Abstract
Plants have been proved as a novel production platform for a wide range of biologically important compounds such as enzymes, therapeutic proteins, antibiotics, and proteins with immunological properties. In this context, plastid genetic engineering can be potentially used to produce recombinant proteins. However, several challenges still remain to be overcome if the full potential of plastid transformation technology is to be realized. They include the development of plastid transformation systems for species other than tobacco, the expression of transgenes in non-green plastids, the increase of protein accumulation and the appearance of pleiotropic effects. In this paper, we discuss the novel tools recently developed to overcome some limitations of chloroplast transformation.
Keywords: plastids, genetic engineering, transplastomic plants, glycosylation, pleiotropic effects, biofortification, molecular farming
Introduction
Higher plants have been proposed as an economic and easily scalable production system for the production of recombinant proteins with pharmaceutical, industrial and agricultural interest with minimal risk of contamination with animal pathogens.1,2 The genetic modification of the plastid genome has recently emerged as an alternative for the expression of different compounds. Compared with nuclear transformation, plastid genetic engineering offers unique advantages such as high and stable production level of proteins, biological containment of transgenes and recombinant products, no gene silencing and position effects, possibility to co-express multiple genes in operons, cellular compartimentalization of compounds harmful to the plant.3,4 Plastid genetic engineering is, therefore, particularly suitable for the use of plants as biofactories. Several recombinant proteins have been expressed using plastid transformation, including therapeutic proteins, antibiotics, proteins with immunological properties and enzymes. Up to date, around 27 vaccine antigens have been expressed in transplastomic plants against 17 different human diseases.3 In addition, plastid genetic engineering has been used for metabolic engineering of numerous pathways as well as for the expression of insecticidal toxins. Recently, we provided a detailed background on plastid transformation and on the biotechnological applications of this technology.4 Here, we will specifically focus on important challenges that plastid transformation technology still need to overcome before a wider application of this system. These issues include the development of plastid transformation systems for species other than tobacco, the expression of transgenes in non-green plastids, the level of protein accumulation and pleiotropic effects.
Plastid Transformation in Different Plant Species
Presently, the plastid transformation technology is available only for a limited number of dicotyledonous plants. Monocot plants seem to be recalcitrant to plastid transformation. In particular, protocols for plastid transformation are available for many solanaceous species such as Nicotiana tabacum, N. plumbaginifolia, N. benthamiana, N. sylvestris, petunia, eggplant, potato and tomato.5 In addition, plastid transformation is available for the crops soybean, lettuce, cabbage and, more recently, for Medicago sativa.6,7
Transgene integration into the plastid is based on two homologous recombination events between the targeting regions of the transformation vector and the wild-type plastome. The importance of the extent of sequence similarity between sequences involved in homologous recombination to obtain high transformation efficiency has been recently demonstrated in several species.8,9 The extent of sequence divergence can influence the frequency of homologous recombination, and thus the integration efficiency, as well as the functionality of recombinant coding sequences, likely affecting the regeneration of viable plants. Plastid transformation efficiency in potato has been recently improved using novel vectors containing potato flanking sequences (Fig. 1).9 Due to sequence variability in different regions, proper sequence analysis before attempting to use vectors developed in non-target species is recommended.10

Figure 1. Production of transplastomic potato plants. (A) Green calli in leaf explants grown on selective medium containing spectinomycin. (B) Shoot regeneration from leaf explants on selective medium. (C) Transplastomic (left) and wild-type (right) potato plants in soil.
In some species, an additional limitation to plastid transformation is the lack of efficient selection/regeneration protocols. To avoid tissue culture, Tungsuchat-Huang et al.5 investigated the possibility of manipulating plastid genome directly in plants using transplastomic tobacco plants in which the visual marker aurea barau gene, that confers a golden leaf color to leaves, was flanked by recombinase target sites. The excision of the marker gene was obtained through delivery of site-specific recombinases on the Agrobacterium T-DNA injected at the axillary bud site of greenhouse grown plants.
Gene Expression in Non-Green Plastids
Due to potential high protein accumulation levels and/or possibility to express multiple genes in operons, the efficient engineering of the plastome of non-green plastids, such as amyloplasts and chromoplasts, is desirable for specific biotechnological applications, including biofortification, pest tolerance, metabolite accumulation and molecular farming, in tubers, fruits and other organs. Nevertheless, the low transgene expression usually achieved in non-green plastids is still a limitation in many cases. For example, Zhou et al.11 showed that in transplastomic tomato plants the fusion protein p24-Nef accumulated in green tomato fruits but not in ripe fruits containing chromoplasts that are less active than chloroplasts in plastid gene expression.11 Very little is known regarding the signals and factors that regulate expression of the plastome in non-green plant tissues.12 Recently, several regulatory sequences that could increase the expression of transgenes in tomato fruit chromoplasts and potato tuber amyloplasts have been identified.9,13,14 Furthermore, Zhang et al.12 developed a quantitative phenotypic method suitable to identify the in vivo activity of cis-acting elements conferring high levels of gene-expression in non-green plastids. The assay described is based on the use of the nptII gene as a visual reporter of gene-activity in non-green plastids and on the analysis of root length upon seedling growth in the presence of the plastid translational inhibitor kanamycin. Testing different promoters and 5′-UTRs, the authors demonstrated, surprisingly, that heterologous elements from maize plastids conferred higher expression levels in root plastids compared with homologous expression elements from tobacco. In particular, the analysis performed by Zhang and colleagues12 revealed that the promoter from the maize clpP gene (encoding the proteolytic subunit of the Clp ATP-dependent protease), in combination with its native translation initiation signals or the Shine-Dalgarno sequence from the T7g10 (gene 10 of bacteriophage T7) leader, mediates the highest gene activity in root-plastids. This is somehow in agreement with the findings from Valkov et al.9 that demonstrated that the plastid gene clpP could be used as a source of novel regulatory sequences to be used in constructs for gene expression in amyloplasts and other non-green plastids.
Protein Accumulation
Due to the existence of hundreds to thousands copies of the plastid genome per cell, plastid transformation usually allows high accumulation of recombinant proteins, up to 70% of TSP in leaves.15 However, there are a number of proteins that accumulated to low levels and that were not expressed to detectable levels. Several factors affect the accumulation levels of foreign proteins in transgenic plastids such as protein type, the transcriptional and translational regulatory elements, plant tissue, plant development stage, the insertion site in the plastome, RNA and protein stability.4 In particular, translation and protein stability are the two regulatory levels that often limit protein accumulation.12
A way to increase the translation efficiency of recombinant proteins is to alter the codon composition of genes to reflect frequent codons used by the expression host.2 Gisby et al.2 compared the expression of the native human coding sequence for the TGF β3 (Human transforming growth factor-β3) with a synthetic sequence containing frequent chloroplast codons. Results showed that the codon-optimized gene raised accumulation of the recombinant protein by 75-fold compared with the native human coding region. Currently, we have a limited knowledge about the rules that govern protein stability in chloroplasts. Recent studies suggest that the N-terminal amino acid sequence harbour important determinants of plastid protein stability.16 The important role of the N-terminus was recently confirmed by the work from Elghabi and colleagues.17 The authors investigated the possibilities to enhance the expression of the HIV fusion inhibitor cyanovirin-N (CV-N), a protein that is difficult to express from the tobacco plastid genome, by protecting its N-terminus and/or C-terminus with polypeptide sequences taken from the highly stable proteins GFP (green fluorescent protein) and PlyGBS (phage endolysin protein). It was possible to solve both mRNA stability and low protein stability by N-terminal fusions to the CV-N coding region.17
The expression cassettes that are used for chloroplast transformation usually consist of a 5′-regulatory region which includes a promoter and a translational control region that may contain a 5′-UTR or a 5′-TCR with the 5′-UTR and the N-terminus of a plastid gene coding region.18 In some cases, the use of a DB (downstream box) was found to be essential for protein accumulation.19 Lenzi et al.19 showed that the HPV 16 L1 capsid protein accumulated up to 1.5% TSP only when vectors carried a TCR formed by the 5′-UTR and first 42 nucleotides of the plastid rbcL gene. The importance of the DB region as regulator of protein production has been confirmed recently by Gray et al.20 that demonstrated that an efficient DB fusion allowed high-level accumulation of bacterial β-glucosidase in transgenic tobacco plastids.
Changes to the 5′-UTR can also greatly affect the accumulation of recombinant proteins. For example, in order to increase the production of the rotavirus VP6 protein in tobacco plastids, Borchers et al.21 altered the 5′-untraslated region and the 5′ end of the coding region in the VP6 transcript. The authors demonstrated that inclusion of the 5′-UTR from T7g10 and a 15-bp sequence encoding 5 additional amino acids residues at the N-terminus increased transcript accumulation and translational efficiency, and resulted in the stable accumulation of the recombinant protein to > 15% total leaf protein.
Lentz et al.22 expressed a camelid antibody fragment in trasplastomic plants using three different strategies: accumulation in the stroma, translational fusion to a stable protein (β-glucuronidase) and redirection of the protein in the thylakoid lumen. Recombinant proteins could be obtained only by accumulation in the thylakoid lumen or as a fusion protein with β-glucuronidase.
Pleiotropic Effects
In the majority of reports, expression of recombinant proteins in plastids generated normal phenotypes. However, few reports indicated that the expression of foreign proteins in plastids caused phenotypic alterations that include male sterility, yellow leaves and reduced growth of transformed plants.3,23-25 For example, we expressed in transplastomic tobacco plants the immunogenic proteins A27L of vaccinia virus that accumulated to about 18% of total soluble protein.23 We observed that in transplastomic lines the accumulation of the Rubisco large and small subunits was reduced. In soil, transplastomic plants grew more slowly and were chlorotic. In agreement with the observed phenotype, analysis by immunofluorescence microscopy performed on protoplasts isolated from transformed or wild-type plants showed altered chlorophyll distribution.23 Moreover, transmission electron microscopy (TEM) analyses of leaf tissues indicated an alteration of chloroplast membrane ultrastructure in transplastomic plants (Fig. 2). Similar results were obtained in tobacco plants expressing the HIV-1 Pr55gag polyprotein at high levels.24 Recently, Whaeed et al.25 expressed in transplastomic plants the modified human papillomavirus-16 L1 antigen (L1_2xCysM) fused with LTB (Escherichia coli heat-labile enterotoxin subunit B) as adjuvant. The authors described that all transplastomic plants showed chlorotic phenotype, male sterility and growth retardation.25 The aberrant phenotypes observed in these studies could be due to metabolite toxicities, interference with photosynthesis or disturbance of the plastid endomembrane system.26 In order to understand the impact of recombinant protein accumulation on resident proteins in transformed plastids, Bally et al.27 used proteomics to characterize tobacco plants accumulating the proteins HPPD (p-hydroxyphenyl pyruvate dioxygenase) or GFP. These proteins accumulated massively in plastids at the expense of Rubisco, however, no obvious modifications in plant phenotype could be observed. The accumulation of the recombinant proteins caused an adjustment of the accumulation levels of enzymes involved in CO2 metabolism such as nuclear-encoded Calvin cycle enzymes of the plastid and mitochondrial glycine decarboxylase.27
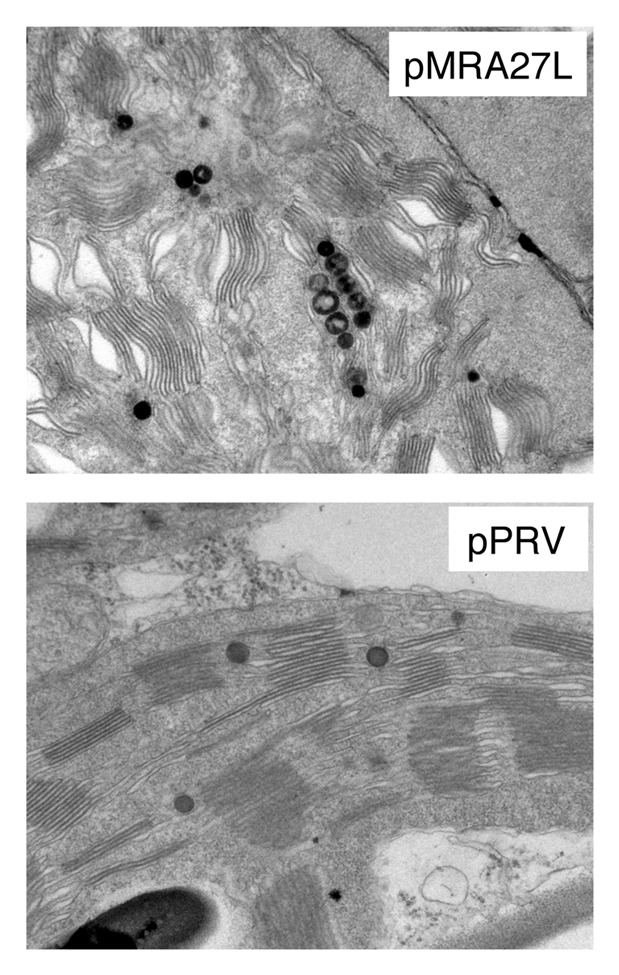
Figure 2. TEM Analyses of leaf tissues performed on tobacco plants transformed with the empty vector pPRV and with the vector pMRA27L containing the gene A27L of vaccinia virus.
In order to minimize the detrimental effects caused by the accumulation of recombinant protein in transformed plastids, it is possible to use several systems for inducible gene expression (for a review, see ref. 3). The approach recently proposed by Verhounig et al.26 is based on a riboswitch that functions as translational regulator of gene expression in plastids in response to its exogenously applied ligand theophilline. However, there are several limitations in the inducible systems described so far, such as the fact that these systems have been performed only in the model plant tobacco and that they yielded low amounts of proteins.
Additional Challenges
Plastids allow post-translational modifications, including protein lipidation, multimerization, N-terminal methionine excision and disulphide bond formation.23,28 Post-translational modifications, such as disulphide bond formations, can influence the stability of recombinant proteins produced in plastids contributing to protein accumulation.28 Moreover, correct folding of the proteins by formation of disulphide bonds is often required for functional tertiary and quaternary structures and, therefore, for antigenicity of the recombinant proteins expressed in transplastomic plants.3 On the other hand, glycosylation is absent in plastids, preventing the production of glycoproteins. The absence of protein glycosylation could represent a possible limitation to the production of vaccine antigens that require glycosylation for their function. However, there are proteins that do not require glycosylation, such as the fragment from camelid single-chain antibodies and many others. Such proteins are therefore compatible with a chloroplast-based production platform.2,22
Another important issue regarding the use of plants for the production of pharmaceutical proteins is transgene containment. Plastids are maternally inherited in 80% of angiosperm species,29 ensuring an efficient bio-containment that prevents undesired transgene transmission to other species. However, in several species, a low-level leakage of plastids in pollen has been reported.29 Recently, work from Elghabi et al.30 showed that using the biolistic approach for plastid transformation, it is possible to achieve occasional unintended co-transformation of the nuclear and plastid genomes with important implications concerning the required molecular characterizations and regulation of transgenic plants.30 Also for these issues the growth of transplastomic plants outdoors is still an argumentative issue.
Conclusion
Considering the high number of recombinant proteins that has been produced in transplastomic plants (more than 50 different recombinant proteins in the last three years) and the recent scientific and technological developments, such as the use of additional marker genes and/or their removal after selection, it can be foreseen that in the next future the plastid transformation technique will be applied for a wider range of purposes both in basic and applied research. In addition, the progresses made in understanding the expression of transgenes in green and non-green plastids, and the improvements in plant regeneration systems and gene transfer methods, will allow to extend the plastid transformation technology to a larger set of plant species, including edible plants and important crops. Only a small number of proteins will remain not expressible in transgenic plastids, such as those requiring glycosylation for their functionality.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/21452
References
- 1.Demain AL, Vaishnav P. Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv. 2009;27:297–306. doi: 10.1016/j.biotechadv.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Gisby MF, Mellors P, Madesis P, Ellin M, Laverty H, O’Kane S, et al. A synthetic gene increases TGFβ3 accumulation by 75-fold in tobacco chloroplasts enabling rapid purification and folding into a biologically active molecule. Plant Biotechnol J. 2011;9:618–28. doi: 10.1111/j.1467-7652.2011.00619.x. [DOI] [PubMed] [Google Scholar]
- 3.Lössl AG, Waheed MT. Chloroplast-derived vaccines against human diseases: achievements, challenges and scopes. Plant Biotechnol J. 2011;9:527–39. doi: 10.1111/j.1467-7652.2011.00615.x. [DOI] [PubMed] [Google Scholar]
- 4.Scotti N, Rigano MM, Cardi T. Production of foreign proteins using plastid transformation. Biotechnol Adv. 2012;30:387–97. doi: 10.1016/j.biotechadv.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Tungsuchat-Huang T, Maliga P. Visual marker and Agrobacterium-delivered recombinase enable the manipulation of the plastid genome in greenhouse-grown tobacco plants. Plant J. 2012;70:717–25. doi: 10.1111/j.1365-313X.2012.04918.x. [DOI] [PubMed] [Google Scholar]
- 6.Cardi T, Lenzi P, Maliga P. Chloroplasts as expression platforms for plant-produced vaccines. Expert Rev Vaccines. 2010;9:893–911. doi: 10.1586/erv.10.78. [DOI] [PubMed] [Google Scholar]
- 7.Wei Z, Liu Y, Lin C, Wang Y, Cai Q, Dong Y, et al. Transformation of alfalfa chloroplasts and expression of green fluorescent protein in a forage crop. Biotechnol Lett. 2011;33:2487–94. doi: 10.1007/s10529-011-0709-2. [DOI] [PubMed] [Google Scholar]
- 8.Ruhlman T, Verma D, Samson N, Daniell H. The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol. 2010;152:2088–104. doi: 10.1104/pp.109.152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valkov VT, Gargano D, Manna C, Formisano G, Dix PJ, Gray JC, et al. High efficiency plastid transformation in potato and regulation of transgene expression in leaves and tubers by alternative 5′ and 3′ regulatory sequences. Transgenic Res. 2011;20:137–51. doi: 10.1007/s11248-010-9402-9. [DOI] [PubMed] [Google Scholar]
- 10.Scotti N, Valkov VT, Cardi T. Improvement of plastid transformation efficiency in potato by using vectors with homologous flanking sequences. GM Crops. 2011;2:89–91. doi: 10.4161/gmcr.2.2.17504. [DOI] [PubMed] [Google Scholar]
- 11.Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers AM, et al. High-level expression of human immunodeficiency virus antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J. 2008;6:897–913. doi: 10.1111/j.1467-7652.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Ruf S, Hasse C, Childs L, Scharff LB, Bock R. Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J. 2012 doi: 10.1111/j.1365-313X.2012.05065.x. [DOI] [PubMed] [Google Scholar]
- 13.Kahlau S, Bock R. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell. 2008;20:856–74. doi: 10.1105/tpc.107.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valkov VT, Scotti N, Kahlau S, Maclean D, Grillo S, Gray JC, et al. Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol. 2009;150:2030–44. doi: 10.1104/pp.109.140483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oey M, Lohse M, Kreikemeyer B, Bock R. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 2009;57:436–45. doi: 10.1111/j.1365-313X.2008.03702.x. [DOI] [PubMed] [Google Scholar]
- 16.Apel W, Schulze WX, Bock R. Identification of protein stability determinants in chloroplasts. Plant J. 2010;63:636–50. doi: 10.1111/j.1365-313X.2010.04268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elghabi Z, Karcher D, Zhou F, Ruf S, Bock R. Optimization of the expression of the HIV fusion inhibitor cyanovirin-N from the tobacco plastid genome. Plant Biotechnol J. 2011;9:599–608. doi: 10.1111/j.1467-7652.2011.00598.x. [DOI] [PubMed] [Google Scholar]
- 18.Maliga P, Bock R. Plastid biotechnology: food, fuel, and medicine for the 21st century. Plant Physiol. 2011;155:1501–10. doi: 10.1104/pp.110.170969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenzi P, Scotti N, Alagna F, Tornesello ML, Pompa A, Vitale A, et al. Translational fusion of chloroplast-expressed human papillomavirus type 16 L1 capsid protein enhances antigen accumulation in transplastomic tobacco. Transgenic Res. 2008;17:1091–102. doi: 10.1007/s11248-008-9186-3. [DOI] [PubMed] [Google Scholar]
- 20.Gray BN, Yang H, Ahner BA, Hanson MR. An efficient downstream box fusion allows high-level accumulation of active bacterial beta-glucosidase in tobacco chloroplasts. Plant Mol Biol. 2011;76:345–55. doi: 10.1007/s11103-011-9743-7. [DOI] [PubMed] [Google Scholar]
- 21.Inka Borchers AM, Gonzalez-Rabade N, Gray JC. Increased accumulation and stability of rotavirus VP6 protein in tobacco chloroplasts following changes to the 5′ untranslated region and the 5′ end of the coding region. Plant Biotechnol J. 2012;10:422–34. doi: 10.1111/j.1467-7652.2011.00675.x. [DOI] [PubMed] [Google Scholar]
- 22.Lentz EM, Garaicoechea L, Alfano EF, Parreno V, Wigdorovitz A. Bravo-Almonacid Translational fusion and redirection to thylakoid lumen as strategies to improve the accumulation of a camelid antibody fragment in transplastomic tobacco. Planta. doi: 10.1007/s00425-012-1642-x. [DOI] [PubMed] [Google Scholar]
- 23.Rigano MM, Manna C, Giulini A, Pedrazzini E, Capobianchi M, Castilletti C, et al. Transgenic chloroplasts are efficient sites for high-yield production of the vaccinia virus envelope protein A27L in plant cellsdagger. Plant Biotechnol J. 2009;7:577–91. doi: 10.1111/j.1467-7652.2009.00425.x. [DOI] [PubMed] [Google Scholar]
- 24.Scotti N, Alagna F, Ferraiolo E, Formisano G, Sannino L, Buonaguro L, et al. High-level expression of the HIV-1 Pr55gag polyprotein in transgenic tobacco chloroplasts. Planta. 2009;229:1109–22. doi: 10.1007/s00425-009-0898-2. [DOI] [PubMed] [Google Scholar]
- 25.Waheed MT, Thönes N, Müller M, Hassan SW, Gottschamel J, Lössl E, et al. Plastid expression of a double-pentameric vaccine candidate containing human papillomavirus-16 L1 antigen fused with LTB as adjuvant: transplastomic plants show pleiotropic phenotypes. Plant Biotechnol J. 2011;9:651–60. doi: 10.1111/j.1467-7652.2011.00612.x. [DOI] [PubMed] [Google Scholar]
- 26.Verhounig A, Karcher D, Bock R. Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci U S A. 2010;107:6204–9. doi: 10.1073/pnas.0914423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bally J, Job C, Belghazi M, Job D. Metabolic adaptation in transplastomic plants massively accumulating recombinant proteins. PLoS One. 2011;6:e25289. doi: 10.1371/journal.pone.0025289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Marchis F, Pompa A, Mannucci R, Morosinotto T, Bellucci M. A plant secretory signal peptide targets plastome-encoded recombinant proteins to the thylakoid membrane. Plant Mol Biol. 2011;76:427–41. doi: 10.1007/s11103-010-9676-6. [DOI] [PubMed] [Google Scholar]
- 29.Thyssen G, Svab Z, Maliga P. Exceptional inheritance of plastids via pollen in Nicotiana sylvestris with no detectable paternal mitochondrial DNA in the progeny. Plant J. 2012 doi: 10.1111/j.1365-313X.2012.05057.x. [DOI] [PubMed] [Google Scholar]
- 30.Elghabi Z, Ruf S, Bock R. Biolistic co-transformation of the nuclear and plastid genomes. Plant J. 2011;67:941–8. doi: 10.1111/j.1365-313X.2011.04631.x. [DOI] [PubMed] [Google Scholar]


