Abstract
Currently, most of the prognostic and predictive gene expression signatures emerging for breast cancer concern the tumor component. In Dedeurwaerder et al. we show that DNA methylation profiling of breast tumors is a particularly sensitive means of capturing features of the immune component of breast tumors. Most importantly, correlation is observed between T-cell marker genes and breast cancer clinical outcome.
Keywords: breast cancer, DNA methylome, epigenomics, lymphocyte infiltration, tumor microenvironment
Breast cancer is a heterogeneous disease, comprising distinct entities with both different biological features and different clinical behaviors. Initially, histological criteria provided the basis of breast cancer classification, but this approach has several limitations.1 Over decades, therefore, much effort has been devoted to finding a better way to classify breast cancers in order to predict disease outcomes and responses to therapy with confidence. The major approach developed has been microarray-based gene expression profiling of breast tumors. Thousands of breast tumors have now been profiled in this way, and this has led to classifying breast tumors into, at least, four groups: triple-negative, HER2-positive, high-proliferative luminal and low-proliferative luminal tumors.2
Cancer was initially thought to arise mainly through genetic alterations, such as mutations (in tumor suppressor genes and oncogenes) and chromosomal abnormalities. Today, however, it is well recognized that cancers are also epigenetic diseases in which the DNA methylome is strongly affected. The genome of cancer cells displays global hypomethylation on the one hand, but local hypermethylation on the other.3 This has prompted researchers to evaluate the epigenetic component of cancers, and notably breast cancers. The emergence of new technologies is making it feasible to assess DNA methylomes at genome-scale.4,5
In the study described in Dedeurwaerder et al.,6 we used Infinium Methylation 27K technology to profile DNA methylomes of 236 human breast tumors. We found that based on their DNA methylation profiles, breast tumors segregate into two major categories linked to their estrogen receptor (ER) status. This provides evidence that whole sets of genes whose expression depends on this status are epigenetically regulated. Such observations are in agreement with other groups7 and with the classification based on gene expression profiles. Beyond these two groups based on ER status, DNA methylation profiling revealed six groups of breast tumors. This approach is thus likely to contribute to refining the classification of breast tumors.
Two of the six groups emerged as particularly interesting, being distinguished from the others by the presence of several hypomethylated immune genes, such as LCK and ITGAL. As we profiled whole tumors, i.e. samples composed mainly of tumoral cells but also of stromal and immune cells from the tumor microenvironment, we hypothesized that these hypomethylated markers might reflect an infiltration of these tumors by immune cells, and particularly by lymphocytes. Histological analysis of these tumors and DNA methylation profiling of ex vivo lymphocytes confirmed this hypothesis. Thus, DNA methylation profiling of breast tumors seems to be able to reveal to some extent the cell type composition of the tumor microenvironment, at least as regards infiltration by immune cells.
In our study, we also evaluated the prognostic value of genes displaying the strongest anti-correlation between their expression and methylation status. We found that 58% (32/55) of these genes to have a high prognostic value for relapse-free survival. Most interestingly, the majority of the identified prognostic markers are involved in immunity, and particularly in T cell biology. Examples include the CD3D, CD3G, CD6, LCK, LAX1, SIT1, UBASH3A and ICOS genes. High expression of these genes appears associated with a better clinical outcome, and with a high level of lymphocyte infiltration. Some of these genes (CD3D, CD3G, ICOS and UBASH3A) furthermore, appeared highly methylated and barely expressed in ex vivo B lymphocytes (as opposed to ex vivo T lymphocytes), indicating that the infiltrating lymphocytes were mostly T lymphocytes.
It is recognized today that the response of a tumor to a given treatment depends on both a tumor component and a host component, the latter comprising a major immune component. To date, unfortunately, the predictive signatures revealed by gene expression profiling concern mainly the tumor component, with markers related principally to tumor-cell proliferation.2 Our study, in contrast, highlights the immune component. Although some studies have linked prognosis and prediction to treatment response to immune-cell expression signatures,8,9 methylation profiling would seem to be a particularly sensitive means of exploring this under-evaluated component. The discovery that DNA methylation plays an important role in Th cell differentiation supports this view.10 By establishing the genome-wide DNA methylation profiles of different types of immune cells liable to be present in the tumor microenvironment, and notably of T lymphocytes, it should be possible to recognize their signatures in breast tumor samples and to correlate these signatures with clinical outcomes and responses to treatment.
Overall, our results thus suggest that DNA methylation profiling could open new avenues to better understand the emerging intricate relationship existing between the tumor cells, the surrounding stroma and the host immune component. Our data also suggest that DNA methylation profiling could be used, in combination with other tools currently available, to improve breast cancer prognosis and prediction of responses to treatment.
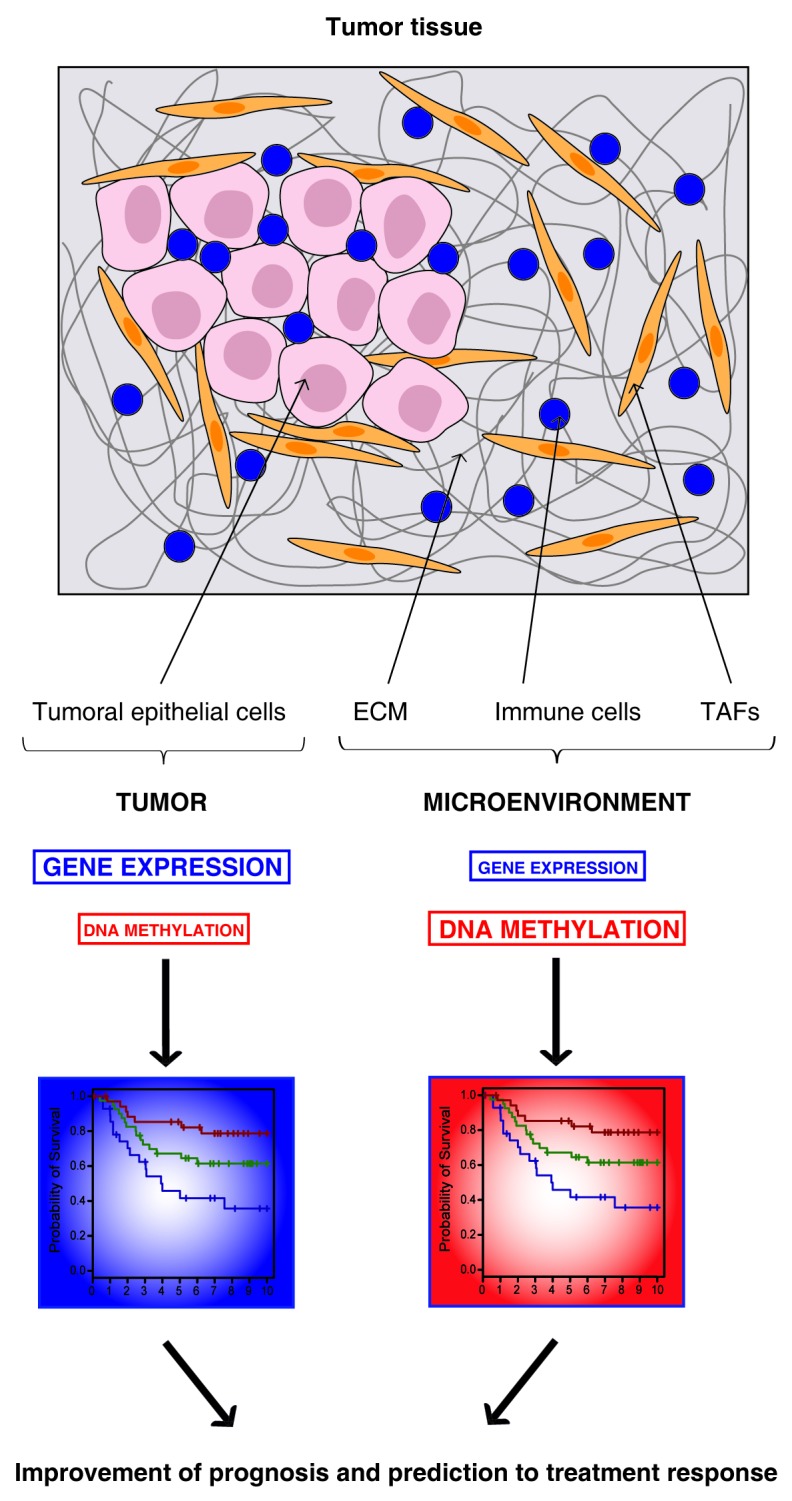
Figure 1.Combination of gene expression and DNA methylation profiling might improve prognosis and prediction to treatment response for breast cancer. Tumor tissue is composed of two major components: the tumoral epithelial cells and the tumor microenvironment that includes the extracellular matrix (ECM), immune cells and tumor-associated fibroblasts (TAFs). While gene expression profiling highlights majorly markers from the tumor component, DNA methylation profiling is more sensitive for the detection of markers from the microenvironment. Therefore, together, gene expression and DNA methylation markers should bring a more complete view of the tumor tissue, this might help for breast cancer patient management.
Glossary
Abbreviations:
- ECM
ExtraCellular Matrix
- ER
Estrogen Receptor
- TAFs
Tumor-Associated Fibroblasts
- Th
T-helper lymphocyte
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/19996
References
- 1.Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–9. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- 2.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, et al. Genome-wide DNA methylation profiling using Infinium(®) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 5.Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–84. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 6.Dedeurwaerder S, Desmedt C, Calonne E, Singhal SK, Haibe-Kains B, Defrance M, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. 2011;3:726–41. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dedeurwaerder S, Fumagalli D, Fuks F. Unravelling the epigenomic dimension of breast cancers. Curr Opin Oncol. 2011;23:559–65. doi: 10.1097/CCO.0b013e32834bd481. [DOI] [PubMed] [Google Scholar]
- 8.Denkert C, Loibl S, Noske A, Roller M, Müller BM, Komor M, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–13. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 9.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 10.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]


