Abstract
Carbonic anhydrase (CA) (EC 4.2.1.1) enzymes catalyze the reversible hydration of CO2, a reaction that is important in many physiological processes. We have cloned and sequenced a full-length cDNA encoding an intracellular β-CA from the unicellular green alga Coccomyxa. Nucleotide sequence data show that the isolated cDNA contains an open reading frame encoding a polypeptide of 227 amino acids. The predicted polypeptide is similar to β-type CAs from Escherichia coli and higher plants, with an identity of 26% to 30%. The Coccomyxa cDNA was overexpressed in E. coli, and the enzyme was purified and biochemically characterized. The mature protein is a homotetramer with an estimated molecular mass of 100 kD. The CO2-hydration activity of the Coccomyxa enzyme is comparable with that of the pea homolog. However, the activity of Coccomyxa CA is largely insensitive to oxidative conditions, in contrast to similar enzymes from most higher plants. Fractionation studies further showed that Coccomyxa CA is extrachloroplastic.
CA (EC 4.2.1.1) is a zinc-containing enzyme that catalyzes the reversible reaction CO2 + H2O ↔ HCO3− + H+. CA is widely distributed throughout nature, from eukaryotes such as vertebrates, invertebrates, and plants, to prokaryotes such as archaeabacteria and eubacteria. The enzyme is classified into three independent CA gene families designated α, β, and γ (Hewett-Emmett and Tashian, 1996). The α-CAs are found primarily in animals (Tashian, 1992), but homologs have also been identified in the bacterium Neisseria gonorrhoeae (Hewett-Emmett and Tashian, 1996) and the green alga Chlamydomonas reinhardtii (Fukuzawa et al., 1990). This is the most extensively studied CA family, and includes the biochemically well-characterized mammalian CA isozymes, the crystal structures of which have been solved to high resolution (Kannan et al., 1975; Eriksson et al., 1988a; Eriksson and Liljas, 1993; Boriack-Sjodin et al., 1995).
Conversely, the γ-CAs are a newly discovered gene family, with the enzyme from Methanosarcina thermophila being the only γ-CA isolated and characterized thus far (Alber and Ferry, 1994). Related sequences have been found in several eubacteria and in Arabidopsis (Hewett-Emmett and Tashian, 1996), but it is not known as yet whether they encode functional CAs. The crystal structure for the γ-CA from M. thermophila is different from that of the α-type enzymes. The γ-CA is trimeric, with the active site situated between the subunits (Kisker et al., 1996).
CAs belonging to the β-CA family have been found in both C3 and C4 monocot and dicot plants, in the mitochondria of C. reinhardtii, and in various eubacteria (Eriksson et al., 1996; Hewett-Emmett and Tashian, 1996). Among the dicot species, the sequence similarity between the different β-CAs is around 80% (60% identity). The homology is slightly lower between the monocot and dicot homologs, but the similarity remains considerable (>70%). In contrast, the β-CAs found in prokaryotes are more variable and exhibit low sequence similarity to the plant homologs (30%). Alignment of all known amino acid sequences from functional β-CAs reveals invariant amino acid residues at 26 positions, of which most are found within two regions. Extended radiographic-absorption fine-structure analysis of spinach CA suggests a Cys-His-Cys-H2O ligand scheme for binding of the zinc ion (Bracey et al., 1994; Rowlett et al., 1994). The first of these invariant Cys residues is found in one conserved region, whereas the His and the other Cys residue are situated in a second conserved region. However, to our knowledge, no three-dimensional structure has been described for any β-CA. Most of the biochemical studies have been done on chloroplastic homologs from C3 dicots and on the Escherichia coli enzyme.
Chloroplastic β-CA is nuclear encoded and synthesized in the cytoplasm with an N-terminal transit peptide that targets the precursor into the chloroplast stroma (Forsman and Pilon, 1995). Subsequent maturation involves removal of the transit peptide, folding, and oligomerization. The native molecular masses of CAs from C3 dicot plants have been reported to vary between 140 and 250 kD, with a subunit mass of 26 to 34 kD, each binding one zinc ion (Reed and Graham, 1981). The β-CA multimeric complex has been shown to consist of eight subunits (Aliev et al., 1986; Björkbacka et al., 1997). CAs from monocot plants have a monomeric mass of around 25 kD and an estimated native mass of 42 kD (Atkins et al., 1972; Atkins, 1974). The CA from E. coli is also reported to be an oligomer, most likely a tetramer or a dimer, depending on the experimental conditions (Guilloton et al., 1992).
The kinetic characteristics of chloroplast β-CA have been studied for both pea (Johansson and Forsman, 1993) and spinach enzymes (Pocker and Ng, 1973; Rowlett et al., 1994). Both possess a high catalytic efficiency, with Kcat values between 105 and 106 s−1 at high pH. The kinetic mechanism for these enzymes is consistent with the general mechanism proposed for the high-activity α-CA isozymes (Silverman and Lindskog, 1988).
The rate of the uncatalyzed interconversion between the two inorganic carbon species, CO2 and HCO3−, is insufficient to cope with the metabolic demand within a plant cell, but this varies among different organisms (for review, see Badger and Price, 1994). Because CA catalyzes the reversible hydration of CO2, it has been proposed that chloroplastic CA is involved in the fixation of CO2 in the Calvin cycle, which involves the carboxylating enzyme Rubisco (Reed and Graham, 1981). CO2 is the only substrate for Rubisco, but HCO3− is the predominant carbon species within the alkaline chloroplast stroma (Makino et al., 1992). In addition, CA appears to play a role in the CCM present in certain cyanobacteria and free-living algae (for review, see Badger, 1987).
Some species of green algae and cyanobacteria that are photosynthetic components of lichens also possess CCMs, but there are some species that appear to lack this mechanism. The green alga Coccomyxa is one such photobiont (Palmqvist et al., 1994). The Coccomyxa genus is composed of many species forming symbiotic relationships in a small but diverse group of lichens (Honegger, 1991). This alga is unusual in that it lacks a CCM but has a very high intracellular CA activity (Hiltonen et al., 1995). The specific biological function of this internal CA in Coccomyxa is unknown at present, as is its subcellular location.
The aim of this study was to characterize the major intracellular β-CA from Coccomyxa. By cloning and sequencing the corresponding cDNA, we were able to overexpress the enzyme in E. coli and purify it to homogeneity. Structural and kinetic studies of this new β-CA demonstrated that the Coccomyxa enzyme possesses several interesting properties distinct from the higher-plant β-CAs. Furthermore, we demonstrated that the Coccomyxa CA is extrachloroplastic and probably located to the cytosol.
MATERIALS AND METHODS
Determination of Internal Amino Acid Sequences
Internal amino acid sequences were determined after SDS-PAGE (12.5% polyacrylamide) of previously semipurified Coccomyxa CA (Hiltonen et al., 1995). The gel was stained with 0.5% Coomassie brilliant blue in 20% methanol and 0.5% acetic acid to visualize the 25-kD polypeptide. The band was excised and subjected to digestion with modified trypsin (Promega) according to the method of Rosenfeld et al. (1992). The collected peptides were subjected to amino acid sequence analysis using a sequencing system (model 476A, Applied Biosystems).
RNA Isolation and cDNA Library Construction
Total RNA was isolated from 2-L Coccomyxa cultures grown according to the method of Hiltonen et al. (1995). Cells were centrifuged at 1,100g for 10 min at 4°C and the pellet (3.7 g) was resuspended in 10 mL of 50 mm Tris-HCl, pH 7.6, and 10 mm EDTA. Cells were lysed in a precooled French pressure cell (Aminco, Silver Spring, MD) at 160 MPa and immediately mixed with 40 mL of 4 m guanidinium isothiocyanate, 50 mm Tris-HCl, pH 7.6, 10 mm EDTA, and 2% sarcosyl. The mixture was centrifuged at 4,000g for 5 min at 4°C. CsCl was added to the supernatant to a final concentration of 40% (w/v), and the supernatant was centrifuged for 10 min at 31,000g at 4°C. The resulting supernatant was laid on a 5.7 m CsCl cushion and centrifuged for 18 h at 150,000g at 20°C. The pellet was resuspended in 200 μL of prewarmed (56°C) RNA-elution buffer (mRNA isolation kit, Stratagene) containing 1% SDS. The RNA sample was phenol extracted once and precipitated with ethanol, then the pellet was resuspended in 100 μL of elution buffer. Polyadenylated RNA was isolated from total RNA using an mRNA isolation kit. A Coccomyxa cDNA library was made from 5 μg of the purified poly(A+) RNA using a cDNA-synthesis kit (ZAP Express, Stratagene).
Screening of the cDNA Library
About 1.5 × 105 plaque-forming units were spread among Escherichia coli XL1-Blue MRF (Stratagene) host cells on agar plates and analyzed according to standard procedures (Sambrook et al., 1989) using a 550-bp DNA probe corresponding to the 3′ end of the Coccomyxa CA. The probe was generated by PCR amplification from the Coccomyxa cDNA library using the degenerate primer 5′-CGGGAATTCGCIGGIGTIACIAA(T/C)(T/C)TITGGAT-3′, corresponding to the internal peptide sequence TAGVTNLWI, and the nondegenerate primer T7 (22-mer, Stratagene). PCR was performed in a thermal cycler (Perkin-Elmer) using 1 × 107 plaque-forming units of the cDNA library. Hybridization to the radiolabeled probe was carried out at 65°C for 15 h in 4× SSPE (1× SSPE = 150 mm NaCl, 1 mm EDTA, and 15 mm sodium phosphate, pH 7.4), 5× Denhardt's solution (0.05% Ficoll 400, 0.05% PVP, and 0.05% BSA), 0.5% SDS, and denatured salmon-sperm DNA (100 μg mL−1). After hybridization the filters were washed at 65°C in 2× SSPE, 0.5% SDS followed by 1× SSPE, 0.1% SDS. Positive plaques were identified and isolated according to the instructions of the manufacturer (Stratagene).
DNA Sequencing
Selected clones were sequenced by the dideoxy chain-termination method using T3 and T7 primers (Stratagene) and the fmol DNA sequencing system (Promega). DNA sequences were analyzed using Genetics Computer Group (Madison, WI) software (Devereux et al., 1984).
Expression of Coccomyxa CA in E. coli and Protein Isolation
A 786-bp fragment containing the complete coding region for the Coccomyxa CA, including an extra 86-bp 3′ untranslated region, was obtained by PCR amplification using the primers 5′-GCGGAATTCATCGAGGGACGCATGTCAGCTAAAGACACTGCC-3′ and 5′-CTCCATCTAGAGTCACCTTGTAGGCA-3′, which contain cleavage sites for EcoRI and XbaI, respectively. The PCR product was digested with EcoRI and XbaI, and then ligated into the expression vector pMAL-c2 (New England Biolabs) just downstream of and in-frame with the malE gene encoding the MBP. The resulting MBP/β-CA was expressed in E. coli and purified using an amylose resin (New England Biolabs). For enzyme production, the cells were grown in Luria-Bertani broth containing 0.2% Glc and 50 μg mL−1 carbenicillin at 37°C to an A600 of 0.6. Isopropylthio-β-galactoside was added to a final concentration of 1.5 mm and the incubation was continued for another 2 h.
The cells were harvested by centrifugation at 4,000g at 4°C, and resuspended in 50 mL of 20 mm Tris-HCl, pH 7.4, 200 mm NaCl, and 1 mm EDTA (column buffer) before being disrupted in a precooled French pressure cell. Intact cells and cell debris were removed by centrifugation at 14,000g. The supernatant was diluted five times in column buffer and loaded on an amylose-resin column preequilibrated with column buffer at a flow rate of 1 mL min−1. After washing with eight column-volumes of column buffer, the fusion protein was eluted with 10 mm maltose in the same buffer. The sample was concentrated using a Centriprep 30 unit (Amicon, Beverly, MA) and incubated for 10 h with factor Xa (Boehringer Mannheim) at a final concentration of 0.5%. The cleaved fusion protein was desalted using a PD-10 column (Pharmacia) before being loaded on a Q-Sepharose FF (Pharmacia) ion-exchange column equilibrated with 20 mm Tris-HCl, pH 8.0, and 1 mm EDTA. Coccomyxa CA was eluted with a stepwise gradient of 20 mm Tris-HCl, pH 8.0, 0.5 m NaCl, and 1 mm EDTA. Fractions containing CA activity were pooled and loaded for one more passage through the affinity column. The isolated protein was concentrated using a Centricon 10 device (Amicon), and fractions containing CA activity throughout the purification steps were analyzed by SDS-PAGE (Laemmli, 1970). The purified CA was sequenced using a sequencing system (model 476A, Applied Biosystems) to verify that the N terminus was correct after cleavage of the fusion protein.
Preparation of Antiserum
The purified Coccomyxa CA, in PBS (10 mm phosphate buffer, pH 7.4, containing 150 mm NaCl and 2.5 mm KCl), was mixed with Freund's complete adjuvant and injected into rabbits (Agrisera AB, Vännäs, Sweden). Every 14 d the immunized rabbits were injected with another 100 μg of CA mixed with incomplete adjuvant.
Protein Concentration
Protein concentrations were determined either according to the method of Bradford (1976) or by determining the A280 for the purified enzyme. The molar extinction coefficient (ε) for Coccomyxa CA was determined as described by Gill and von Hippel (1989), giving a value of ε280 = 37,500 ± 3,500 m−1 cm−1. All stated enzyme concentrations are subunit concentrations.
Protein Structure Analyses
The native molecular mass of the purified Coccomyxa CA was estimated by gel-filtration chromatography performed on a Sephacryl S-300H column (Pharmacia) equilibrated with 20 mm Tris-HCl and 0.1 m NaCl, pH 7.4. Ovalbumin (45 kD), aldolase (158 kD), catalase (240 kD), and ferritin (450 kD) (Combithek, Boehringer Mannheim) were used as protein standards. Purified Coccomyxa CA and soluble cell proteins were analyzed by 9% polyacrylamide native-PAGE. The proteins were blotted onto a nitrocellulose filter for immunoreaction tests with antiserum directed against CA from Coccomyxa in conjunction with horseradish peroxidase-conjugated secondary antibodies and an enhanced-chemiluminescence detection system (Amersham).
CD spectra for Coccomyxa CA were measured on a spectropolarimeter (model J-7720, Jasco, Easton, MD) at 23°C. Each spectrum shown was the result of three scans using a bandwidth of 1 nm. For far-UV-region analysis the protein concentration was 0.25 mg mL−1 and the path length was 1 mm. For near-UV analysis the protein concentration was 1.0 mg mL−1 and the path length was 4 mm. Spectra recorded for pea CA were obtained using the same protocol except that the far-UV region was scanned in a spectrodichrograph (model CD6, Jobin-Yvon Instruments SA, Longjumeau, France) using a sample concentration of 0.5 mg mL−1 and a 0.5-mm path length. The samples contained 10 mm potassium phosphate buffer, pH 7.5. The observed ellipticities were converted to mean residue ellipticities (θ) on the basis of a molecular mass of 24.7 kD and 227 amino acids for Coccomyxa CA, and 24.2 kD and 221 amino acids for pea CA.
CA Activity Measurements
During enzyme purification, fractionation, activation, and inhibition studies, CO2-hydration activity was assayed at 2°C using the colorimetric method of Rickli et al. (1964). Initial rates of CO2 hydration were measured at 578 nm using a sequential stopped-flow spectrofluorimeter (model DX-17MV, Applied Photophysics, Leatherhead, UK) at 25°C by the changing-pH-indicator method (Khalifah, 1971; Steiner et al., 1975). The buffer/indicator pair was Taps (3-[{2-hydroxy-1,1-bis(hydroxymethyl)etyl}amino]-1-proparesulfonic acid)/m-cresol purple with 10 μm EDTA. The initial rates, calculated by fitting data from the first part of the trace to a first-order rate equation, were fitted by nonlinear regression to the Michaelis-Menten equation using the GraFit program (Erithacus Software Ltd., London, UK).
Modification of Free Cys Residues with DTNB
The free thiol content was estimated from the increase in A412 caused by formation of a 2-nitro-5-thiobenzoate anion caused by cleavage of DTNB upon reaction with a thiolate anion. A molar extinction coefficient of 14,150 m−1 cm−1 was used in the calculations (Riddles et al., 1983). The reactions were carried out in 0.1 m potassium phosphate buffer, pH 7.3, 1 mm EDTA, in a spectrophotometer (model 320, Perkin-Elmer). Enzyme concentrations were 0.5 to 3 μm, and DTNB was added in a molar excess of 1000. Reduced enzyme samples were obtained by incubation with 10 mm DTT or 100 mm 2-mercaptoethanol for 1 h, followed by extensive dialysis against degassed phosphate buffer under N2.
Subcellular Fractionation
A 10% to 80% linear Percoll gradient was generated by mixing 100% Percoll with 2× breaking medium (1× breaking medium: 35 mm Hepes-KOH, pH 7.7, 375 mm sorbitol, 10 mm EDTA, 1 mm MnCl2, and 5 mm MgCl2) in a 1:1 ratio and centrifuging the solution at 40,000g for 1 h. One liter of Coccomyxa culture (5 μg mL−1 chlorophyll) was centrifuged at 1,500g for 10 min at 4°C and resuspended in 20 mL of 1× breaking medium. Cells were disrupted for 30 s in a precooled Bead Beaker (Biospec Products, Bartlesville, OK) filled with 0.5-mm-diameter glass beads and cell suspension (1:1 [v/v]). The disrupted cells were carefully layered onto the Percoll gradient and centrifuged at 3,000g for 20 min at 4°C. Four distinct fractions were taken, each characterized with a light microscope.
Marker enzyme activities were measured at 25°C in all subcellular fractions. The chloroplast stromal marker NADP-GAPDH was measured according to the method of Winter et al. (1982), except that 4 mm DTT rather than GSH was included in the assay medium to ensure full activation of the enzyme. PEPC was measured as a marker for the cytosol (Gardeström and Edwards, 1983). For all marker-enzyme-activity measurements, changes in A340 resulting from NAD(P)H cleavage were monitored on a spectrophotometer (model DU-8, Beckman). Protein samples from the fractionation step were separated on a 15% SDS-PAGE gel and blotted onto a nitrocellulose filter (MSI, Westboro, MA). Antiserum directed against Coccomyxa CA was used, and the antibody-antigen conjugate was detected using horseradish peroxidase-linked secondary antibodies and enhanced chemiluminescence (Amersham).
RESULTS
Cloning and Sequencing of β-CA cDNA
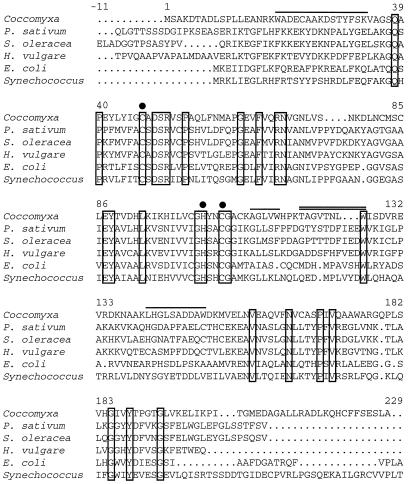
Semipurified intracellular CA isolated from Coccomyxa cells as previously described (Hiltonen et al., 1995) was digested with trypsin and the resulting peptides were separated by HPLC. Five different internal amino acid sequences were determined (Fig. 1). One of the sequences, TAGVTNLW, was used to design a degenerate 18-base primer. Along with a 22-base primer specific for the T7 promoter, the degenerate oligonucleotide was used to amplify a 550-bp fragment from the Coccomyxa cDNA library. Subsequent sequencing of the fragment confirmed that it was derived from a cDNA encoding part of a β-CA gene.
Figure 1.
Comparison of the deduced amino acid sequence of Coccomyxa CA with Synechococcus sp. PCC 7942 IcfA and E. coli CynT proteins and the chloroplast-located CAs from the higher plants pea (Pisum sativum), spinach (Spinacia oleracea), and barley (Hordeum vulgare). The overlined amino acids represent the trypsin-cleaved peptide fragments identified by amino acid sequencing. The internal peptide sequence used to design a degenerated primer and PCR amplification of the 550-bp cDNA screening probe is double overlined. The boxed residues represent amino acids that are conserved among all functional β-CAs. The transit peptides from pea, spinach, and barley have been deleted, and putative zinc-ligands are marked with filled circles. Numbers refer to the Coccomyxa sequence starting at position 1 with the gaps excluded. The alignment was generated using the PILEUP program (Genetics Computer Group).
The 550-bp fragment was used as a specific β-CA probe to screen the Coccomyxa cDNA library, from which 15 positive clones were obtained. Three of the longest cDNAs were completely sequenced in both directions, and all three were identical except for different amounts of truncation at the 5′ end. The longest of the three selected clones was 1137 bp, consisting of an open-reading frame of 681 bp, with 198 and 258 untranslated nucleotides in the noncoding 5′ and 3′ regions, respectively. The cDNA library was rescreened with a probe corresponding to the 5′ end of the CA cDNA. This fragment starts at amino acid position 23 and stops 570 bp downstream. The second screening gave an additional set of seven positive clones. These clones, together with five of the unsequenced clones from the first screening, were sequenced from the 5′ end and 500 bp downstream and were all found to be identical to the full-length cDNA. The 5′-untranslated region contained stop codons in all three reading frames. No Met codon was found in this region upstream of the start Met codon. The deduced protein of 227 amino acid residues has a predicted molecular mass of 24.7 kD and compared with other β-CAs (Fig. 1) is about 50% similar (26%–30% identical) to higher-plant and eubacterial homologs.
Overexpression of Coccomyxa CA in E. coli
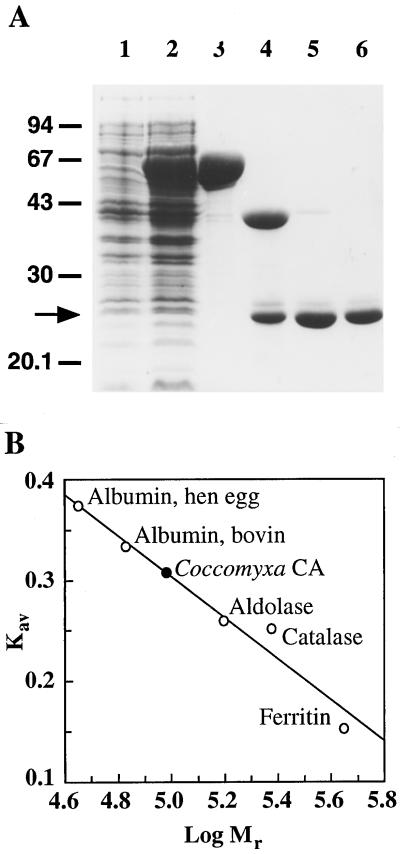
To characterize the Coccomyxa CA in more detail, the corresponding cDNA was overexpressed in E. coli as a fusion to the C terminus of the MBP. After purification of the fusion protein, the Coccomyxa CA was removed by factor Xa digestion, and then isolated by ion-exchange chromatography. The purified protein migrated as a single discrete band with a molecular mass of 25 kD on a SDS-PAGE gel (Fig. 2A). The CA has an apparent native molecular mass of approximately 100 kD, as indicated by gel-filtration chromatography (Fig. 2B). Western-blot analysis was performed to determine whether the oligomeric state of the overexpressed protein matched that of the Coccomyxa endogenous enzyme. Under nondenaturing conditions the overexpressed protein separated as a single complex, and corresponded to a similar-sized band in the crude extract (data not shown). Thus, this algal enzyme seems to be homotetrameric.
Figure 2.
Purification and native molecular mass estimation of Coccomyxa CA expressed in E. coli. The Coccomyxa CA was overexpressed in E. coli as a fusion protein with MBP. A, Samples from the purification steps were analyzed by SDS-PAGE. The gel was stained with Coomassie blue G-250. Lane 1, Uninduced cell extract (10 μg); lane 2, induced cell extract (10 μg); lane 3, first amylose resin chromatography (10 μg); lane 4, factor Xa digestion (5 μg); lane 5, Q-Sepharose chromatography (2 μg); and lane 6, second amylose resin chromatography (2 μg). B, Purified Coccomyxa CA analyzed by size-exclusion chromatography using a Sephacryl S-300H column (Pharmacia). Kav = (Ve − Vo)/(Vt − Vo); where Ve is the elution volume, Vo is the column void volume, and Vt is the total bed volume.
Structural Analysis of Coccomyxa CA
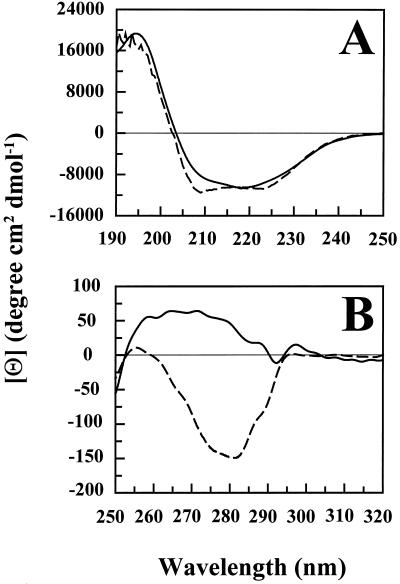
In the far-UV region (Fig. 3A) the CD spectrum of the Coccomyxa enzyme has an intense positive band at 194 nm and two slightly weaker negative bands at 208 and 218 nm. This band pattern suggests that Coccomyxa CA has a high α-helix structure content (Johnson, 1990). The higher-plant homolog from pea also seems to be dominated by α-helix structures, and the spectra for the two enzymes suggest a similar content of various secondary structure elements (Fig. 3A). In the near-UV region there are extensive differences: Coccomyxa CA has a predominantly positive CD spectrum, whereas the spectrum for pea CA is dominated by a negative band at around 280 nm (Fig. 3B).
Figure 3.
CD spectra of Coccomyxa and pea CA. A, Far-UV region; B, near-UV region. Solid line, Coccomyxa CA; dashed line, pea CA.
Kinetic Properties
The Coccomyxa CA was found to have a high catalytic activity. Kinetic parameters for the CO2-hydration reaction were determined using the stopped-flow technique. Values of Kcat = (3.8 ± 0.1) × 105 s−1 and Km = 4.7 ± 0.3 mm were obtained in 50 mm Taps buffer, pH 8.7, at 25°C. These are somewhat higher than the values reported for the pea CA (Johansson and Forsman, 1993), whereas the Kcat:Km ratio of (8.0 ± 0.5) × 107 m−1 s−1 is almost identical to that reported for the higher-plant CA. The differences are comparatively small and could reflect subtle differences in buffer and pH dependencies. Levels of inhibition of the CO2-hydration activity of Coccomyxa CA caused by specific inhibitors are presented in Table I, together with Ki values for the pea homolog. A relatively large difference was observed between the algal and pea enzymes in their sensitivity to the sulfonamide inhibitors. The binding affinity of ethoxyzolamide is almost 30 times higher for the pea enzyme than for the Coccomyxa CA, whereas the binding affinity of acetazolamide is more than 10 times higher for the algal protein. The inhibition by anions showed only minor variations between the two CAs.
Table I.
Inhibition of Coccomyxa CA by sulfonamides and anions
| Inhibitor |
Ki
|
|
|---|---|---|
| Coccomyxa CA | Pea CA | |
| μm | ||
| Ethoxyzolamide | 11 ± 1 | 0.4 |
| Acetazolamide | 2.1 ± 0.5 | 28 |
| SCN− | 6.6 ± 0.7 | 20 |
| N3− | 17 ± 3 | 6 |
| Cl− | 48,000 ± 5,000 | 40,000 |
CO2-hydration activity was assayed by colorimetry according to the method of Rickli et al. (1964) with a pH change from 8.2 to 6.5. The reactions were followed at 2°C in 10 mm barbital buffer in the presence of inhibitor at different concentrations. Data for pea CA are from Johansson and Forsman (1993).
Oxidation/Reduction of Coccomyxa CA
β-CAs localized to higher plant chloroplasts have been reported to be sensitive to oxidation and, therefore, are dependent on a reducing environment to retain catalytic activity (Tobin, 1970; Atkins et al., 1972; Cybulsky et al., 1979; Johansson and Forsman, 1993). Oxidized pea CA required a reducing agent for maximal activation of the enzyme (Johansson and Forsman, 1993). In contrast, the activity of Coccomyxa CA was found to be independent of the presence of a reducing agent. The CO2-hydration activity of enzyme purified without reductant in the isolation buffers was high, and it remained constant when the enzyme was incubated for 10 min in 100 mm 2-mercaptoethanol or 1 mm DTT (data not shown). Thus, the catalytic activity of Coccomyxa CA is not significantly affected by the oxidation state. Analysis of the amino acid sequence for Coccomyxa CA (Fig. 1) shows the presence of nine Cys residues in each subunit. Of these, only the two Cys residues thought to act as zinc ligands are conserved.
The accessibility of the Cys residues was determined by the addition of DTNB to oxidized and reduced enzyme. The overexpressed enzyme purified from E. coli was assumed to be oxidized because of the absence of reductants in the isolation buffers. The enzyme was reduced by incubation with 10 mm DTT or 100 mm 2-mercaptoethanol for 1 h, followed by dialysis under a cushion of N2 to remove excess reducing agent. Reacting the oxidized Coccomyxa CA with DTNB gave a molar ratio of modified Cys residues per subunit of 3.9 ± 0.1. Assuming that two Cys residues are zinc ligands, and thus are inaccessible to DTNB, it follows that three Cys residues within the oxidized enzyme did not react with DTNB. In the reduced enzyme, the molar ratio of modified Cys residues per subunit was 5.6 ± 0.3, indicating that one to two extra Cys residues are accessible after reduction, although at least one residue remains inaccessible in the reduced state. These results suggest that the oxidized Coccomyxa CA contains a disulfide bridge that can be broken upon reduction. Moreover, the formation of this disulfide does not affect the enzymatic activity. The mobility of Coccomyxa CA in gel electrophoresis under denaturing conditions is not affected by the presence or absence of reducing agents in the sample buffer, indicating that no disulfide bridges are formed between subunits (data not shown).
Cell-Fractionation Studies
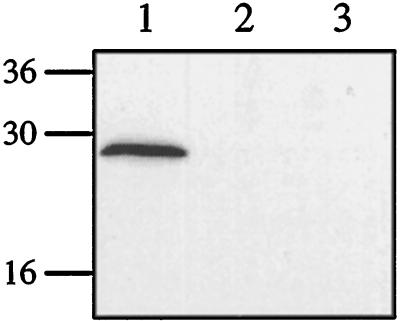
Microscopic studies of the fractions generated by separation in a Percoll gradient showed an enrichment of seemingly intact chloroplasts in fraction 3, although it also contained aggregated material, probably derived from thylakoid membranes. Fraction 1 contained soluble proteins, derived from both the cytosol and broken organelles; fraction 2 contained thylakoid membranes; and fraction 4, the lowest fraction, contained intact cells. Measurements of marker-enzyme activities supported the microscopic observations (Table II). Most of the activity of both NADP-GAPDH (95%) and PEPC (100%) were detected in fraction 1, whereas no enzyme activity was detected in fractions 2 and 4. Fraction 3 contained activity for the chloroplast marker enzyme NADP-GAPDH (5%) but no PEPC activity. This confirms the presence of intact chloroplasts in fraction 3. Of the four fractions, CA activity was found only in fraction 1. Western-blot analysis confirmed the CA activity measurements, with CA protein detected only in fraction 1 (Fig. 4). No protein corresponding to Coccomyxa CA was detected in fraction 3, the fraction containing intact chloroplasts.
Table II.
Distribution of marker-enzyme activities in a soluble fraction (fraction I) and an enriched chloroplast fraction (fraction 3)
| Fraction | NADP-GAPDH | PEPC | CA | PEPC:NADP-GAPDH | CA:NADP-GAPDH |
|---|---|---|---|---|---|
| pmol cell−1 h−1 | microunitsa cell−1 | ||||
| Soluble | 43.9 | 5.8 | 45.1 | 0.13 | 1.03 |
| Chloroplast | 2.6 | <0.02b | <0.05b | <0.01 | <0.02 |
The values are the means from two fractionation experiments.
1 unit= 1 activity unit as decribed by Rickli et al. (1964).
Activities of PEPC and CA were below the detection limit.
Figure 4.
Western-blot subcellular fractions separated by SDS-PAGE and analyzed by immunoblotting using antisera specific for Coccomyxa CA. Lane 1, Soluble fraction (5 μg of protein); lane 2, thylakoid fraction (5 μg); and lane 3, chloroplast fraction (5 μg).
DISCUSSION
We have isolated and characterized a cDNA encoding a β-CA from the alga Coccomyxa. Enzymes from the β-CA family share some common features with members of the other two CA families, α-CAs and γ-CAs. They are all zinc enzymes that catalyze the reversible dehydration of HCO3− to CO2, and they are all sensitive to similar kinds of chemical inhibitors, including sulfonamides and monovalent anions. Nevertheless, the α-, β-, and γ-CAs clearly belong to three distinct gene families according to sequence homology (Hewett-Emmett and Tashian, 1996). On the same basis, β-CAs can be further divided into three subgroups: those originating from eubacteria, dicot plants, and monocot plants (Hewett-Emmett and Tashian, 1996). Alignment of the deduced amino acid sequence for the Coccomyxa CA with other known β-CAs, however, shows that this enzyme cannot readily be classed with any of these subgroups.
According to both subcellular fractionation and western-blot analysis, it appears that the subcellular localization of Coccomyxa CA is extrachloroplastic. The marked decrease in the PEPC:NADP-GAPDH ratio indicates little or no contamination of cytoplasm in the chloroplast fraction. If CA were chloroplastic, the CA:NADP-GAPDH ratio would increase in the chloroplast fraction compared with the soluble fraction (Table II). Instead, a distinct decrease was observed, clearly indicating an extrachloroplastic localization of CA. This is supported by western-blot analysis, in which no CA protein could be detected in the chloroplast fraction (Fig. 4, lane 3). Because of the lack of an obvious transit peptide, there is no indication at present of any other localization for the Coccomyxa CA than in the cytosol. Furthermore, the isolated cDNA is almost certainly full length, since the 200-bp 5′-untranslated region upstream from the putative start Met does not contain any additional Met codons before the stop codons in any of the three frames. The 5′ ends of 12 positive clones were also analyzed and all sequences were found to be identical except for different amounts of truncation, which strongly indicates that the sequence was reliably identified. In summary, the majority of CA activity in Coccomyxa is located in the cytosol, although the presence of as-yet-unidentified chloroplastic or mitochondrial CAs cannot be excluded.
The specific function of a cytosolic CA in Coccomyxa is unclear at this time. In a previous study Palmqvist et al. (1995) suggested that this CA was chloroplast located and that it had a role similar to that of CA in C3 plants. A cytosolic CA could also facilitate the diffusion of inorganic carbon from the inner surface of the plasmalemma to the chloroplast envelope (Badger and Price, 1994). Moreover, the absence of a CCM in Coccomyxa has previously been correlated with the relatively more efficient Rubisco of this alga than that of algae possessing a CCM (Palmqvist et al., 1995). Palmqvist et al. (1995) also suggested that there was an extracellular CA, but so far we have been unable to measure any periplasmic CA activity from intact Coccomyxa cells. Another possibility is that the CA in Coccomyxa may not be directly involved in photosynthesis. As suggested by Fett and Coleman (1994), cytosolic CA may instead be required to catalyze the formation of HCO3−, the substrate for cytosolic PEPC, in a role similar to that suggested for the CA localized in the mesophyll cells of C4 plants.
The tricarboxylic acid cycle is the source of carbon skeletons for many growth processes. If the pool of intermediates in the cycle undergoes a net loss, oxaloacetate will not be regenerated. However, the mechanism whereby oxaloacetate is formed by carboxylation of PEP allows the tricarboxylic acid cycle to be replenished for continued operation. A cytoplasmic CA was recently identified in potato leaves (Rumeau et al., 1996), suggesting that the existence of cytosolic β-CA may be common to both algae and higher plants.
The CA from Coccomyxa is at least as efficient a catalyst as higher-plant CAs such as those from pea (Johansson and Forsman, 1993) and spinach (Rowlett et al., 1994). Similarly, we observed structural features common to the different β-CAs. The primary structures contain a high degree of identity. The content of secondary structure elements seems to be similar, because the general outline of the CD spectra in the far-UV region is very similar, suggesting a domination of α-helix structure. This highlights one of the structural differences between the α-, β-, and γ-CAs: the α- and γ-CAs are composed mainly of β-sheet structures (Kannan et al., 1975; Eriksson et al., 1988a; Eriksson and Liljas, 1993; Boriack-Sjodin et al., 1995; Kisker et al., 1996). However, Coccomyxa CA also shows several distinct properties that imply certain differences between the algal and higher-plant β-CAs.
There are differences in quaternary structure; the Coccomyxa CA is a homotetramer, whereas CAs from C3 dicots apparently are homooctamers (Aliev et al., 1986; Björkbacka et al., 1997). Furthermore, the near-UV CD spectra of CAs from Coccomyxa and pea differ extensively; the CD bands in this wavelength region arise mainly from immobilized aromatic side chains located in an asymmetric environment (Strickland, 1974). This region is generally assumed to be indicative of the tertiary structure of the protein. However, because the Coccomyxa and pea enzymes differ in Trp content (having five and two Trp residues, respectively), and the enzymes apparently possess distinct quaternary structures, the different shapes of the near-UV CD spectra do not necessarily imply different overall folding of the individual subunits. Furthermore, we have studied pea CA mutants that assemble into tetramers rather than wild-type octamers, and these mutants have predominantly positive CD spectra in the near-UV region, with shapes and intensities very similar to those of Coccomyxa CA (Björkbacka et al., 1997).
The enzymatic activity of the Coccomyxa CA was found to be independent of a reducing environment. CAs from the two prokaryotes E. coli and Synechococcus sp. PCC 7942 are similarly insensitive to oxidation (Guilloton et al., 1992; Price et al., 1992), whereas the CAs in pea and other C3 dicots are dependent on a reducing environment to retain catalytic activity. Of the nine Cys residues in the Coccomyxa CA, only the two proposed zinc ligands are conserved. Under oxidizing conditions, the Coccomyxa CA apparently forms a single disulfide bond. Only four Cys residues were modified by DTNB in the oxidized enzyme, whereas five to six Cys residues were modified in the reduced Coccomyxa CA. This bond is probably not formed within the active-site region. Therefore, the catalytic activity of the Coccomyxa CA remains unchanged whether the enzyme is oxidized or reduced.
The Coccomyxa CA activity is inhibited by generally recognized CA inhibitors. However, the relative sensitivities of the Coccomyxa and pea CAs to the two sulfonamides used in this study differ. The more hydrophilic sulfonamide acetazolamide binds more strongly to the Coccomyxa protein, whereas the more hydrophobic ethoxyzolamide has a stronger affinity for the pea CA. Because both CAs are equally efficient catalysts, it is unlikely that there are any significant structural differences involving the catalytically active residues. In the human CA II, the sulfonamide nitrogen ion has been shown by crystallographic studies to bind to the zinc ion by replacing the hydroxide ion, resulting in the aromatic or heterocyclic part of the sulfonamide being oriented toward the hydrophobic side of the active site (Eriksson et al., 1988b). Assuming that the sulfonamide coordination to the zinc is similar in the β-CAs, the weaker binding of the more hydrophobic (and the stronger binding of the more hydrophilic) sulfonamide to the Coccomyxa CA than to the pea enzyme may reflect differences in the hydrophobicity of the surfaces near the active site. This could be limited to differences in one or a few residues interacting with the aromatic part of the inhibitor.
At present the only β-CAs that have been catalytically investigated are from the higher plants pea (Johansson and Forsman, 1993) and spinach (Pocker and Ng, 1973; Rowlett et al., 1994). In general, the enzymatic characteristics for these β-CAs are consistent with the zinc-hydroxide mechanism proposed for α-CAs (Steiner et al., 1975), and it seems likely that the Coccomyxa CA also follows the same general mechanism. The work presented here will provide a foundation for a more detailed characterization of the physiological function of CA in Coccomyxa under different growth conditions, especially in comparison with the CA homologs in algae possessing a CCM.
ACKNOWLEDGMENT
We thank Dr. Bo Ek (Department of Cell Research, Swedish University of Agricultural Sciences, Uppsala, Sweden) for skillful determination of the amino acid sequences.
Abbreviations:
- CA
carbonic anhydrase
- CCM
inorganic carbon-concentrating mechanism
- CD
circular dichroism
- DTNB
5′,5′-dithiobis(2-nitrobenzoic azid)
- MBP
maltose-binding protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PEPC
PEP carboxylase
Footnotes
This research was supported by grants from the Swedish Natural Science Research Council and Magn Bergvalls Stiftelse.
The accession number for the Coccomyxa β-CA cDNA sequence reported in this article is U49976.
LITERATURE CITED
- Alber B, Ferry JG. A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc Natl Acad Sci USA. 1994;91:6909–6913. doi: 10.1073/pnas.91.15.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliev DA, Guliev NM, Mamedov TG, Tsuprun VL. Physicochemical properties and quaternary structure of chick pea leaf carbonic anhydrase. Biokhimiya. 1986;51:1785–1794. [Google Scholar]
- Atkins CA. Occurrence and some properties of carbonic anhydrase from legume root nodules. Phytochemistry. 1974;13:93–98. [Google Scholar]
- Atkins CA, Patterson BD, Graham D. Plant carbonic anhydrases. Plant Physiol. 1972;50:218–223. doi: 10.1104/pp.50.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR (1987) The CO2-concentrating mechanism in aquatic phototrophs. In MD Hatch, NK Boardman, eds, The Biochemistry of Plants: A Comprehensive Treatise. Vol 10, Photosynthesis. Academic Press, New York, pp 219–274
- Badger MR, Price GD. The role of carbonic anhydrase in photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:369–392. [Google Scholar]
- Björkbacka H, Johansson I-M, Skärfstad E, Forsman C. The sulfhydryl groups of Cys 269 and Cys 272 are critical for the oligomeric state of chloroplast carbonic anhydrase from Pisum sativum. Biochemistry. 1997;36:4287–4294. doi: 10.1021/bi962825k. [DOI] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Heck RW, Laipis PJ, Silverman DN, Christianson DW. Structure determination of murine mitochondrial carbonic anhydrase V at 2.45 Å resolution: implications for catalytic proton transfer and inhibition design. Proc Natl Acad Sci USA. 1995;92:10949–10953. doi: 10.1073/pnas.92.24.10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracey MH, Christiansen J, Tovar P, Cramer SP, Barlett SG. Spinach carbonic anhydrase: investigation of the zinc-binding ligands by site-directed mutagenesis, elemental analysis, and EXAFS. Biochemistry. 1994;33:13126–13131. doi: 10.1021/bi00248a023. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cybulsky DL, Nagy A, Kandel SI, Kandel M, Allen GG. Carbonic anhydrase from spinach leaves: chemical modification and affinity labelling. J Biol Chem. 1979;254:2032–2039. [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson AE, Jones TA, Liljas A. Refined structure of human carbonic anhydrase II at 2.0 Å resolution. Proteins Struct Funct Genet. 1988a;4:274–282. doi: 10.1002/prot.340040406. [DOI] [PubMed] [Google Scholar]
- Eriksson AE, Kylsten PM, Jones TA, Liljas A. Crystallographic studies of inhibitor binding sites in human carbonic anhydrase II: a pentacoordinated binding of the SCN− ion to the zinc at high pH. Proteins Struct Funct Genet. 1988b;4:283–293. doi: 10.1002/prot.340040407. [DOI] [PubMed] [Google Scholar]
- Eriksson AE, Liljas A. Refined structure of bovine carbonic anhydrase III at 2.0 Å resolution. Proteins Struct Funct Genet. 1993;16:29–42. doi: 10.1002/prot.340160104. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Karlsson J, Ramazanov Z, Gardeström P, Samuelsson G. Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1996;93:12031–12034. doi: 10.1073/pnas.93.21.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett JP, Coleman JR. Characterization and expression of two cDNAs encoding carbonic anhydrase in Arabidopsis thaliana. Plant Physiol. 1994;105:707–713. doi: 10.1104/pp.105.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsman C, Pilon M. Chloroplast import and sequential maturation of pea carbonic anhydrase: the roles of various parts of the transit peptide. FEBS Lett. 1995;358:39–42. doi: 10.1016/0014-5793(94)01391-d. [DOI] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Yamamoto Y, Dionisio-Sese ML, Miyachi S. cDNA cloning, sequence and expression of carbonic anhydrase in Chlamydomonas reinhardtii: regulation by environmental CO2 concentration. Proc Natl Acad Sci USA. 1990;87:4383–4387. doi: 10.1073/pnas.87.11.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardeström P, Edwards GE. Isolation of mitochondria from leaf tissue of Panicum miliaceum, a NAD-malic enzyme type C4 plant. Plant Physiol. 1983;71:24–29. doi: 10.1104/pp.71.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Guilloton MB, Korte JJ, Lamblin AF, Fuchs JA, Anderson PM. Carbonic anhydrase in Escherichia coli: a product of the cyn operon. J Biol Chem. 1992;267:3731–3734. [PubMed] [Google Scholar]
- Hewett-Emmett D, Tashian RE. Functional diversity, conservation and convergence in the evolution of the α-, β-, and γ-carbonic anhydrase gene families. Mol Phys Evol. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- Hiltonen T, Karlsson J, Palmqvist K, Clarke AK, Samuelsson G. Purification and characterisation of an intracellular carbonic anhydrase from the unicellular green alga Coccomyxa. Planta. 1995;195:345–351. doi: 10.1007/BF00202591. [DOI] [PubMed] [Google Scholar]
- Honegger R. Functional aspects of the lichen symbiosis. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:553–578. [Google Scholar]
- Johansson I-M, Forsman C. Kinetic studies of pea carbonic anhydrase. Eur J Biochem. 1993;218:439–446. doi: 10.1111/j.1432-1033.1993.tb18394.x. [DOI] [PubMed] [Google Scholar]
- Johnson WC., Jr Protein secondary structure and circular dichroism: a practical guide. Proteins Struct Funct Genet. 1990;7:205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- Kannan KK, Notstrand B, Fridborg K, Lövfren S, Ohlsson A, Petef M. Crystal structure of human carbonic anhydrase B: three-dimensional structure at a nominal 2.2 Å resolution. Proc Natl Acad Sci USA. 1975;72:51–55. doi: 10.1073/pnas.72.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem. 1971;246:2561–2573. [PubMed] [Google Scholar]
- Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC. A left-handed β-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 1996;15:2323–2330. [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Makino A, Sakashita H, Hidema J, Tadahiko M, Kunohiko O, Osmond B. Distinctive responses of ribulose-1,5-bisphosphate carboxylase and carbonic anhydrase in wheat leaves to nitrogen nutrition and their possible relationships to CO2-transfer resistance. Plant Physiol. 1992;100:1737–1743. doi: 10.1104/pp.100.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist K, Ögren E, Lernmark U. The CO2-concentrating mechanism is absent in the green alga Coccomyxa: a comparative study of photosynthetic CO2 and light responses of Coccomyxa, Chlamydomonas reinhardtii and barley protoplasts. Plant Cell Environ. 1994;17:65–72. [Google Scholar]
- Palmqvist K, Sültemeyer D, Baldet P, Andrews TJ, Badger MR. Characterisation of inorganic carbon fluxes, carbonic anhydrase(s) and ribulose-1,5-biphosphate carboxylase-oxygenase in the green unicellular alga Coccomyxa: comparisons with low-CO2 cells of Chlamydomonas reinhardtii. Planta. 1995;197:352–361. [Google Scholar]
- Pocker Y, Ng SY. Plant carbonic anhydrase: properties and carbon dioxide hydration kinetics. Biochemistry. 1973;12:5127–5134. doi: 10.1021/bi00749a016. [DOI] [PubMed] [Google Scholar]
- Price GD, Coleman JR, Badger MR. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ML, Graham D. Carbonic anhydrase in plants: distribution, properties and possible physiological roles. In: Reinhold L, Harborne JB, Swain T, editors. Progress in Phytochemistry, Vol 7. Oxford, UK: Pergamon Press; 1981. pp. 47–94. [Google Scholar]
- Rickli EE, Ghazanfar SAS, Gibbons BH, Edsall JT. Carbonic anhydrase from human erythrocytes: preparation and properties of two enzymes. J Biol Chem. 1964;239:1065–1078. [PubMed] [Google Scholar]
- Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Rowlett RS, Chance MR, Wirt MD, Sidelinger DE, Royal JL, Woodroffe M, Wang Y-FA, Saha RP, Lam MG. Kinetic and structural characterization of spinach carbonic anhydrase. Biochemistry. 1994;33:13967–13976. doi: 10.1021/bi00251a003. [DOI] [PubMed] [Google Scholar]
- Rumeau D, Cuiné S, Fina L, Gault N, Nicole M, Peltier G. Subcellular distribution of carbonic anhydrase in Solanum tuberosum L. leaves: characterization of two compartment-specific isoforms. Planta. 1996;199:79–88. doi: 10.1007/BF00196884. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Silverman DN, Lindskog S. The catalytic mechanism of carbonic anhydrase: implications of a rate-limiting proteolysis of water. Acc Chem Res. 1988;21:30–36. [Google Scholar]
- Steiner H, Jonsson B-H, Lindskog S. The catalytic mechanism of carbonic anhydrase, hydrogen-isotope effects on the kinetic parameters of the human C isoenzyme. Eur J Biochem. 1975;59:253–259. doi: 10.1111/j.1432-1033.1975.tb02449.x. [DOI] [PubMed] [Google Scholar]
- Strickland EH. Aromatic contributions to circular dichroism spectra of proteins. CRC Rev Biochem. 1974;3:113–175. doi: 10.3109/10409237409105445. [DOI] [PubMed] [Google Scholar]
- Tashian RE. Genetics of the mammalian carbonic anhydrases. Adv Genet. 1992;30:321–356. doi: 10.1016/s0065-2660(08)60323-5. [DOI] [PubMed] [Google Scholar]
- Tobin AJ. Carbonic anhydrase from parsley leaves. J Biol Chem. 1970;245:2656–2666. [PubMed] [Google Scholar]
- Winter K, Foster JG, Edwards GE, Holtum JAM. Intracellular localization of enzymes of carbon metabolism in Mesembryanthemum crystallinum exhibiting C3 photosynthetic characteristics or performing Crassulacean acid metabolism. Plant Physiol. 1982;69:300–307. doi: 10.1104/pp.69.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]