Abstract
Systemic acquired resistance is an important component of the disease-resistance arsenal of plants, and is associated with an enhanced potency for activating local defense responses upon pathogen attack. Here we demonstrate that pretreatment with benzothiadiazole (BTH), a synthetic activator of acquired resistance in plants, augmented the sensitivity for low-dose elicitation of coumarin phytoalexin secretion by cultured parsley (Petroselinum crispum L.) cells. Enhanced coumarin secretion was associated with potentiated activation of genes encoding Phe ammonia-lyase (PAL). The augmentation of PAL gene induction was proportional to the length of pretreatment with BTH, indicating time-dependent priming of the cells. In contrast to the PAL genes, those for anionic peroxidase were directly induced by BTH in the absence of elicitor, thus confirming a dual role for BTH in the activation of plant defenses. Strikingly, the ability of various chemicals to enhance plant disease resistance correlated with their capability to potentiate parsley PAL gene elicitation, emphasizing an important role for defense response potentiation in acquired plant disease resistance.
Upon infection with necrotizing pathogens many plants develop an enhanced resistance to further pathogen attack not only in the area of primary infection but also in distal, uninoculated organs. This phenomenon strongly depends on the accumulation of SA (Durner et al., 1997) and is named SAR (for recent reviews, see Ryals et al., 1996; Wobbe and Klessig, 1996; Sticher et al., 1997). In tobacco and Arabidopsis establishment of SAR is accompanied by the activation of SAR genes (Ward et al., 1991), including those encoding some of the PR proteins (Cutt and Klessig, 1992; Stintzi et al., 1993). As some PR proteins have been identified as chitinases (PR-3) and β-1,3-glucanases (PR-2), which can hydrolyze microbial cell wall components, their accumulation has initially been assumed to be responsible for SAR. However, SAR is also effective against bacterial and viral pathogens in addition to fungi (Ryals et al., 1996). Antibacterial or antiviral activity has not been found for any PR protein nor has enhanced resistance to any of these two types of pathogens been reported from transgenic plants overexpressing PR genes. Thus, it is obvious that accumulation of PR proteins cannot per se explain the SAR phenomenon. During the past few decades there has been increasing evidence for augmentation of locally induced defense responses upon pathogen infection of systemically protected plants (for review, see Sticher et al., 1997). Although this conditioning phenomenon has been known for quite a while, not much attention was paid to it when studying SAR and, therefore, little is known so far about the molecular and biochemical mechanism(s) that mediate(s) conditioning.
In recent years the Kauss laboratory demonstrated that tissue conditioning and the resulting potentiation of local defense responses can be studied in the well-described parsley (Petroselinum crispum L.) cell culture/Pmg elicitor model system: preincubation with the natural or synthetic SAR inducers SA or INA in a time-dependent manner primed parsley cells for augmented elicitation of the early oxidative burst (Kauss and Jeblick, 1995), the secretion of both cell wall phenolics (Kauss et al., 1993) and coumarin phytoalexins (Kauss et al., 1992) and the associated activation of some defense-related phenylpropanoid genes (Kauss et al., 1992; Thulke and Conrath, 1998). In an extension of these studies to whole plants, Mur et al. (1996) recently verified SA-mediated potentiation of local defense gene activation in systemically protected tobacco plants. Thus, the findings with SA-pretreated, cultured parsley cells might reflect at least in part the situation in systemically resistant plant tissue.
Since it was discovered that SA is an endogenous signal for the activation of SAR (for review, see Durner et al., 1997), there has been increased characterization of synthetic chemicals that are able to mimic SA. BTH, which induces disease resistance in various host/pathogen systems, is the most attractive among these plant activators (Friedrich et al., 1996; Görlach et al., 1996; Lawton et al., 1996). In tobacco, Arabidopsis, and wheat, BTH does not cause SA biosynthesis but induces the same set of SAR genes as is induced by SA (Ward et al., 1991; Friedrich et al., 1996; Görlach et al., 1996; Lawton et al., 1996). However, little is known about how BTH induces SAR. This led us to investigate in a parsley model system the influence of pretreatment with BTH on the elicitation of various defense responses as a first step toward elucidating the mode of action of BTH in SAR induction.
MATERIALS AND METHODS
Parsley Cell Culture
The parsley (Petroselinum crispum L.) cell culture was grown and kept as a callus on modified B5 medium as described previously (Kauss et al., 1992). Suspension cultures were initiated about twice a year by transferring small aliquots of well-grown callus tissue to 150 mL of fresh B5 medium in 500-mL Erlenmeyer flasks and subsequent agitation at 100 rpm. The cell suspension was kept by transferring 20 mL (about 3.5 g cell fresh weight) of a 6-d-old cell suspension to 150 mL of fresh B5 medium.
Parsley Cell Suspension Treatment
Parsley cell suspensions were used for the experiments 3 d after subculturing. Aliquots of 25 mL (about 1.5 g cell fresh weight) were transferred to 100-mL Erlenmeyer flasks containing 1% (v/v) DMSO or BTH dissolved in DMSO at the indicated concentrations. The low level of DMSO had no effect on the defense responses under study or on cell viability (data not shown). If not stated otherwise, cell suspensions were incubated under normal growth conditions for 24 h before addition, on the 4th d after subculturing, of either water or Pmg elicitor to 4 μg mL−1, representing a low, suboptimal dose. Three hours later, 15 mL of cell suspension was harvested by filtration, and the cells were quick frozen in liquid nitrogen and stored at −80°C until northern analysis. To assay coumarin phytoalexin secretion the remainder of cell suspension was incubated for another 21 h before extraction and quantification of coumarins from the culture medium as described below.
Plasmids/cDNA Inserts
Bacterial cells harboring the parsley PAL and POX cDNA-containing plasmids were grown overnight and harvested by centrifugation at 4°C. Plasmid isolation was performed with a commercial plasmid midi kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. cDNA inserts were cut out of the plasmids by restriction digestion and separated by agarose-gel electrophoresis. After extraction from melted gel slices, the cDNA inserts were redissolved in Tris-EDTA buffer (pH 8.0) and stored at −20°C until use.
A 1.2-kb EcoRI fragment of a PAL clone, which likely detects products of several members of a small gene family (Lois et al., 1989; Somssich et al., 1989), was used to detect PAL transcripts, whereas a 1.5-kb EcoRI fragment of a POX clone, which also seems to detect transcripts of a small gene family (Kawalleck et al., 1995), served to detect POX mRNA.
RNA Extraction
Total RNA was extracted from frozen suspension-cultured parsley cells according to the method of Glick and Thompson (1993). RNA was precipitated by the addition of 1 volume of isopropanol and incubation at −20°C overnight. RNA was pelleted by centrifugation for 15 min at 4°C in a minifuge at maximum speed. The RNA pellet was washed twice in 500 and 200 μL of 75% (v/v) ethanol, dried, redissolved in water, and then stored at −80°C until northern analysis. RNA yield was about 0.4 to 1.0 mg of total RNA g−1 cell fresh weight.
Northern Analysis
Total RNA (10 μg) was fractionated on a 1.2% (w/v) agarose-2.5% (w/v) formaldehyde gel as described previously (Ausubel et al., 1987). Ethidium bromide (0.07 mg mL−1) was included in the sample loading buffer to facilitate confirmation of equal sample loading and transfer. The RNA was blotted to positively charged nylon membranes (Hybond N+, Amersham) by downstream capillary transfer (Ausubel et al., 1987) using 10× SSC (1.5 m sodium chloride and 0.15 m sodium citrate, pH 7.0). After transfer, RNA was cross-linked to the membrane using a UV cross-linker (model RPN 2500, Amersham). Prehybridization was done for 3 h at 65°C in 5× SSC, 0.05 m sodium phosphate buffer (pH 6.8), 5× Denhardt's solution (1% [v/v], of each PVP, BSA, and Ficoll [type 400]), 1 mm EDTA (pH 8.0), 100 μg mL−1 sheared salmon-sperm DNA, and 1% (w/v) SDS. Hybridization to random-primed 32P-labeled cDNAs (Feinberg and Vogelstein, 1983) was under same conditions except that incubation was for at least 16 h. After hybridization the blot was washed once in 5× SSC, 0.1% (w/v) SDS at 65°C for 15 min and then once in 2× SSC, 0.1% (w/v) SDS at 65°C for 30 min. Blots were exposed to x-ray film at −80°C.
Coumarin Determination
Secreted coumarins were extracted from the culture medium by washing 24 h after elicitation twice with 1 mL of chloroform. Estimation of coumarins in the combined chloroform phases was based on their A320 and on the assumption that all of the chloroform-soluble and UV-absorbing material in the culture medium represented coumarins. In fact, we have found that coumarins make up at least 90% of the total UV (320 nm)-absorbing compounds in the chloroform extracts (data not shown). As the extracted coumarins represent a complex mixture of various benzopyran derivatives, their quantification was based on an average molar extinction coefficient of 12,000 L mol−1 cm−1, according to the method of Hauffe et al. (1985).
All experiments shown in this manuscript were repeated at least three times with same results.
Materials
Pure BTH, INA, and its derivative #1 were generously provided by H. Kessmann, J. Ryals, and T. Staub (Novartis Crop Protection, Inc., Research, Triangle Park, NC [J.R.], and Basel, Switzerland [H.K. and T.S.]). SA, 5-Cl-SA, and 3-OH-BA were from Sigma, and the crude cell wall elicitor from Pmg, race 1, was prepared as described by Lozoya et al. (1991).
RESULTS
BTH Pretreatment Augmented the Elicitation of PAL mRNA and the Induction of Coumarin Secretion
As a first step toward investigating the influence of pretreatment with BTH on the elicitation of defense responses in suspension-cultured parsley cells, the culture was pretreated with various concentrations of BTH before addition of either a low dose of Pmg elicitor or water. The cells then were analyzed for accumulation of PAL transcripts, as the encoded enzyme catalyzes the first key step in phenylpropanoid metabolism, which leads to the production and secretion of coumarin phytoalexins (Hahlbrock and Scheel, 1989). These can easily be extracted from the culture medium.
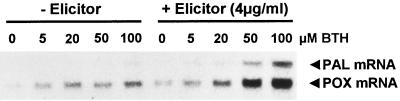
As shown in Figure 1, preincubating parsley cells in BTH augmented the low-dose elicitation of PAL mRNA accumulation (Fig. 1A) and coumarin secretion (Fig. 1B) at any dose applied. Although there was some variation between single experiments (for example, compare Figs. 1A and 4), potentiation of both of these responses was maximal at 50 μm BTH in all of the experiments performed and, thus, the compound was thereafter employed at 50 μm for cell priming. It should be noted that BTH, even at high doses, in most cases only slightly induced PAL gene activation at the 27-h time point (Fig. 1A) or coumarin secretion at the 48-h time point (Fig. 1B) after its application.
Figure 1.
Potentiation of elicitor-induced PAL gene activation (A) and coumarin secretion (B) upon pretreatment with BTH. Cultured parsley cells were pretreated for 24 h with the indicated concentrations of BTH and then incubated in the absence or presence of a low dose of elicitor (4 μg mL−1). Three hours after elicitor application, total RNA was extracted from an aliquot of cells and assayed for accumulation of PAL mRNA by northern analysis (A). The remainder of the cell suspension was agitated for another 21 h before extraction and quantification of coumarins from the suspension medium (B). To assess the extent of potentiation, the cell's response to a saturating dose of elicitor (40 μg mL−1) was also monitored.
Figure 4.
Pretreatment with BTH has different effects on the accumulation of POX and PAL mRNAs. Upon pretreatment for 24 h at various BTH concentrations, cultured parsley cells were incubated for another 3 h in the absence (−) or presence (+) of Pmg elicitor (4 μg mL−1). Cells were analyzed for accumulation of PAL and POX mRNAs by northern analysis simultaneously using parsley PAL- and POX-specific cDNA probes for hybridization. The band obtained with each of the two probes corresponds to the size of the respective mRNAs.
Augmentation by BTH Was Best at Low Elicitor Doses
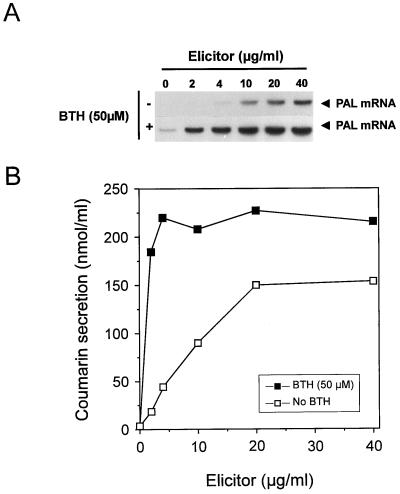
To investigate whether pretreatment of cultured parsley cells with BTH also augments their response to other concentrations of elicitor, a BTH-pretreated parsley cell culture was supplied with various elicitor doses and then assayed for accumulation of PAL mRNA and coumarin secretion. The result of this experiment (Fig. 2) demonstrates that potentiation of elicited PAL transcript accumulation was especially striking at the low elicitor doses (2–10 μg mL−1), which caused only faint transcript accumulation in the absence of BTH pretreatment. At higher elicitor doses, PAL gene activation was greatly induced in the absence of BTH during preincubation, so PAL gene potentiation was less apparent (Fig. 2A). Essentially consistent with these results at the PAL mRNA level are those at the coumarin level (Fig. 2B): enhancement by BTH preincubation was about 10-fold at 2 μg mL−1 and about 5-fold at 4 μg mL−1 elicitor. At apparently saturating elicitor doses (20–40 μg mL−1), augmentation upon BTH priming of coumarin secretion was only about 1.5-fold (Fig. 2B).
Figure 2.
PAL gene activation (A) and coumarin secretion (B) by various elicitor concentrations upon pretreatment of cultured parsley cells in the absence or presence of BTH. Cell suspensions were pretreated for 24 h in the absence or presence of BTH (50 μm) and then supplied with the indicated concentrations of Pmg elicitor. Three hours later, an aliquot of cells was monitored for accumulation of PAL mRNA (A). Coumarins were extracted from the culture medium of the remainder of the cell suspension and quantified 24 h after elicitation.
Priming by BTH Depended on Preincubation
BTH may cause augmentation of elicited PAL transcript accumulation and coumarin secretion either as a result of a synergistic action due to simultaneous presence of the chemical and the elicitor or due to mediate priming of the cells in a time-dependent manner. Therefore, parsley cell cultures were pretreated with BTH for different time periods from 0 to 24 h before addition of a low dose of Pmg elicitor and subsequent analysis of PAL gene activation and coumarin secretion. The results of this experiment (Fig. 3) demonstrate that the longer the period of BTH pretreatment the better the potentiation of the elicitor response. Of all of the times tested, enhancement of PAL transcript accumulation was best after 24 h of pretreatment and could still be detected, although to a lower degree, upon a 10-h preincubation period (Fig. 3A). When BTH was given only 3 h or less prior to low-dose elicitation, potentiation of the elicited PAL gene response was less apparent. Augmentation was also best after 24 h of preincubation, and dropped with decreased length of pretreatment when coumarin secretion was assayed (Fig. 3B). However, in contrast to monitoring PAL mRNA, some potentiation of coumarin secretion was still detected upon simultaneous addition at the zero time point of BTH and elicitor (Fig. 3B). This augmentation of the elicitor response is likely due to some BTH-induced priming that still occurred during the 24-h time period between addition of BTH/elicitor and the extraction of coumarins.
Figure 3.
Length of BTH preincubation determines the extent of potentiation of PAL gene activation (A) and coumarin secretion (B). A parsley cell culture was pretreated in the absence or presence of BTH (50 μm) for the indicated time periods prior to addition of elicitor (4 μg mL−1) or water (no elicitor) on the 4th d after subculturing (time zero). A, Cells were analyzed for accumulation of PAL gene transcripts by northern analysis 3 h after elicitation. B, Coumarins were extracted from the culture medium 24 h after elicitor application.
BTH Has a Dual Role at Defense Gene Activation
Using SA for cell priming we have recently shown that different groups of defense-related parsley genes differ in their response to SA treatment. One group of genes, including those for POX, were directly responsive to SA, whereas activation of another group of genes, including those encoding PAL, was potentiated by pretreatment in SA (Thulke and Conrath, 1998). To see whether this dual role in the activation of defense genes also holds true for BTH, the influence of BTH pretreatment on the activation of the above two genes was tested. As shown in Figure 4, there was direct induction of POX genes by BTH in the absence of elicitor, which was dose dependent and started at 5 μm BTH. Application of an elicitor dose (4 μg mL−1), which only faintly activated the POX genes, had no appreciable effect on the BTH activation of these genes, resulting in about additive signal intensities (Fig. 4, bottom line). In contrast to POX genes, there was poor activation of PAL genes by BTH in the absence of elicitor, yet we detected BTH-induced potentiation of PAL gene elicitation (Fig. 4, top line).
Biological Activity Correlated with Capability for Augmented Activation of PAL Genes
If BTH-mediated potentiation of defense gene activation plays a major role in SAR, one could expect potentiation as a common mode of action of all known inducers of enhanced disease resistance. To test this possibility, we assayed several disease-resistance-inducing, as well as two noninducing, chemicals for their ability to potentiate PAL gene elicitation in our parsley cell-culture model system.
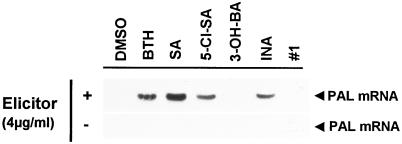
BTH, INA, and SA have been shown to induce disease resistance in a variety of host/pathogen systems (for review, see Ryals et al., 1996). In addition, the SA derivative 5-Cl-SA enhances resistance to tobacco mosaic virus infection and induces PR-1 genes in tobacco, whereas the INA derivative no. 1 (for the chemical structure of the compound, see Conrath et al., 1995) and the SA analog 3-OH-BA were found inactive in these two assays (Conrath et al., 1995). As shown in Figure 5, there was good correlation between biological activity of these chemicals to induce plant disease resistance and their ability to potentiate elicited PAL gene activation in parsley cells. This result strongly suggests that plant activators may act in part by augmenting the activation of certain defense-related genes in plants.
Figure 5.
Potentiation of elicited PAL gene activation upon pretreatment of cultured parsley cells with various chemicals. Cells were preincubated for 24 h with the indicated compounds at 50 μm (BTH) or 250 μm (SA, 5-Cl-SA, 3-OH-BA, INA, or INA analog no. 1) and then incubated in the absence (−) or presence (+) of a low dose of elicitor (4 μg mL−1). Three hours later, the cells were assayed for accumulation of PAL mRNA as in Figure 3A. All of the compounds tested were dissolved in DMSO (1% [v/v] final concentration) and, thus, preincubation with 1% (v/v) DMSO served as a control.
DISCUSSION
In the present study we investigated the effect of pretreatment with BTH on the subsequent elicitation of coumarin secretion and the associated activation of defense-related genes in parsley cells as a first step toward elucidating the mode of action of BTH in the establishment of SAR. By doing so we found that in cultured parsley cells the effect of BTH on defense gene activation depends on the gene under investigation. Thus, parsley POX genes, the products of which cross-hybridized with the probe we used, were found to be directly responsive to BTH treatment (Fig. 4) and, hence, to behave as classical SAR genes. In contrast, the PAL gene transcripts did not accumulate after BTH treatment in the absence of elicitor, yet in BTH-pretreated cells these latter genes became augmentedly induced upon low-dose elicitation (Figs. 1A, 2A, and 4), being associated with an enhanced production of coumarin phytoalexins (Figs. 1B and 2B). Since neither BTH nor the elicitor caused production of SA in cultured parsley cells (K. Nau, O.U. Thulke, and U. Conrath, unpublished results), we can exclude the possibility that pretreatment with BTH allows a critical level of endogenous SA to be reached, thus leading to augmented defense response activation at the low elicitor concentrations (Kauss et al., 1992; Thulke and Conrath, 1998). Rather, as the augmentation of PAL gene activation and coumarin secretion was proportional to the length of BTH pretreatment (Fig. 3), we assume that the plant acquired resistance activator, in a time-dependent process, induces the synthesis of one or more signal transduction components that shift the cells to an alerted state. Some of these factors then might activate certain defense genes, such as those encoding POX, whereas others may act in concert with elicitor-inducible signaling component(s) leading to augmented elicitation of certain other defense responses, such as PAL gene activation and coumarin secretion.
The present results with BTH are consistent with our earlier studies in which we employed SA and INA as the priming compounds (Kauss et al., 1992; Thulke and Conrath, 1998), supporting the assumption that SA, INA, and BTH may all bind to the same receptor and act via the same signal transduction mechanism in the activation of SAR (Friedrich et al., 1996; Lawton et al., 1996; Ryals et al., 1996). The structural similarities between these three SAR inducers (Görlach et al., 1996) adds further support to this possibility. Du and Klessig (1997) recently identified a novel SA-binding protein 2 from tobacco. BTH competed approximately 15-fold better than SA for SA-binding protein 2 binding, consistent with its greater potency to activate SAR genes (Görlach et al., 1996) in wheat and to prime cultured parsley cells for augmented defense gene activation (Fig. 5). However, whether SA-binding protein 2 is functioning as a receptor for SA and BTH during defense signaling in tobacco and whether the protein, or a homolog of it, is also present in parsley cells remains to be elucidated.
The BTH-mediated conditioning of cultured parsley cells for augmented defense response activation is intriguingly analogous to the priming by granulocyte-macrophage colony-stimulating factor or γ-interferon of human monocytes for potentiated lipopolysaccharide elicitation of α-interferon, tumor necrosis factor, and interleukin-12 biosynthesis (Hayes et al., 1991, 1995b; Hayes and Zoon, 1993). Monocyte priming for enhanced tumor necrosis factor induction by γ-interferon has been shown to require several hours of pretreatment and to be manifested at the level of mRNA accumulation (Hayes et al., 1995a). In addition, enhanced accumulation of tumor necrosis factor transcripts was associated with augmented activity of nuclear factor-κ B (Hayes at al., 1995a). It is interesting that the nim1 and npr1 gene products from Arabidopsis, which play a crucial role in SAR and gene-for-gene disease resistance, have been shown to share sequence homology to the IκBα class of transcription regulators, suggesting interaction of the nim1/npr1 proteins with a nuclear factor-kB-related transcription factor in SAR induction (Cao et al., 1997; Ryals et al., 1997).
Because of the strong correlation between the ability to induce enhanced plant disease resistance and the capability to augment the elicitation of PAL genes in parsley cells (Fig. 5), it is probable that the resistance-inducing activity of natural and synthetic plant activators, in addition to direct activation of the SAR genes, is also based on their capability to potentiate the induction of plant defense responses. This might explain why the accumulation of PR proteins, although a reliable molecular marker for the systemically resistant state, is not sufficient to explain the whole phenomenon of SAR (see the introduction). Draper and co-workers (Mur et al., 1996) recently extrapolated the potentiation experiments to the level of whole plants by demonstrating, in systemically protected transgenic tobacco plants, augmented activation of chimeric AoPR-1:uidA (Gus) and PAL-3:uidA (Gus) genes upon challenge infection with pathogenic pseudomonads. Similarly, the elicitation of a class III chitinase gene in hypocotyls of etiolated cucumber seedlings has recently been shown to be augmented upon long-term pretreatment of the seedlings with INA (Kästner et al., 1998). The important role of defense response potentiation in SAR is also supported by a study by Siegrist et al. (1994), who reported on increased deposition of various cell wall phenolics and augmented chitinase and peroxidase activity upon Colletotrichum lagenarium infection of cucumber seedlings exhibiting enhanced disease resistance upon pretreatment with SA, 5-Cl-SA, or INA.
Because of the strong correlation between activity of the various compounds to induce enhanced plant disease resistance and their ability to augment PAL gene elicitation in parsley cell cultures (Fig. 5), our parsley model system, in addition to studying cell priming and the resulting defense response potentiation, also seems to be useful to screen for potential plant activators.
ACKNOWLEDGMENTS
We would like to thank Klaus Hahlbrock and Imre Somssich for supporting us with cDNA clones for parsley PAL and POX. Elmon Schmelzer, Dierk Scheel, and Klaus Hahlbrock are thanked for providing us with the parsley and Pmg cultures. We are also grateful to John Ryals, Helmut Kessmann, and Theo Staub for providing BTH, INA, and its derivative no. 1. We also greatly appreciate critical review of this manuscript and continuous support and interest in this project by Heinrich Kauss. Christa Jung is thanked for help with preparing the figures.
Abbreviations:
- 3-OH-BA
3-hydroxybenzoic acid
- 5-Cl-SA
5-chloro-SA
- INA
2,6-dichloroisonicotinic acid
- BTH
benzothiadiazole (benzo [1,2,3] thiadiazole-7-carbothioic acid S-methyl ester)
- PAL
Phe ammonia-lyase
- Pmg
Phytophthora megasperma f. sp. glycinea
- POX
anionic peroxidase
- PR
pathogenesis-related
- SA
salicylic acid
- SAR
systemic acquired resistance
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Stuhl K (1987) Current Protocols in Molecular Biology. John Wiley and Sons, New York
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis npr1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Conrath U, Chen Z, Ricigliano JR, Klessig DF. Two inducers of plant defense responses, 2,6-dichloroisonicotinic acid and salicylic acid, inhibit catalase activity in tobacco. Proc Natl Acad Sci USA. 1995;92:7143–7147. doi: 10.1073/pnas.92.16.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutt JR, Klessig DF. Pathogenesis-related proteins. In: Boller T, Meins F, editors. Plant Gene Research: Genes Involved in Plant Defense. Wien, Austria: Springer; 1992. pp. 209–243. [Google Scholar]
- Du H, Klessig DF. Identification of a soluble, high-affinity salicylic acid-binding protein in tobacco. Plant Physiol. 1997;113:1319–1327. doi: 10.1104/pp.113.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J, Shah J, Klessig DF. Salicylic acid and disease resistance in plants. Trends Plant Sci. 1997;2:266–274. [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radio-labelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Friedrich L, Lawton K, Ruess W, Masner P, Specher N, Gut-Rella M, Meier B, Dincher S, Staub T, Uknes S and others. A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 1996;10:61–70. [Google Scholar]
- Glick BR, Thompson JE (1993) Methods in Plant Molecular Biology and Biotechnology. CRC Press, Inc., Boca Raton, FL
- Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Ostendorp M, Staub T, Ward E, Kessmann H and others. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- Hauffe KD, Hahlbrock K, Scheel D. Elicitor-stimulated furanocoumarin biosynthesis in cultured parsley cells: S-adenosyl-l-methionine:bergaptol and S-adenosyl-l-methionine:xanthotoxol O-methyltransferases. Z Naturforsch. 1985;41:228–239. [Google Scholar]
- Hayes MP, Enterline JC, Gerrard TL, Zoon KC. Regulation of interferon production in human monocytes: requirements for priming for lipopolysaccharide-induced production. J Leukoc Biol. 1991;50:176–181. doi: 10.1002/jlb.50.2.176. [DOI] [PubMed] [Google Scholar]
- Hayes MP, Freeman SL, Donnelly RP. IFN-gamma priming of monocytes enhances LPS-induced TNF production by augmenting both transcription and mRNA stability. Cytokine. 1995a;7:427–435. doi: 10.1006/cyto.1995.0058. [DOI] [PubMed] [Google Scholar]
- Hayes MP, Wang J, Norcross MA. Regulation of interleukin-12 expression in human monocytes: selective priming by interferon-gamma of lipopolysaccharide-inducible p35 and p40 genes. Blood. 1995b;86:646–650. [PubMed] [Google Scholar]
- Hayes MP, Zoon KC. Priming of human monocytes for enhanced lipopolysaccharide responses: expression of alpha interferon, interferon regulatory factors, and tumor necrosis factor. Infect Immun. 1993;61:3222–3227. doi: 10.1128/iai.61.8.3222-3227.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kästner B, Tenhaken R, Kauss H. Chitinase in cucumber hypocotyls is induced by germinating fungal spores and by fungal elicitor in synergism with inducers of acquired resistance. Plant J. 1998;13:447–454. [Google Scholar]
- Kauss H, Franke R, Krause K, Conrath U, Jeblick W, Grimmig B, Matern U. Conditioning of parsley (Petroselinum crispum L.) suspension cells increases elicitor-induced incorporation of cell wall phenolics. Plant Physiol. 1993;102:459–466. doi: 10.1104/pp.102.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H, Jeblick W. Pretreatment of parsley suspension cultures with salicylic acid enhances spontaneous and elicited production of H2O2. Plant Physiol. 1995;108:1171–1178. doi: 10.1104/pp.108.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H, Theisinger-Hinkel E, Mindermann R, Conrath U. Dichloroisonicotinic and salicylic acid, inducers of systemic acquired resistance, enhance fungal elicitor responses in parsley cells. Plant J. 1992;2:655–660. [Google Scholar]
- Kawalleck P, Schmelzer E, Hahlbrock K, Somssich I. Two pathogen-responsive genes in parsley encode a tyrosine-rich hydroxyproline rich glycoprotein (hrgp) and an anionic peroxidase. Mol Gen Genet. 1995;247:444–452. doi: 10.1007/BF00293146. [DOI] [PubMed] [Google Scholar]
- Lawton K, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- Lois R, Dietrich A, Hahlbrock K, Schulz W. A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 1989;8:1641–1648. doi: 10.1002/j.1460-2075.1989.tb03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozoya R, Bock A, Lois R, Hahlbrock K, Scheel D. Transcriptional repression of light-induced flavonoid synthesis by elicitor treatment of cultured parsley cells. Plant J. 1991;1:227–234. [Google Scholar]
- Mur LAJ, Naylor G, Warner SAJ, Sugars JM, White RF, Draper J. Salicylic acid potentiates defense gene expression in tissue exhibiting acquired resistance to pathogen attack. Plant J. 1996;9:559–571. [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner HY, Johnson J, Delaney TP, Jesse T, Vos P and others. The Arabidopsis nim protein shows homology to the mammalian transcription factor IκB. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist J, Jeblick W, Kauss H. Defense responses in infected and elicited cucumber (Cucumis sativus L.) hypocotyl segments exhibiting acquired resistance. Plant Physiol. 1994;105:1365–1374. doi: 10.1104/pp.105.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich IE, Bollmann J, Hahlbrock K, Kombrink E, Schultz W. Differential early activation of defense-related genes in elicitor-treated parsley cells. Plant Mol Biol. 1989;12:227–234. doi: 10.1007/BF00020507. [DOI] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Metraux JP. Systemic acquired resistance. Annu Rev Phytopathol. 1997;35:235–270. doi: 10.1146/annurev.phyto.35.1.235. [DOI] [PubMed] [Google Scholar]
- Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoghi S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Plant “pathogenesis-related” proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- Thulke OU, Conrath U. Salicylic acid has a dual role in the activation of defense-related genes in parsley. Plant J. 1998;14:35–42. doi: 10.1046/j.1365-313X.1998.00093.x. [DOI] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe KK, Klessig DF. Salicylic acid: an important signal in plants. In: Verma DPS, editor. Plant Gene Research: Signal Transduction and Development. Wien, Austria: Springer; 1996. pp. 167–196. [Google Scholar]