Abstract
Nitrogen (N) and sulfur (S) have inter-related and distinct impacts on microalgal metabolism; with N starvation having previously been reported to induce elevated levels of the biodiesel feedstock material triacylglycerol (TAG), while S deprivation is extensively studied for its effects on biohydrogen production in microalgae.1,2 We have previously demonstrated that N- and S-starved cells of Chlamydomonas reinhardtii display different metabolic trends, suggesting that different response mechanisms exist to compensate for the absence of those two elements.3 We used C. reinhardtii CC-124 mt(-) and CC-125 mt(+) strains to test possible metabolic changes related to TAG accumulation in response to N and S deprivation, considering that gamete differentiation in this organism is mainly regulated by N.4 Our findings contribute to the understanding of microalgal response to element deprivation and potential use of element deprivation for biodiesel feedstock production using microalgae, but much remains to be elucidated on the precise contribution of both N and S starvation on microalgal metabolism.
Keywords: Chlamydomonas reinhardtii, biodiesel, nitrogen, sulfur, triacylglycerol
Due to high bioenergy outturn and the synthesis of high value added products associated with this group, microalgae are frequently investigated as a potential means for various biotechnological applications.5 Biofuel production from microalgae has emerged as a promising way of partially remediating the dependency of global energy demand on fossil fuels. Compared with fossil fuels, biofuel production from microalgae is currently not cost-effective; however, continued increases in oil prices, together with a potential decrease in the cost of biodiesel from microalgae, are expected to make biofuels a viable alternative in the near future. A great variety of microalgae show distinct metabolic properties and are able to switch their metabolic output levels in response to different abiotic stress factors. Levels of TAG, a principal biodiesel feedstock, vary widely across microalgae, depending on their species-specific nature and environmental factors such as changes in element concentration or presence, light intensity and temperature.6,7
N vs. S Deprivation: Differential Survival Strategy of Microalgae
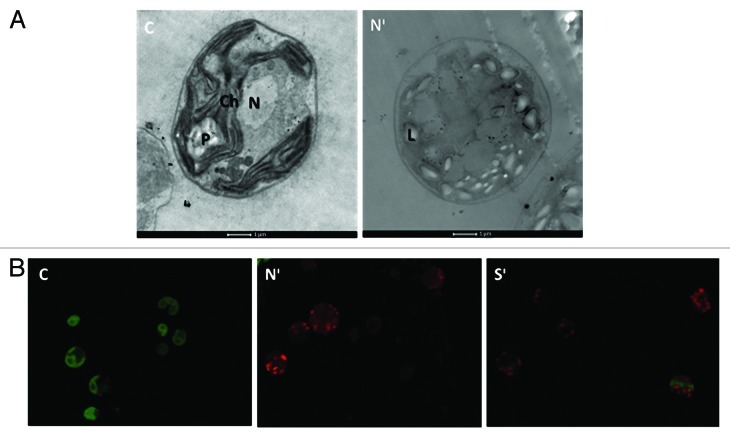
As a macroelement, N has a profound importance for microalgal metabolism and the limitation of this element is compensated by radical changes in several key metabolic pathways. In the process of acclimation to N deficiency, microalgae have been reported to degrade ribosomes and decrease enzyme activities involved in photosynthesis, glyoxylate cycle, gluconeogenesis and photosynthetic carbon fixation cycle while simultaneously inducing carotenoid production to protect against oxidative stress, increasing the expression levels of TAG synthesis related genes in significant quantities, and differentiating into gametes, considered a potential survival strategy since zygotes can withstand adverse conditions.8,9Figure 1 shows that nitrogen deprivation causes dramatic anatomical changes in C. reinhardtii. Most notably the chloroplast is degraded into smaller sphere-like sub-compartments and cytoplasmic lipid droplets are formed (Fig. 1A). Besides these, chloroplast degradation is not as fast as N deprivation in S-deprived cells (Fig. 1B) supporting the data reported previously.3 Microalgae have been reported to synthesize arylsulphatase to recover SO4−2 from SO4−2 esters, upregulate acyltransferases and ATP sulfurylase expression, downregulate proteins involved in translation and folding, and decrease chloroplast ribosomal polypeptides greatly.10,11 Furthermore, cessation of cell cycle, upregulation of the enzymes of the oxidative pentose phosphate pathway, N- or S- scavenging proteins, activation of various mechanisms for reactive oxygen species removal, repression of Calvin cycle enzymes and decrease of photosynthetic activity are observed during both types of deprivations.12,13 Such changes represent attempts to survive the increased oxidative stress associated with nutrient deprivation and to recover N or S from the environment by highly selective uptake processes for those elements.
Figure 1. Transmission electron microscopy (A) and Confocal fluorescence microscopy (B) images of control, N-starved and S-starved C. reinhardtii cells sampled on fifth day of incubation. In the fluorescence images green represents chlorophyll autofluorescence and light red represents Nile Red fluorescence. Abbreviations: C, Control; Nᶦ, N-deprived cells; Sᶦ, S-deprived cells; Ch, Chloroplast; P, Pyrenoid; N, Nucleus; L, lipid bodies.
Work in our lab recently demonstrated that S-deprived samples of C. reinhardtii mt(+) and mt(-) strains display increased growth rates, cell volumes, neutral lipid and TAG accumulation compared with their N-deprived equivalents, while a more rapid decrease in chlorophyll content was observed in C. reinhardtii cultures under N deprivation (Table 1). Furthermore, the metabolic changes associated with nutrient starvation occurred in a time-dependent manner, generally reaching a maximum on the fourth and fifth days of starvation and decreasing or remaining stable afterwards.3 This trend may indicate that vegetative cells of C. reinhardtii can mitigate the effects of N and S starvation for four to five days before the stress associated with long-term nutrient deprivation leads to autophagy to recycle part of the cytoplasm including organelles. N deficiency was reported to induce autophagy, which is a self-degrading process common in eukaryotes that provides needed energy and raw materials for cellular repair, in many organisms14 including C. reinhardtii.15 Longer starvation period that autophagy response is insufficient would lead cell death. Dead cells may then be scavenged by their conspecifics for their N or S content, allowing limited growth and a stable cell count.
Table 1. Changes in growth and biochemical parameters in wild type C. reinhardtii CC-124 and CC-125 strains after four days of N or S deprivation. Detailed information can be found in our recent paper3.
| Parameters tested |
C. reinhardtii CC-124 (mt -) |
C. reinhardtii CC-125 (mt +) |
||
|---|---|---|---|---|
| N deprivation | S deprivation | N deprivation | S deprivation | |
| Cell Growth |
83% decrease |
65% decrease |
66% decrease |
49% decrease |
| Total biovolume |
62.6% decrease |
220% increase |
54.6% decrease |
310% increase |
| Relative dry weight |
32% decrease |
27% decrease |
23% decrease |
20% decrease |
| Protein level |
88% decrease |
89% decrease |
87% decrease |
89% decrease |
| Chlorophyll content |
61% decrease |
26% decrease |
89% decrease |
74% decrease |
| Carotenoid content |
3.6-fold increase |
2.8-fold increase |
1.9-fold increase |
2.3-fold increase |
| Cell biovolume |
2.9-fold increase |
6.1-fold increase |
1.7-fold increase |
5.8-fold increase |
| Starch level |
2.3-fold increase |
3.4-fold increase |
4.3-fold increase |
4.7-fold increase |
| Relative polisaccharide level |
8.1-fold increase |
9.9-fold increase |
13.1-fold increase |
8.6-fold increase |
| Total neutral lipid level |
2.4-fold increase |
2.6-fold increase |
1.7-fold increase |
3-fold increase |
| Relative TAG level | 6.9-fold increase | 15.3-fold increase | 29.1-fold increase | 16.5-fold increase |
Our studies showed that N starvation generally yielded similar effects as S starvation, but the negative impacts on cell count, total protein and chlorophyll levels were much more severe (Table 1). This result is likely caused by the relative importance and abundance of N compared with S, such that while S can be salvaged from dead cells or obtained from intracellular stores, N must be supplied constantly for adequate growth. N content of dry C. reinhardtii biomass is known to be over 10-fold greater than the S content.2 As such, a much greater mass of N is necessary for C. reinhardtii, while a comparatively lesser amount of S, such as that found in the initial cells inoculated into the S-free medium, may be enough to partially facilitate growth. Compared with S-starved cells, N-starved C. reinhardtii cells also displayed a lower amount of enlargement, which may also be correlated with the greater metabolic stress N-starved samples undergo (Table 1). We have observed that cellular functions are affected more rapidly in N-starved microalgal cells. N starvation leads to an almost instantaneous cell growth arrest due to the obligatory presence of N in every protein and most metabolites cannot be compensated by autophagy or other recycling pathways. On the other hand, S deprivation leads to less sudden but again severe responses in overall metabolism and cellular functions. This temporal delay in response probably corresponds to a period of cellular recycling by autophagy and better accumulation of stress marker molecules (carotenoid, TAG, etc.).
S Deprivation May Be Used as a Potential Means for TAG Production from Microalgae
As previously reported,3 chlorophyll content decreased rapidly upon both S and N starvation, while a corresponding increase in carotenoid content was also observed. C. reinhardtii is known to restructure its photosynthetic machinery upon S deprivation, resulting in a decrease in the expression of many of the proteins making up the photosystem complexes I and II within 24 h.1 Such adjustments occur to minimize oxidative stress, as reactive oxygen species (ROS) are generated during photosynthesis and the shutdown of the latter may afford a measure of control over their levels.16 As such, the decrease in chlorophyll synthesis is interpreted to be a part of the alteration, deactivation and disassembly of photosynthetic complexes as a response to oxidative stress resulting from S starvation. Likewise, N deprivation is closely associated with the degradation of ribulose-1,5-bisphosphate carboxylase oxygenase to recycle the latter’s N content17 and the depletion of this protein may necessitate alterations in the mechanism of photosynthesis, leading to the decrease in chlorophyll content observed in N-deficient C. reinhardtii.18,19 An increase in carotenoid content, observed in both N- and S-starved samples, is a response to the stress conditions brought about by nutrient deficiency and is consistent with previous studies.20 As such, their accumulation may be a stress response intended to prevent oxidative damage. Our recent investigation showed that both starch and neutral lipids greatly accumulate in S-deprived C. reinhardtii and that those increases correspond to the rapid decrease in protein levels observed during the first day of starvation. Production of starch took priority over lipid synthesis, suggesting that the two metabolites may compete and that the disturption of starch metabolism may increase lipid production capacity, as has been suggested previously.2,21 While both metabolites were found to increase greatly upon starvation with no apparent antagonistic effects, this is likely the result of cell enlargement caused by S deprivation instead of a true lack of competition between lipid and starch synthesis. Our results suggest that a global shutdown in energetic functions may occur upon S deprivation. Flagella are almost always lost after third day of S deprivation and chlorophyll levels drop considerably, leading to the conclusion that anabolic reactions are severely reduced in that particular cell. Herein, we propose that upon reduction of energy consumption, the trend of metabolism favors storage of energetic denser molecules. Lipids are highly energetic molecules having a higher energy yield per gram than sugars, while starch is known to be the densest form of sugars that is usually used for storage in plants.
Records detailing the use of N starvation to increase lipid production for biodiesel production exist in literature,2 our study suggests that S starvation is the preferable approach due to the lack of adequate cell growth and biovolume attainment upon N exposure.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/21427
References
- 1.Zhang Z, Shrager J, Jain M, Chang CW, Vallon O, Grossman AR. Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot Cell. 2004;3:1331–48. doi: 10.1128/EC.3.5.1331-1348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell. 2009;8:1856–68. doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cakmak T, Angun P, Demiray YE, Ozkan AD, Elibol Z, Tekinay T. Differential effects of nitrogen and sulfur deprivation on growth and biodiesel feedstock production of Chlamydomonas reinhardtii. Biotechnol Bioeng. 2012;109:1947–57. doi: 10.1002/bit.24474. [DOI] [PubMed] [Google Scholar]
- 4.Sager RGS, Granick S. Nutritional control of sexuality in Chlamydomonas reinhardi. J Gen Physiol. 1954;37:729–42. doi: 10.1085/jgp.37.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spolaore P, Joannis-Cassan C, Duran E, Isambert A. Commercial applications of microalgae. J Biosci Bioeng. 2006;101:87–96. doi: 10.1263/jbb.101.87. [DOI] [PubMed] [Google Scholar]
- 6.Converti A, Casazza AA, Ortiz EY, Perego P, Del Borghi M. Effect of temperature and nitrogen concentration on the growth and lipid content of Nannochloropsis oculata and Chlorella vulgaris for biodiesel production. Chem Eng Process. 2009;48:1146–51. doi: 10.1016/j.cep.2009.03.006. [DOI] [Google Scholar]
- 7.Deng X, Fei X, Li Y. The effects of nutritional restriction on neutral lipid accumulation in Chlamydomonas and Chlorella. Afr J Microbiol Res. 2011;5:260–70. [Google Scholar]
- 8.Dean AP, Sigee DC, Estrada B, Pittman JK. Using FTIR spectroscopy for rapid determination of lipid accumulation in response to nitrogen limitation in freshwater microalgae. Bioresour Technol. 2010;101:4499–507. doi: 10.1016/j.biortech.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 9.Beck C, Haring M. Gametic differentiation of Chlamydomonas. Int Rev Cytol. 1996;168:259–302. doi: 10.1016/S0074-7696(08)60886-4. [DOI] [Google Scholar]
- 10.Zhang L, Happe T, Melis A. Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga) Planta. 2002;214:552–61. doi: 10.1007/s004250100660. [DOI] [PubMed] [Google Scholar]
- 11.Yıldız FH, Davies JP, Grossman AR. Characterizatıon of sulfate transport in Chlamydomonas reınhardtii durıng sulfur-limited and sulfur-sufficient growth. Plant Physiol. 1994;104:981–7. doi: 10.1104/pp.104.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman AR, Croft M, Gladyshev VN, Merchant SS, Posewitz MC, Prochnik S, et al. Novel metabolism in Chlamydomonas through the lens of genomics. Curr Opin Plant Biol. 2007;10:190–8. doi: 10.1016/j.pbi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Toepel J, Albaum SP, Arvidsson S, Goesmann A, la Russa M, Rogge K, et al. Construction and evaluation of a whole genome microarray of Chlamydomonas reinhardtii. BMC Genomics. 2011;12:579. doi: 10.1186/1471-2164-12-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–52. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-Pérez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010;152:1874–88. doi: 10.1104/pp.109.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001;20:5587–94. doi: 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Ferris C, Moreno J. Redox regulation of enzymatic-activity and proteolytic susceptibility of ribulose-1,5-bisphosphate carboxylase oxygenase from Euglena gracilis. Photosynth Res. 1993;35:55–66. doi: 10.1007/BF02185411. [DOI] [PubMed] [Google Scholar]
- 18.Plumley FG, Schmidt GW. Nitrogen-dependent regulation of photosynthetic gene expression. Proc Natl Acad Sci U S A. 1989;86:2678–82. doi: 10.1073/pnas.86.8.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siaut M, Cuine S, Cagnon C, Fessler B, Nguyen M, Carrier P, et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011;11:11. doi: 10.1186/1472-6750-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salguero A, de la Morena B, Vigara J, Vega JM, Vilchez C, León R. Carotenoids as protective response against oxidative damage in Dunaliella bardawil. Biomol Eng. 2003;20:249–53. doi: 10.1016/S1389-0344(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Han D, Hu G, Sommerfeld M, Hu Q. Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol Bioeng. 2010;107:258–68. doi: 10.1002/bit.22807. [DOI] [PubMed] [Google Scholar]