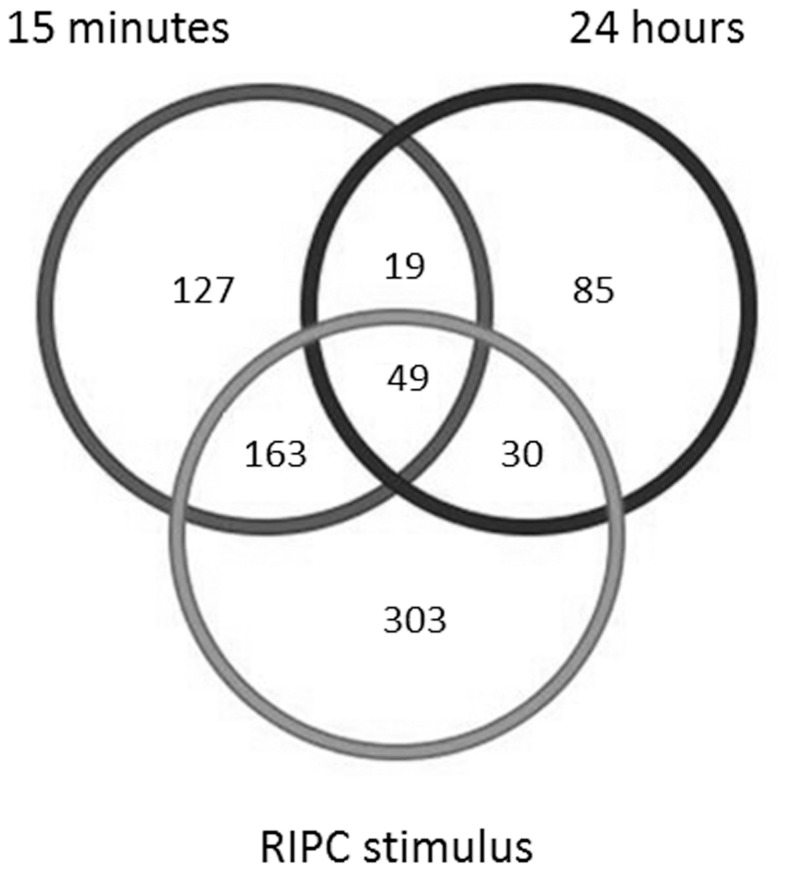
The number of peptides that changed significantly compared to the baseline sample during the RIPC protocol as well as at 15 minutes and 24 hours after the RIPC stimulus is presented in Figure 4 . Proteins that were differencially expressed at the various timepoints are shown in Table 3 , 4 and 5 .
Figure 4. Venn diagram demonstrating the number of peptides that changed significantly during transient limb ischemia (combining all R1, R2, and R3 reperfusion periods), as well as at 15 minutes and 24 hours after the RIPC stimulus, compared with the baseline sample.
Table 3. Differentially expressed proteins coming from the ischemic arm demonstrating up regulation and down regulation.
| Proteins up regulated | Proteins down regulated |
| α2-antiplasmin precursor | Alpha-1-microglobulin/Bikunin precursor |
| Albumin | Antithrombin Iii Complex, Chain A |
| Albumin pre-proprotein | Apolipoprotein H |
| Alpha-1-antitrypsin | Complement C1r |
| Amyloidogenic Transthyretin Variants | Complement C4B |
| Apolipoprotein A-I | Complement C8 Gamma |
| Apolipoprotein A-Ii | Gelsolin precursor |
| Apolipoprotein D | Histidine-rich glycoprotein precursor |
| Beta-globin | Immunoglobulin heavy chain constant region |
| Fibrin beta | Immunoglobulin light chain variable region |
| Fibronectin 1 | Immunoglobulin M |
| Haemoglobin | Immunoglobulin M heavy chain |
| Haemoglobin alpha-2 globin mutant | N-acetylmuramoyl-L-alanine amidase precursor |
| Haemoglobin beta | Neuropilin-1 B1 Domain In Complex With A Vegf-Blocking Fab, Chain L |
| Hemopexin, isoform | P14-Fluorescein-N135q-S380c-Antithrombin-Iii Chain I, |
| Haptoglobin | Protein Len, Bence-Jones |
| Haptoglobin 2-alpha | Vitronectin precursor |
| Immunoglobulin A Light chain | |
| Immunoglobulin G-Aptamer Complex | |
| Immunoglobulin G-1 (Fc Fragment) | |
| Immunoglobulin G-1 heavy chain constant region | |
| Immunoglobulin G-2 heavy chain constant region | |
| Immunoglobulin kappa chain variable region | |
| Inter-alpha (globulin) inhibitor H2 | |
| Lipoprotein B100 | |
| Protein Rei, Bence-Jones | |
| Protein Tro alpha 1 H | |
| Sry-related HMG box gene | |
| Transferrin | |
| Vitamin D Binding Protein | |
| Vitamin D-binding protein precursor | |
| Zinc-Alpha-2-Glycoprotein |
Table 4. Differentially expressed proteins in response to the RIPC stimulus demonstrating up regulation and down regulation during the early response (15 min).
| Proteins up regulated | Proteins down regulated |
| Albumin | Alpha-1-microglobulin/Bikunin precursor |
| Albumin pre-proprotein | Antithrombin Iii Complex, Chain A |
| Alpha-1-antitrypsin | Apolipoprotein H |
| Amyloidogenic Transthyretin Variants | Complement C4B |
| Apolipoprotein A-I | Complement C8 |
| Apolipoprotein A-Ii | Complement C1r |
| Apolipoprotein D | Gelsolin precursor |
| Beta-globin | Histidine-rich glycoprotein precursor |
| Complement C3 | Immunoglobulin heavy chain constant region |
| Fibrinogen gamma | Immunoglobulin light chainvariable region |
| Fibrin beta | Immunoglobulin kappa chain variable region |
| Fibronectin 1 | Immunoglobulin M |
| Haemoglobin | Immunoglobulin M heavy chain |
| Haemoglobin alpha-2 globin mutant | Lipoprotein B100 |
| Hemopexin, isoform | N-acetylmuramoyl-L-alanine amidase precursor |
| Haptoglobin | Neuropilin-1 B1 Domain In Complex With A Vegf-Blocking Fab, Chain L |
| Haptoglobin 2-alpha | P14-Fluorescein-N135q-S380c-Antithrombin-Iii, Chain I |
| Haemoglobin beta | Protein Len, Bence-Jones |
| Immunoglobulin A Light chain | |
| Immunoglobulin G-Aptamer Complex | |
| Immunoglobulin G-1 (Fc Fragment) | |
| Immunoglobulin G-1 heavy chain constant region | |
| Immunoglobulin G-2 heavy chain constant region | |
| Inter-alpha (globulin) inhibitor H2 | |
| Protein Rei, Bence-Jones | |
| Protein Tro alpha 1 H | |
| Sry-related HMG box gene | |
| Transferrin | |
| Vitamin D Binding Protein | |
| Vitamin D-binding protein precursor | |
| Zinc-Alpha-2-Glycoprotein |
Table 5. Differentially expressed proteins in response to the RIPC stimulus demonstrating up regulation and down regulation during the late response (24 h).
| Proteins up regulated | Proteins down regulated |
| Amyloidogenic Transthyretin Variants | Albumin |
| Apolipoprotein A-Ii | Albumin pre-proprotein |
| Apolipoprotein D | Alpha-1-antitrypsin |
| Fibrinogen gamma | Alpha-1-microglobulin/Bikunin precursor |
| Fibrin beta | Antithrombin-Iii Complex, Chain A, |
| Hemopexin, isoform CRA_a | Apolipoprotein H |
| Haptoglobin 2-alpha | Apolipoprotein A1 |
| Immunoglobulin A Light chain | Complement C4B |
| Protein Rei, Bence-Jones | Complement C8 |
| Protein Tro alpha 1 H | Complement C1r |
| Sry-related HMG box gene | Fibronectin 1 |
| Transferrin | Gelsolin precursor |
| Zinc-Alpha-2-Glycoprotein | Histidine-rich glycoprotein precurso |
| Haemoglobin | |
| Immunoglobulin heavy chain constant region | |
| Immunoglobulin light chain variable region | |
| Immunoglobulin G-1 (Fc Fragment) | |
| Immunoglobulin G-1 heavy chain constant region | |
| Immunoglobulin G-2 heavy chain constant region | |
| Immunoglobulin kappa chain variable region | |
| Immunoglobulin M | |
| Immunoglobulin M heavy chain | |
| Inter-alpha (globulin) inhibitor H2 | |
| Lipoprotein B100 | |
| N-acetylmuramoyl-L-alanine amidase precursor | |
| Neuropilin-1 B1 Domain In Complex With A Vegf-Blocking Fab, Chain L, | |
| P14-Fluorescein-N135q-S380c-Antithrombin-Iii, Chain I | |
| Protein Len, Bence-Jones | |
| Vitamin D Binding Protein | |
| Vitamin D-binding protein precursor |
Three of the proteins were identified using both experimental approaches. These proteins were fibrin beta, fibrinogen gamma and apolipoprotein A. The main pathway involved in the RIPC response was acute phase response signalling.
Discussion
A multi-organ protection by RIPC can be transferred to the target organs via humoral factors in plasma [6], [15]. Those factors may or may not be proteins. However, proteomic changes in plasma are an important component of the inflammatory response to IR injury. Thus, it appeared logical to examine proteomic changes in plasma induced by transient limb ischemia.
To the best of our knowledge, this is the first study to examine the global proteomic changes in plasma induced by brief forearm ischemia. Arrell et al., demonstrated in a rabbit model that pharmacologically induced preconditioning evoked proteomic changes in the myocardium [21]. Although proteomic changes in the target organ are of great interest, we focused on describing proteomic changes in plasma, in light of evidence that transfer of plasma from the transiently ischemic limb induced RIPC in the target organs [15].
Proteomic evaluation of plasma is challenging due to the high abundance proteins that obscure lower abundance proteins [22], [23]. We depleted two most abundant proteins – albumin and IgG. Although we attempted to deplete the samples of these high abundance proteins, residual albumin and IgG were still present. Since neither albumin nor IgG binds to heparin, further separation by fractionation based on the ability to bind to heparin was helpful. Thus, we used two different, but complementary methodological proteomic approaches in order to better define the proteomic changes in plasma. Fractionation and LC permitted a better evaluation of the heparin bound fraction and effectively cleared the albumin and IgG to further unmask lower abundance proteins. By analysing both fractions, we ensured that the majority of the plasma proteome was assessed.
The results of this study demonstrated that plasma proteome changes occurred during the RIPC and were cumulative with each episode of IR. The number of peptides in plasma coming from the ischemic arm increased with each episode of transient arm ischemia. These peptides were predominantly up-regulated ( Figure 3 ). In contrast, at 15 minutes and 24 hours after the RIPC stimulus the peptides were predominantly down-regulated. The latter finding is consistent with our previous genomic study that demonstrated predominant down-regulation of pro-inflammatory gene expression early and late after the RIPC stimulus [9].
We identified 51 proteins which were differentially expressed in response to the RIPC protocol compared to baseline when the results of the two approaches were combined. The proteins identified, play a role in a range of cellular functions including immune response, haemostasis, haemoglobin binding and synthesis, protease inhibition, acute phase response, iron binding, lipid transport, oxygen binding, heme binding, vitamin D transport, protein binding, maintenance of osmotic pressure, trypsin inhibition, molecular transport and protein signalling, endothelial cell activation, actin binding, peptidoglycan biosynthesis and DNA binding. This suggests that the mechanisms involved in RIPC may involve a complex interaction of multiple redundant pathways such that there is regulation of cells surviving or yielding to ischemic damage. Many proteins identified in our study are biomarkers of cardiovascular disease [24].
Alpha-1-antitrypsin is one such protein that has been shown to contribute to protection of the kidney in a mouse model of IR injury through the initiation of the acute phase response to injury and exerting anti-apoptotic and anti-inflammatory effects [25]. We found this protein to be up regulated during the RIPC protocol as well as 15 minutes later consistent with its known role as an initiator of the acute phase response during injury. Haptoglobin is another acute phase protein with apparent involvement in IR injury. The level of haptoglobin is decreased during IR injury and normalized by preconditioning, attenuating the IR injury [26]. We found consistently higher levels of haptoglobin at all time points analysed compared to baseline.
Apolipoproteins were shown to be predominantly up-regulated during the RIPC stimulus as well as during the early and late after it. Apolipoproteins prevent endothelial dysfunction and inhibit lipid oxidation in models of myocardial and renal IR injury [27], [28] and may play a role in protection against IR injury. In particular, apolipoprotein A1 is involved in IR injury and has anti-inflammatory activity [28]. Apolipoprotein A1 protected against IR injury through suppression of intercellular adhesion molecule-1 and p-selectin expression, thus, decreasing neutrophil adhesion and subsequent tissue injury that resulted in improved cardiac contractility. It also reduced release the of creatnine kinase, tumor necrosis factor-alpha and other inflammatory cytokines and myeloperoxidase serum levels post ischemic insult [28]–[30]. Arrhythmias (ventricular tachycardia and ventricular fibrillation) associated with IR can be attenuated by lipoproteins [31].
Two complement proteins (C1r and C8) were down-regulated in our study during and after the RIPC stimulus. This is consistent with previous studies that demonstrated the gene expression of these proteins to be down regulated in the myocardium of rabbits in vivo [32] and in isolated rabbit hearts [33] in response to a preconditioning stimulus.
Haemostatic proteins have also been implicated in ischemic preconditioning [34]. They activate fibrinolysis and reduce inflammation through mechanisms involving fibrinogen gamma [34]. In addition, fibrin beta decreases myocardial infarct size, scar formation, inflammation and the levels of cytokines (interleukin 1 beta, tumor necrosis factor-alpha and interleukin 6) in plasma [35]. Intravenous administration of fibrin-derived peptides is cardioprotective and reduces infarct size in rodents and pigs and appears to be as effective as preconditioning [35], [36]. Administration of fibrin beta to humans is reported to be safe with potential to protect against IR injury [36].
Transferrin was up-regulated during and after the RIPC. Although the exact involvement of transferrin in protection against IR injury is unknown, transferrin regulates production of reactive oxygen species via iron regulation and appears to have a protective role in IR injury [37], [38].
Although our analysis revealed proteins that are known to have a role during IR injury, there were also proteins whose role during IR injury is unknown. Our discussion is therefore centered on the proteins with known involvement in the IR injury. We were unable to obtain MS/MS data for all peptides that were changed significantly and these peptides require further analysis. The analysis involved matching peptide sequences against the sequence data of known proteins in the NCBInr human database. It is possible that there are proteins that have not been mapped in this database and therefore the origin of some of the detected peptides is not known. Peptides not identified in existing database searches may reflect programmed frame shifts or DNA sequencing errors [39].
A few studies suggested that a blood borne factor thought to be responsible for the ability of RIPC serum to transfer protection appears to have a molecular weight below 30 kDa [40]–[43]. Recently, Shimizu et al [40] demonstrated for the first time that transient limb ischemia liberates protective factors with molecular masses below 15 kDa, resistant to freezing and thawing, which is hydrophobic and not easily denatured. Serejo et al [42] have recently reported that the blood effluent from preconditioned rat hearts which was dialyzed to retain molecules above a molecular mass of 3500 Da had protective properties. On the other hand, Lang et al [43] reported that no differentially abundant proteins from RIPC with a known signalling function could be found above molecular mass of 8 kDa, the lower molecular mass limit of their proteomic study. Interestingly, Serejo et al [42] concluded that their finding “excludes the participation of adenosine (267.24 Da), opioids (500–800 Da), bradykinin (1060.22 Da), and other substances with molecular weights below the dialysis cutoff (3.5 kDa) as putative mediators of preconditioning”. These results still remain controversial and should be interpreted cautiously. At the present study, we found that some plasma proteins with molecular mass below 30 kDa coming from ischemic arm were up-regulated, for example, amiloidogenic transthyretin varients (15887 Da), apolipoprotein D (21276 Da), beta globin (2104 Da), haemoglobin alpha 2 (15258 Da), haemoglobin beta (15998 Da), vitamin D binding protein (2905 Da), while complement 8 gamma (22277 Da) was down-regulated. Proteomic assessment of the plasma taken from ischemic arms needs further scrutiny.
In the current study, the global proteomic responses to the RIPC stimulus reflected the genomic responses to the RIPC stimulus demonstrated in our previous study [9], in which there was a predominance of down-regulation of gene expression both early (at 15 minutes) and late (at 24 hours) after transient limb ischemia. We observed an increased number of down-regulated proteins in the early and even more so, during the late response to the RIPC stimulus.
Further research needs to be carried out to identify the pathways implicated in the RIPC response as well to identify the peptides and other metabolites that may be involved. This could be achieved by further depleting plasma of high abundance proteins to investigate those found in plasma at lower abundance. Furthermore, a proteomic assessment of plasma dialysate might be useful to assess the proteins with molecular weight of less than 15–30 kDa. If an effector of the RIPC stimulus is identified and its potency is properly enhanced, the application of such augmented RIPC could be immense, including all fields of cardiac surgery, organ transplantation, protection against stroke and post-cardiopulmonary bypass renal failure.
The study was designed to assess a global proteomic response to the RIPC and not to determine the proteins that may confer the protection. As such the study did not specifically assess the protein of low molecular weight.
Conclusions
In summary, the results of this study demonstrate that the RIPC stimulus evokes a global proteomics response early and late, with predominant decrease in protein expression. There was an overall trend of up-regulation of protein expression in blood taken from the transiently ischemic limb during the RIPC protocol and this increase in the number of up-regulated peptides was cumulative with each cycle of the IR of the limb.
Acknowledgments
The authors acknowledge the support of Ms Robyn Summerhayes, Dr Nori Oka and Dr Matt Liava’a and Ms Chantal Attard during the processing of the samples.
Funding Statement
This project was funded by the National Health and Medical Research Council (NHMRC) grant (# 628756) and NHMRC postgraduate scholarship (# 1017734). Yves d’Udekem is a Career Development Fellow of The National Heart Foundation of Australia (CR 10M 5339). This study was supported by the Victorian Government’s Operational Infrastructure Support Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Murry CE, Jennings RB, Reimer KA (1986) Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation 74: 1124–1136. [DOI] [PubMed] [Google Scholar]
- 2. Kanoria S, Jalan R, Seifalian AM, Williams R, Davidson BR (2007) Protocols and mechanisms for remote ischemic preconditioning: a novel method for reducing ischemia reperfusion injury. Transplantation. 84: 445–458. [DOI] [PubMed] [Google Scholar]
- 3. Kharbanda RK, Nielsen TT, Redington AN (2009) Translation of remote ischaemic preconditioning into clinical practice. Lancet (374) 1557–1565. [DOI] [PubMed] [Google Scholar]
- 4. Gho BC, Schoemaker RG, van den Doel MA, Duncker DJ, Verdouw PD (1996) Myocardial protection by brief ischemia in noncardiac tissue. Circulation. 94: 2193–2200. [DOI] [PubMed] [Google Scholar]
- 5. Botker HE, Kharbanda R, Schmidt MR, Bottcher M, Kaltoft AK, et al. (2010) Remote ischaemic conditioning before hospital admission, as a complement to angioplasty, and effect on myocardial salvage in patients with acute myocardial infarction: a randomised trial. Lancet. 375: 727–734. [DOI] [PubMed] [Google Scholar]
- 6. Konstantinov IE, Li J, Cheung MM, Shimizu M, Stokoe J, et al. (2005) Remote ischemic preconditioning of the recipient reduces myocardial ischemia-reperfusion injury of the denervated donor heart via a Katp channel-dependent mechanism. Transplantation. 79: 1691–1695. [DOI] [PubMed] [Google Scholar]
- 7. Kharbanda RK, Li J, Konstantinov IE, Cheung MM, White PA, et al. (2006) Remote ischaemic preconditioning protects against cardiopulmonary bypass-induced tissue injury: a preclinical study. Heart. 92: 1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmidt MR, Smerup M, Konstantinov IE, Shimizu M, Li J, et al. (2007) Intermittent peripheral tissue ischemia during coronary ischemia reduces myocardial infarction through a KATP-dependent mechanism: first demonstration of remote ischemic perconditioning. Am J Physiol Heart Circ Physiol (292) H1883–1890. [DOI] [PubMed] [Google Scholar]
- 9. Konstantinov IE, Arab S, Kharbanda RK, Li J, Cheung MM, et al. (2004) The remote ischemic preconditioning stimulus modifies inflammatory gene expression in humans. Physiol Genomics (19) 143–150. [DOI] [PubMed] [Google Scholar]
- 10. Konstantinov IE, Arab S, Li J, Coles JG, Boscarino C, et al. (2005) The remote ischemic preconditioning stimulus modifies gene expression in mouse myocardium. J Thorac Cardiovasc Surg. 130: 1326–1332. [DOI] [PubMed] [Google Scholar]
- 11. Konstantinov IE, Coles JG, Boscarino C, Takahashi M, Goncalves J, et al. (2004) Gene expression profiles in children undergoing cardiac surgery for right heart obstructive lesions. J Thorac Cardiovasc Surg (127) 746–754. [DOI] [PubMed] [Google Scholar]
- 12. Saxena P, Shaw OM, Misso NL, Naran A, Shehatha J, et al. (2011) Remote ischemic preconditioning stimulus decreases the expression of kinin receptors in human neutrophils. J Surg Res. 171: 311–316. [DOI] [PubMed] [Google Scholar]
- 13. Shimizu M, Saxena P, Konstantinov IE, Cherepanov V, Cheung MM, et al. (2010) Remote ischemic preconditioning decreases adhesion and selectively modifies functional responses of human neutrophils. J Surg Res (158) 155–161. [DOI] [PubMed] [Google Scholar]
- 14. Cheung MMH, Kharbanda RK, Konstantinov IE, Shimizu M, Frndova H, et al. (2006) Randomized controlled trial of the effects of remote ischemic preconditioning on children undergoing cardiac surgery: first clinical application in humans. J Am Coll Cardiol. 47: 2277–2282. [DOI] [PubMed] [Google Scholar]
- 15. Shimizu M, Tropak M, Diaz RJ, Suto F, Surendra H, et al. (2009) Transient limb ischaemia remotely preconditions through a humoral mechanism acting directly on the myocardium: evidence suggesting cross-species protection. Clin Sci (Lond). 117: 191–200. [DOI] [PubMed] [Google Scholar]
- 16. Ignjatovic V, Lai C, Summerhayes R, Mathesius U, Tawfilis S, et al. (2011 ) Age-related differences in plasma proteins: how plasma proteins change from neonates to adults. PLoS One. 6: e17213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kjellberg M, Ikonomou T, Stenflo J (2006) The cleaved and latent forms of antithrombin are normal constituents of blood plasma: a quantitative method to measure cleaved antithrombin. J Thromb Haemost. 4: 168–176. [DOI] [PubMed] [Google Scholar]
- 18.Andrew SM, Titus JA, Zumstein L (1997) Current protocols in immunology: John Wiley and sons Inc;. [DOI] [PubMed]
- 19.Stone KL, Williams KR (2002) Enzymatic Digestion of Proteins in Solution and in SDS Polyacrylamide Gels. In: Walker JM, ed. The protein protocols handbook. Totowa, New Jersey: Humana Press Inc : 511–523.
- 20. Ang CS, Binos S, Knight MI, Moate PJ, Cocks BG, et al. (2011) Global survey of the bovine salivary proteome: integrating multidimensional prefractionation, targeted, and glycocapture strategies. J Proteome Res. 10: 5059–5069. [DOI] [PubMed] [Google Scholar]
- 21. Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, et al. (2006) Proteomic analysis of pharmacological preconditioning: novel protein targets converge to mitochondrial metabolism pathways. Circ Res. 99: 706–714. [DOI] [PubMed] [Google Scholar]
- 22. Anderson NL, Anderson NG (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 1: 845–867. [DOI] [PubMed] [Google Scholar]
- 23. Darde VM, Barderas MG, Vivanco F (2007) Depletion of high-abundance proteins in plasma by immunoaffinity subtraction for two-dimensional difference gel electrophoresis analysis. Methods Mol Biol. 357: 351–364. [DOI] [PubMed] [Google Scholar]
- 24. Anderson L (2005) Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol.563(Pt 1): 23–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Daemen MA, Heemskerk VH, van’t Veer C, Denecker G, Wolfs TG, et al. (2000) Functional protection by acute phase proteins alpha(1)-acid glycoprotein and alpha(1)-antitrypsin against ischemia/reperfusion injury by preventing apoptosis and inflammation. Circulation (102) 1420–1426. [DOI] [PubMed] [Google Scholar]
- 26. Fernandez V, Castillo I, Tapia G, Romanque P, Uribe-Echevarria S, et al. (2007) Thyroid hormone preconditioning: protection against ischemia-reperfusion liver injury in the rat. Hepatology. 45: 170–177. [DOI] [PubMed] [Google Scholar]
- 27. Calabresi L, Gomaraschi M, Rossoni G, Franceschini G (2006) Synthetic high density lipoproteins for the treatment of myocardial ischemia/reperfusion injury. Pharmacol Ther (111) 836–854. [DOI] [PubMed] [Google Scholar]
- 28. Gu SS, Shi N, Wu MP (2007) The protective effect of ApolipoproteinA-I on myocardial ischemia-reperfusion injury in rats. Life Sci (81) 702–709. [DOI] [PubMed] [Google Scholar]
- 29. Gomaraschi M, Calabresi L, Rossoni G, Iametti S, Franceschini G, et al. (2008) Anti-inflammatory and cardioprotective activities of synthetic high-density lipoprotein containing apolipoprotein A-I mimetic peptides. J Pharmacol Exp Ther (324) 776–783. [DOI] [PubMed] [Google Scholar]
- 30. Shi N, Wu MP (2008) Apolipoprotein A-I attenuates renal ischemia/reperfusion injury in rats. J Biomed Sci (15) 577–583. [DOI] [PubMed] [Google Scholar]
- 31. Imaizumi S, Miura S, Nakamura K, Kiya Y, Uehara Y, et al. (2008) Antiarrhythmogenic effect of reconstituted high-density lipoprotein against ischemia/reperfusion in rats. J Am Coll Cardiol (51) 1604–1612. [DOI] [PubMed] [Google Scholar]
- 32. Tanhehco EJ, Yasojima K, McGeer PL, McGeer EG, Lucchesi BR (2000) Preconditioning reduces myocardial complement gene expression in vivo. Am J Physiol Heart Circ Physiol (279) H1157–1165. [DOI] [PubMed] [Google Scholar]
- 33. Tanhehco EJ, Yasojima K, McGeer PL, Washington RA, Kilgore KS, et al. (1999) Preconditioning reduces tissue complement gene expression in the rabbit isolated heart. Am J Physiol (277) H2373–2380. [DOI] [PubMed] [Google Scholar]
- 34. Warzecha Z, Dembinski A, Ceranowicz P, Dembinski M, Cieszkowski J, et al. (2007) Influence of ischemic preconditioning on blood coagulation, fibrinolytic activity and pancreatic repair in the course of caerulein-induced acute pancreatitis in rats. J Physiol Pharmacol. 58: 303–319. [PubMed] [Google Scholar]
- 35. Zacharowski K, Zacharowski PA, Friedl P, Mastan P, Koch A, et al. (2007) The effects of the fibrin-derived peptide Bbeta(15–42) in acute and chronic rodent models of myocardial ischemia-reperfusion. Shock (27) 631–637. [DOI] [PubMed] [Google Scholar]
- 36. Roesner JP, Petzelbauer P, Koch A, Mersmann J, Zacharowski PA, et al. (2007) The fibrin-derived peptide Bbeta15–42 is cardioprotective in a pig model of myocardial ischemia-reperfusion injury. Crit Care Med (35) 1730–1735. [DOI] [PubMed] [Google Scholar]
- 37. Cairo G, Tacchini L, Recalcati S, Azzimonti B, Minotti G, et al. (1998) Effect of reactive oxygen species on iron regulatory protein activity. Ann N Y Acad Sci (851) 179–186. [DOI] [PubMed] [Google Scholar]
- 38. Tacchini L, Fusar Poli D, Bernelli-Zazzera A, Cairo G (2002) Transferrin receptor gene expression and transferrin-bound iron uptake are increased during postischemic rat liver reperfusion. Hepatology. 36: 103–111. [DOI] [PubMed] [Google Scholar]
- 39. Gupta N, Tanner S, Jaitly N, Adkins JN, Lipton M, et al. (2007) Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 17: 1362–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shimizu M, Tropak M, Diaz RJ, Suto F, Surendra H, et al. (2009) Transient limb ischemia remotely preconditions through a humoral mechanism acting directly on the myocardium: evidence suggesting croass-species protection. Clin Sci (Lond) 117: 191–200. [DOI] [PubMed] [Google Scholar]
- 41. Dickson EW, Blehar DJ, Carraway RE, Heard SO, Steinberg G, et al. (2001) Naloxone blockes transferred preconditioning in isolated rabbit hearts. J Mol Cell Cardiol 33: 1751–1756. [DOI] [PubMed] [Google Scholar]
- 42. Serejo FC, Rodrigues LF, da Silva Tavares KC, de Carvalho AC, Nascimento JH (2007) Myocardial injury is decreased by late remote ischaemic preconditioning and aggravated by tramadol in patients undergoing cardiac surgery: a randomised controlled trial. J Cardiovasc Pharmacol 49: 214–220.17438406 [Google Scholar]
- 43. Lang SC, Elsasser A, Scheler C, Vetter S, Tiefenbacher CP, et al. (2006) Myocardial preconditioning and remote renal preconditioning: identifying a protective factor using proteomic methods? Basic Res Cardiol 101: 149–158. [DOI] [PubMed] [Google Scholar]